Abstract
Mitogen Activated Protein Kinase (MAPK) pathways form the backbone of signal transduction within the mammalian cell. Here, we apply a systematic experimental and computational approach to map 2,269 interactions between human MAPK-related proteins and other cellular machinery and to assemble these data into functional modules. A core network of 641 interactions is supported by multiple lines of evidence including conservation with yeast. Using siRNA knockdowns, we reveal that a significant number of novel interactors can modulate MAPK mediated signaling. We uncover the Na-H exchanger NHE1 as a scaffold for a novel set of MAPKs, link HSP90 chaperones to MAPK pathways, and identify MUC12 as the human analogue to the yeast signaling mucin Msb2. This study makes available a large resource of MAPK interactions along with the accompanying clone libraries. It illustrates a methodology for probing signaling networks based on functional refinement of experimentally-derived protein interaction maps.
Keywords: Two-hybrid, conservation, interactome, scaffold, NHE1
INTRODUCTION
The MAPK pathways are a collection of protein signaling cascades stimulated by a wide variety of extra-cellular signals, including growth factors, cytokines, and environmental stress1, 2. Upon activation, MAPK pathways regulate a large number of fundamental cellular functions, including differentiation, proliferation, and apoptosis, through the activation of specific transcription factors and other regulatory proteins3. Because of this central role in signal transduction, MAPK proteins have been repeatedly implicated in the pathogenesis of cancer and autoimmune diseases, leading to their selection as targets for drug development4. MAPK pathways are also well conserved over the eukaryotic kingdom from yeast to man, enabling study of their structure and kinetics through genetic analysis of model organisms2.
The canonical MAPK pathway consists of three sequentially-activated MAPK family members: a MAPK kinase kinase (MAP3K) which activates a MAPK kinase (MAP2K) which, in turn, activates a MAP kinase (MAPK). However, it is clear that this canonical pathway is embedded in a rich network of interactions with a wide variety of other components, including membrane receptors, transcription factors, kinase scaffolds, and extensive cross-talk with other activators and inhibitors of signaling5. It has been suggested that a systems-level approach will ultimately be necessary to map these additional components and to understand their MAPK-related functions3, 6.
Towards this goal, we pursued a combined experimental and computational approach to assemble a large resource of MAPK protein interactions and to illustrate methods to functionally explore this network. This work involved four steps: (1) A screen for physical interactions between MAPK proteins and the rest of the proteome, (2) An assessment of network quality and functional assessment of MAPK interactors through siRNA screening, (3) A specific analysis of the MAPK network to identify kinase scaffolds, illustrating how the network can be used as a resource to guide the discovery of novel protein functions, and (4) Identification of interaction modules and evolutionary conservation to aid in functional interpretation. Based on the MAPK network resource, we find evidence that the Na+/H+ exchanger NHE1 functions as a kinase scaffold, and we identify a conserved signaling cascade mediated by the signaling mucin MUC12.
RESULTS AND DISCUSSION
Characterization of a MAPK interactome
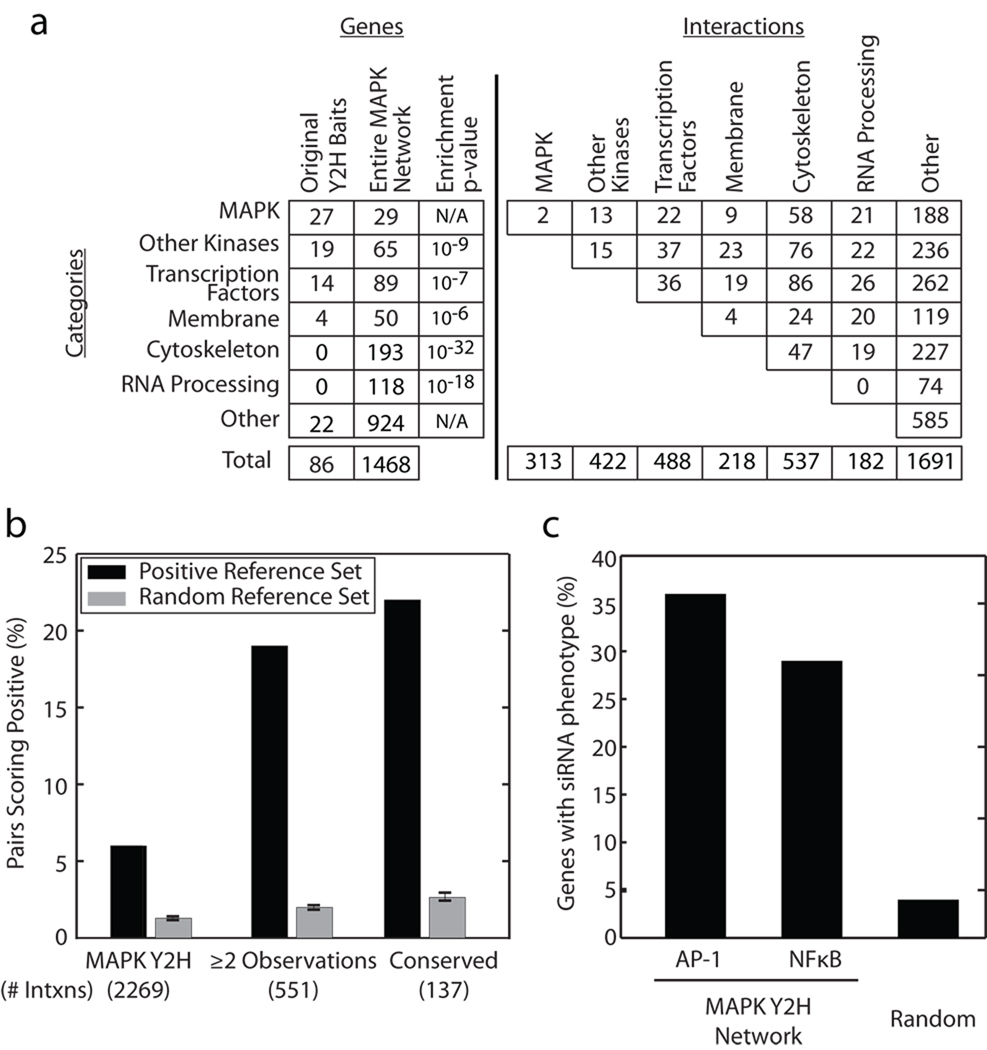
We assembled a human MAPK network comprised of protein interactions identified through a two-stage yeast two-hybrid (Y2H) screen. A primary Y2H screen was performed using 86 MAPK-related bait proteins (Supplementary Fig. 1) selected by literature curation (see Methods). Included in these baits were 46 kinases (of which 27 were known MAP-family kinases), 14 transcription factors downstream of MAPK signals, and four proteins associated with membrane receptors (Fig. 1a). This effort yielded 1,496 protein-protein interactions among 1,096 proteins. As follow-up to this primary screen, we examined 2`1 secondary baits that had been detected as preys in the first stage (based on their availability as bait cDNA clones). This round identified an additional 786 interactions for a total of 2,269 unique interactions among 1,468 proteins (termed the MAPK Y2H network) (Supplementary Table 1). Bait and prey cDNA clones are available upon reqest.
Figure 1.
Functional properties of the MAPK network. (a) Breakdown of network proteins (columns 1–3) and protein interactions (columns 4–10) by functional category. Enrichment p-value is based on the probability of identifying an equal or greater number of proteins in the same category at random assessed via the hypergeometric test with a background of 30,000 genes. (b) The percentage of Positive Reference Set or Negative Reference Set pairs overlapping with each network is shown. Error bars are s.e.m. (see Methods). (c) Percentage of MAPK network genes which are able to knockdown AP-1 or NFκB activity. Random represents the percentage of forty-five randomly selected proteins which knockdown NFκB levels.
Analysis of the MAPK Y2H network revealed 313 interactions involving MAP-family kinases and 422 involving other kinases. After removing the original baits, the MAPK Y2H network was found to be enriched for protein families known to be critical to MAPK signaling such as membrane proteins and transcription factors, suggesting numerous possible upstream and downstream components of MAPK cascades (Fig. 1a). The network was also highly enriched for proteins involved in cytoskeletal organization and RNA binding and processing. The significant number of cytoskeletal proteins identified in this study suggests the active regulation of microtubule dynamics by MAP kinases or the use of the cytoskeleton as an organizational scaffold in MAPK signaling; it has been postulated that up to one third of MAP-family kinases are associated with the microtubule cytoskeleton7.
Many members of the MAPK Y2H network had been implicated in previous screens for genes that function in MAPK signaling. We found that the network (with 86 original baits removed) was highly enriched for proteins that are phosphorylated in response to stimulation of HeLa cells with epidermal growth factor8 (EGF; 429 of 2,089 phosphorylated proteins; P < 10−170). We also found substantial overlap with a screen for kinase activation in fly: the MAPK Y2H network contained 734 genes with Drosophila homologs, 92 of which were shown to be required for the activation of extracellular signal-regulated kinases (ERK) by an unbiased RNAi screen9 (P < 10−13).
Assessment of MAPK network quality and function
To estimate the sensitivity of the interaction screen (the percentage of true interactions recovered), we assembled a Positive Reference Set (PRS) of interactions through curation of prior literature. This set contained 1,453 previously-reported interactions among proteins covered by the MAPK Y2H network (Methods). Of the interactions in the PRS, we found that 93 were recovered by the MAPK Y2H network, yielding a sensitivity of 6% (Fig. 1b). To assess the significance of this number, we next assembled a Random Reference Set (RRS) of interactions as in (10). The RRS was constructed by permutation of the PRS to form a non-overlapping set of random interactions, which approximates a set of non-interacting protein pairs10. The MAPK Y2H network contained approximately 1% of these randomly-chosen interactions, indicating a ~6-fold enrichment for the PRS over random.
We found that sensitivity could be increased substantially based on two criteria: 1) Observation of an interaction multiple times during the Y2H screen, and 2) Whether the interaction was conserved with yeast. An interaction may be observed multiple times because, during Y2H screening, protein preys are pooled with a particular bait and successful bait-prey interactions are identified by individual colony selection and sequencing (Methods). A very large number of interactions in our study (551 interactions) were identified multiple times through multiple sequenced clones. Similarly, because MAPK pathways are well-conserved among eukaryotes2, evidence that orthologs of the bait and prey can interact in yeast increases the confidence in interaction. For interactions that were multiply sampled or conserved, respectively, the sensitivity was 19% or 22%, representing a > 3-fold increase in comparison to the entire MAPK Y2H network (Fig. 1b). Based on these results, we combined the multiply-sampled interactions and conserved interactions to form a “Core MAPK Network” of 641 high-confidence interactions (including 47 interactions selected by both criteria) (Supplementary Table 2).
Evidence for physical interaction may or may not mean that interaction is functional. Although network function is difficult to assess systematically, classic studies of multiple MAPK pathways have shown that AP-1 or NFκB transcription factors are critical effectors of these pathways under appropriate stimulation11, 12. We therefore pursued a functional validation strategy based on guilt-by-association: given MAPK proteins that affect AP-1 or NFκB activation, siRNA knockdown of their functional interactors should also modulate these phenotypes. Using this strategy, we selected a panel of 14 MAPK interacting proteins from the MAPK Y2H Network and systematically knocked these down using four independent siRNAs per gene to mitigate possible off target effects. The response to each knockdown was assessed using AP-1 or NFκB transcriptional activation in response to phorbol-12-myristate-13-acetate (PMA) or tumor necrosis factor (TNF), respectively. We found that 36% of targeted genes had two or more independent siRNAs that were able to significantly reduce AP-1 levels (Fig. 1c, Supplementary Fig. 2). This rate was similarly high in the case of NFκB, with a rate of 29%. In contrast, siRNAs targeting of a set of 45 random genes reduced NFκB levels for two of these (4%), roughly a tenth the rate of the tested MAPK interactors (Fig. 1c, Supplementary Fig. 2). This siRNA panel is not a true gold standard, such that failure to affect AP-1 or NFκB activity does not negate the corresponding protein-protein interaction, which might be involved in unrelated aspects of MAPK signaling. However, the analysis does show that the Core MAPK Network recovers approximately 20% of known interactions among these proteins and that approximately a third of interactions can be functionally validated using siRNA.
Kinase subnetworks identify NHE1 as a novel kinase scaffold
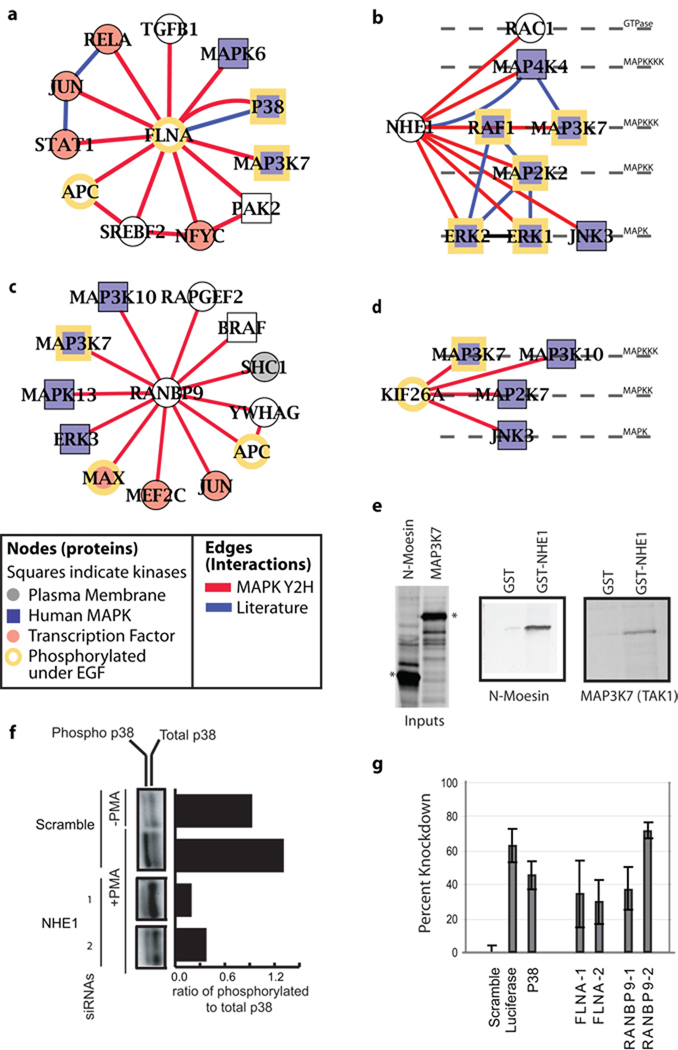
As an example of how the MAPK network can be used as a resource for identification of novel protein functions, we analyzed the network for evidence of MAPK scaffolds. MAPK scaffolds form a signaling apparatus through the simultaneous binding of kinases and their substrates13. To identify such scaffolds, we selected proteins that interact with multiple MAPK levels and have at least 40% of their interactions with kinases, yielding a total of 10 candidate scaffolds (four of which are shown in Fig. 2a–d; Supplementary Table 3). As a positive control, this strategy detected a well-established human MAPK scaffold, the actin-binding protein FLNA, which has been postulated to organize kinase signaling between the membrane and cytoskeleton and to regulate transcription factors such as AP-114. Of the 11 interactions involving FLNA, seven are with kinases or transcription factors (Fig. 2a).
Figure 2.
Protein subnetworks reveal known and putative MAPK scaffolds. Network neighborhoods are shown for (a) Filamin protein FLNA, (b) the Na-H exchanger NHE1, (c) RAN binding protein RANBP9, and (d) the kinesin family member KIF26A. Newly-identified Y2H interactions (red) and interactions from literature are shown (blue). Proteins are colored based on their annotation as membrane (grey), MAPK (blue), transcription factor (red), or phosphorylated under EGF stimulation (yellow border)8. (e) Binding of in vitro-translated MAP3K7 (TAK1) to GST tagged C-terminus of NHE1 or GST alone. N-moesin, a known NHE1 binding partner is used as a positive control31. Expressed input proteins used for in vitro binding assays are marked with asterisks. (f) Phosphorylated (pp38) and total levels of p38, assayed with and without PMA stimulation and two different siRNA knockdowns of NHE1. The bars quantify the pp38 / p38 ratio by image analysis of western blot. As a negative control, we observed nearly equal amounts of pp38 to p38 in the absence of PMA stimulation (−PMA). As a positive control, PMA stimulation with a scrambled siRNA induced p38 phosphorylation (Scramble, +PMA). (g) Using a luciferase reporter fused to the AP-1 gene, we tested the ability of various siRNAs to reduce AP-1 activation when stimulated with PMA32. As a negative control, scrambled siRNAs resulted in maximal AP-1 transcription (“Scramble”). As positive controls, siRNAs targeted to luciferase showed a large reduction of luminescence (“Luciferase”), and siRNAs directed directly to p38, upstream of AP-1, reduced signal intensity by 50%. Error bars represent s.e.m. with 6 replicates.
We found that the plasma membrane Na-H exchanger NHE1 interacted with a total of seven MAPK family proteins, spanning all four levels of the MAPK hierarchy including MAP4K4, two MAP3Ks (MAP3K7 and RAF1), MAP2K2, and three MAPKs (ERK1, ERK2, and JNK3) (Fig. 2b). NHE1 also interacted with the Rho GTPase Rac1, which can modulate NHE1 activity15. In addition to its known role in ion exchange, we postulated that NHE1 may function as a plasma-membrane scaffold for the assembly of signaling complexes16. Using tagged NHE1, we confirmed the interaction between the C-terminal cytoplasmic domain of NHE1 and MAP3K7 (TAK1) in vitro (Fig. 2e). We also identified two independent siRNAs targeted to NHE1 that were able to significantly reduce phosphorylation levels of p38 in response to PMA, an established assay of MAPK pathway function (Fig. 2f). Thus, based on the new protein interactions and functional siRNA screening data, it is likely that the cytoplasmic tail of NHE1 promotes the assembly of a GTPase and MAP-family kinases into (possibly multiple) signaling complexes. As we previously showed that NHE1 binds to and is phosphorylated by MAP4K4 in response to external growth factors17, a similar mechanism might govern the activity of the other interactions identified here. Since MAP3K7 is activated by the transforming growth factor β (TGF-β) receptor18 and we have previously identified an NHE1-immune complex containing the type II TGF-β receptor, the new interaction data suggest that NHE1 can act as a regulatory molecule for processing various cell stimuli in union with the TGF-β receptor, MAP3K7 and other MAPKs19.
A number of novel interactions involving RANBP9 were also detected (Fig. 2c). Of particular interest is an interaction between RANBP9 and the RAPGEF2 guanine exchange factor, which is a member of the Ras subfamily of GTPases. RANBP9 was originally characterized as binding to the Ras GTPase-binding protein RAN and has been shown to activate the Ras signaling pathway20. Based on the Y2H evidence, we postulated that RANBP9 functions as a scaffold for RAPGEF2 and various MAPK kinases, including ERK3, and promotes activation of the transcription factors MAX, MEF2C, and JUN (a component of the AP-1 transcription factor). We found that siRNAs directed to RANBP9 significantly reduced AP-1 transcription in response to PMA, in a manner similar to knockdown of the known scaffold FLNA (Fig. 2g). These experiments are consistent with a model whereby RANBP9 organizes MAPK signaling upstream of AP-1.
Network modules link MUC12 and HSP90 to MAPK proteins
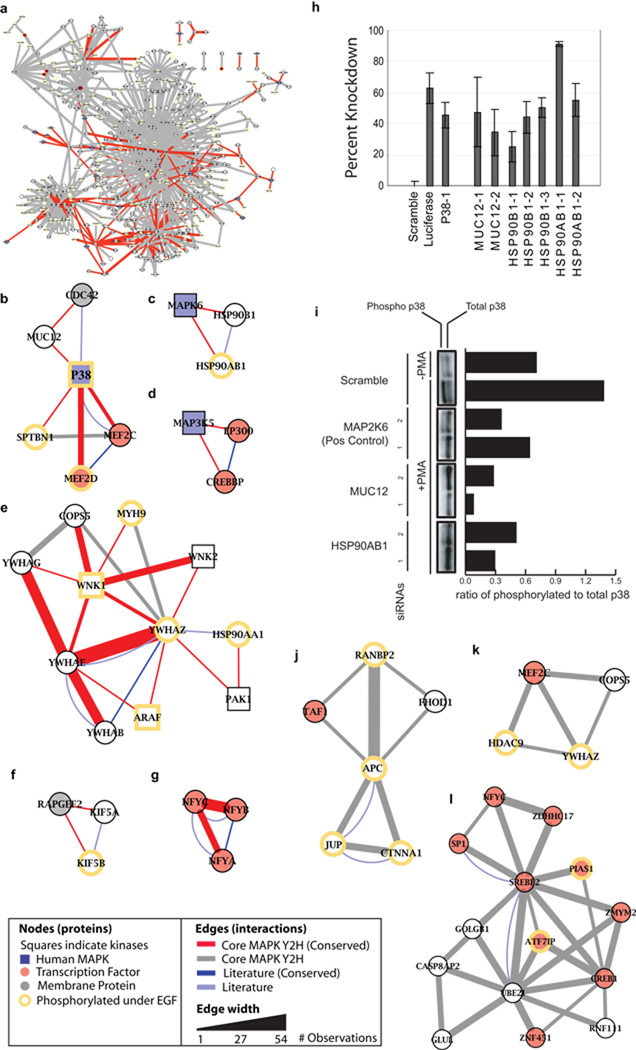
To shed further light on the structure and function of the Core MAPK Network (Fig. 3a), we analyzed this network to extract conserved and/or species-specific modules (Methods). To form these modules, we first combined the Core MAPK Network with the Positive Reference Set to capture novel as well as literature-curated MAPK interactions. Network “modules” were identified as all complete interacting triplets of proteins (i.e., triangles) for which at least two interactions were from the Core MAPK Network. Triangles and other dense regions of interactions have been shown to be indicative of protein complexes, signaling pathways, and other functionally cohesive groups of proteins21. In total, 134 triplets were found covering 195 core MAPK Y2H interactions (Supplementary Table 5). We next identified a subset of 19 “Conserved modules”, which were modules for which at least two of the interactions were conserved with yeast. We found that such modules fell into six connected components, highlighting potential conserved mechanisms of signaling and regulation (Fig. 3b–g).
Figure 3.
Functional modules in the core network. (a) Bird’s eye view of the core MAPK Y2H network. (b–g) High confidence conserved functional modules. Red edges correspond to core MAPK Y2H interactions which were conserved with yeast. Grey edges indicate core interactions not conserved with yeast. Thickness of the edge increases with the number of observations. (h) AP-1 luciferase activation assay for various siRNAs targeting members of conserved modules. (i) p38 phosphorylation levels are decreased with siRNAs targeting members of conserved modules. (j–l) Novel modules not conserved with yeast.
One of these conserved modules included interactions of the MUC12 protein with p38 and Cdc42 (Fig. 3b), suggesting that MUC12 might function in a CDC42-responsive signaling cascade. This hypothesis is supported through conservation with yeast, in which Cdc42 is found to interact with mucin family member Msb2 whose function in kinase signaling has already been established22. In support of a role for MUC12 in human MAPK signaling, we found that multiple distinct siRNA constructs against MUC12 could significantly knockdown both AP-1 activation (Fig. 3h) and p38 phosphorylation (Fig. 3i) in response to PMA. Together, the Y2H interaction and siRNA results suggest the existence of a novel human signaling mucin acting in an analogous fashion to the yeast mucin Msb2.
Another conserved module suggested a role for HSP90 members HSP90B1 and HSP90AB1 in the function of MAPK6 (ERK3, Fig. 3c), perhaps to stabilize this protein in much the same way as HSP90B1 has been shown to stabilize ERBB223. These interactions are plausible since HSP90 proteins are known to associate with kinases that mediate multiple inputs24. To further investigate this hypothesis, we screened multiple siRNA constructs directed to HSP90AB1 and HSP90B1. We observed a reduction in levels of AP-1 transcription (Fig. 3h) for HSP90AB1 and HSP90B1 as well as phosphorylated p38 (Fig. 3i) for HSP90AB1, suggesting a critical role for these HSP90 members in MAPK-mediated signaling.
We also identified three distinct network modules (Fig. 3j–l) which were not conserved with yeast. These modules either indicate interactions missing from the current yeast interactome map or machinery that may be present only in higher eukayotes. The modules highlight evidence for an interaction between RANBP2, the GTP-binding protein at the nuclear pore, and the APC tumor suppressor gene which is known to promote the association of the nuclear pore with microtubules25. Furthermore, these modules reveal a novel high-confidence interaction between APC and the catenin CTNNA1 (Fig. 3j). Since interaction of APC with other β-catenin pathway members has been shown to be critical for WNT signaling, interactions of CTNNA1 with APC may play a role in this pathway as well. Because WNT signaling is conserved among metazoans but is not found in yeast, these examples illustrate the power of network mapping on a species-specific basis.
SUMMARY AND CONCLUSIONS
We have demonstrated a combined experimental and computational approach for the systematic expansion and refinement of protein networks based on high-throughput interaction mapping followed by functional validation and refinement. There have been a great many studies reported recently that start with a systematic siRNA screen and then interpret the top hits by projecting them onto a protein interaction network curated from literature26–28. Here, we reverse this workflow and start with a systematic interactome screen in preparation for targeted siRNA to interpret the high priority novel interactions. This methodology may be particularly useful for the study of cell functions that are not well covered by existing protein interaction maps published in prior literature. Moreover, such an approach adds functional value to basic interactome mapping efforts, a point which is particularly important given that only a very few previous protein interaction screens have addressed which interactions are functional29, 30.
Beyond the methodological aspects, this work has led to the discovery of more than 2,000 protein interactions related to MAPK proteins which, along with the associated cDNA clone libraries, constitute a significant resource for the research community. The quality of the network is comparable to previously-published Y2H datasets and can be substantially increased by techniques such as multiple sampling and cross species comparison. We have illustrated how this network can be analyzed to suggest novel kinase scaffolds, demonstrating that, in principle, numerous discoveries might be mined from these data. Other large-scale technologies such as gene expression profiling, co-affinity purification, and phospho-proteomics may add complementary facets of information, yielding a more complete understanding of MAPK signaling in humans and yeast.
MATERIALS AND METHODS
Y2H screening
Yeast strains and expression vectors for Y2H screening are as previously described33. Human protein baits were screened against preys derived from 22 different human cDNA libraries. The original 86 baits were selected based on BioCarta (http://www.biocarta.com/pathfiles/m_p38mapkPathway.asp) as well as a review by Qi et al.34. Full experimental and computational details are provided in the Supplementary Methods. Interactions have been deposited into the BIOGRID database (www.thebiogrid.org). The yeast clones which harbor a specific bait-prey interaction pair are available upon request from the authors and can be used to confirm the bait ID, prey ID, amplify and clone bait or prey inserts, or verify an interaction by another method.
Literature-curated reference sets
To form the Positive Reference Set (PRS), a total of 23,978 literature-curated interactions were obtained from databases including BIND, HPRD and BioCarta (Supplementary Table 4). To estimate sensitivity for each Y2H human protein interaction network the PRS was restricted to interactions involving proteins for which at least one interaction was observed in that network data set (Fig. 1b)10. To generate a Random Reference Set (RRS), the PRS was randomly permuted and any overlapping PRS interactions removed. One hundred such RRS datasets were generated and compared to each network dataset. For each Y2H network, the percent overlap with the RRS is summarized by its average and standard deviation of overlap.
Identification of conserved interactions
A yeast MAPK network was assembled from both literature-curated and experimental sources (for a listing see Supplementary Methods). A human protein interaction was considered “conserved” if both proteins had homologs that interacted in yeast. Human/yeast homologs were defined using a strict BLAST E-value < 10−10, and to avoid spurious matches due to large gene families we allowed no more than 10 yeast homologs for each human protein (10 best E-values). Of the 137 conserved interactions observed by this criteria, 21 (15%) were between sequence orthologs (Supplementary Table 2)35.
AP-1 and NFκB luciferase siRNA assay
The HEK293 cell line stably expresses an AP-1 or NFκB responsive luciferase reporter as previously described32. An aliquot of 8×103 HEK293 cells was plated into 96-well tissue culture plates and each well transfected with 25 ng of indicated siRNA by using Lipofectamine2000 reagent (Invitrogen). After 72 h of transfection, cells were stimulated with 10ng/ml of PMA for 8 hours for the case of AP-1 or 10ng/ml of TNF for 8 hours in the case of NFκB. Luciferase activity was measured by using Bright Glow (Promega) according to the manufacturer instructions. Cell titer was measured by the CellTiter-Glo Luminescent Cell Viability Assay (Promega). Both cell titer and luciferase activity counts were normalized by the median of the scramble siRNAs36. All siRNA sequences are given (Supplementary Table 7).
Western blotting for phospho-p38 and in vitro binding assays
HeLa cells were transfected using RNAiMAX according to standard protocol (Invitrogen Life Technologies). After 72 h of transfection, cells were stimulated with 10ng/ml of PMA for 8 hours. Cells were lysed in buffer containing 25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% Sodium deoxycholate, 0.1 % SDS, and phosphatase inhibitors (Sigma Aldrich). A 10 ng amount of cell lysate was resolved by SDS-PAGE, and blots were immunoblotted with the antibodies detecting the phosphorylated form of p38 (Promega). All blots were developed with HRP-conjugated secondary antibodies and ECL (Amersham Biosciences). In-vitro binding assays for N-terminal tagged NHE1 were performed as described previously31.
Supplementary Material
ACKNOWLEDGEMENTS
We thank R. Kelley, S. Suthram and G. Warner for assistance on various aspects of this work. This work was generously supported by funds from the National Institutes of Health (R01-GM070743, R01-GM47413 and P30-MH062261) and Unilever PLC.
Footnotes
CONFLICT OF INTERESTS
R.B. is a shareholder of Prolexys Pharmaceuticals, Inc. S.S. is an employee, scientific founder, and shareholder of Prolexys Pharmaceuticals Inc.
AUTHOR CONTRIBUTIONS
S.B., R.B., S.S., and T.I. conceived the study; C.C., J.S., S.W., R.B., C.K., and C.H.M. performed the experiments; S.B., M.G., and M.S. analyzed the data; S.B., T.I., S.S., D.L.B., and S.K.C. wrote the paper and guided the study.
REFERENCES
- 1.Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 2.Widmann C, Gibson S, Jarpe MB, Johnson GL. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol. Rev. 1999;79:143–180. doi: 10.1152/physrev.1999.79.1.143. [DOI] [PubMed] [Google Scholar]
- 3.Kolch W, Calder M, Gilbert D. When kinases meet mathematics: the systems biology of MAPK signalling. FEBS Lett. 2005;579:1891–1895. doi: 10.1016/j.febslet.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298:1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- 5.Kolch W. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat. Rev. Mol. Cell Biol. 2005;6:827–837. doi: 10.1038/nrm1743. [DOI] [PubMed] [Google Scholar]
- 6.Johnson SA, Hunter T. Kinomics: methods for deciphering the kinome. Nat. methods. 2005;2:17–25. doi: 10.1038/nmeth731. [DOI] [PubMed] [Google Scholar]
- 7.Reszka AA, Seger R, Diltz CD, Krebs EG, Fischer EH. Association of mitogen-activated protein kinase with the microtubule cytoskeleton. Proc. Natl. Acad. Sci. USA. 1995;92:8881–8885. doi: 10.1073/pnas.92.19.8881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olsen JV, et al. Global, In Vivo, and Site-Specific Phosphorylation Dynamics in Signaling Networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 9.Friedman A, Perrimon N. A functional RNAi screen for regulators of receptor tyrosine kinase and ERK signalling. Nature. 2006;444:230–234. doi: 10.1038/nature05280. [DOI] [PubMed] [Google Scholar]
- 10.Venkatesan K, et al. An empirical framework for binary interactome mapping. Nat. methods. 2009;6:83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karin M. The regulation of AP-1 activity by mitogen-activated protein kinases. J. Biol. Chem. 1995;270:16483–16486. doi: 10.1074/jbc.270.28.16483. [DOI] [PubMed] [Google Scholar]
- 12.Rothe M, Sarma V, Dixit VM, Goeddel DV. TRAF2-mediated activation of NF-kappa B by TNF receptor 2 and CD40. Science. 1995;269:1424–1427. doi: 10.1126/science.7544915. [DOI] [PubMed] [Google Scholar]
- 13.Whitmarsh AJ, Davis RJ. Structural organization of MAP-kinase signaling modules by scaffold proteins in yeast and mammals. Trends Biochem. Sci. 1998;23:481–485. doi: 10.1016/s0968-0004(98)01309-7. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi K, Altman A. Filamin A is required for T cell activation mediated by protein kinase C-theta. J. Immunol. 2006;177:1721–1728. doi: 10.4049/jimmunol.177.3.1721. [DOI] [PubMed] [Google Scholar]
- 15.Hooley R, Yu CY, Symons M, Barber DL. G alpha 13 stimulates Na+-H+ exchange through distinct Cdc42-dependent and RhoA-dependent pathways. J. Biol. Chem. 1996;271:6152–6158. doi: 10.1074/jbc.271.11.6152. [DOI] [PubMed] [Google Scholar]
- 16.Baumgartner M, Patel H, Barber DL. Na(+)/H(+) exchanger NHE1 as plasma membrane scaffold in the assembly of signaling complexes. Am. J. Physiol. Cell Physiol. 2004;287:C844–C850. doi: 10.1152/ajpcell.00094.2004. [DOI] [PubMed] [Google Scholar]
- 17.Yan W, Nehrke K, Choi J, Barber DL. The Nck-interacting kinase (NIK) phosphorylates the Na+-H+ exchanger NHE1 and regulates NHE1 activation by platelet-derived growth factor. J. Biol. Chem. 2001;276:31349–31356. doi: 10.1074/jbc.M102679200. [DOI] [PubMed] [Google Scholar]
- 18.Yamaguchi K, et al. Identification of a member of the MAPKKK family as a potential mediator of TGF-beta signal transduction. Science. 1995;270:2008–2011. doi: 10.1126/science.270.5244.2008. [DOI] [PubMed] [Google Scholar]
- 19.Karydis A, Jimenez-Vidal M, Denker SP, Barber DL. Mislocalized scaffolding by the Na-H exchanger NHE1 dominantly inhibits fibronectin production and TGF-beta activation. Mol. Biol. Cell. 2009;20:2327–2336. doi: 10.1091/mbc.E08-08-0842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang D, Li Z, Messing EM, Wu G. Activation of Ras/Erk pathway by a novel MET-interacting protein RanBPM. J. Biol. Chem. 2002;277:36216–36222. doi: 10.1074/jbc.M205111200. [DOI] [PubMed] [Google Scholar]
- 21.Kelley BP, et al. Conserved pathways within bacteria and yeast as revealed by global protein network alignment. Proc. Natl. Acad. Sci. USA. 2003;100:11394–11399. doi: 10.1073/pnas.1534710100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cullen PJ, et al. A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous growth pathway in yeast. Genes Dev. 2004;18:1695–1708. doi: 10.1101/gad.1178604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu W, Mimnaugh EG, Kim JS, Trepel JB, Neckers LM. Hsp90, not Grp94, regulates the intracellular trafficking and stability of nascent ErbB2. Cell Stress Chaperones. 2002;7:91–96. doi: 10.1379/1466-1268(2002)007<0091:hngrti>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Citri A, et al. Hsp90 recognizes a common surface on client kinases. J. Biol. Chem. 2006;281:14361–14369. doi: 10.1074/jbc.M512613200. [DOI] [PubMed] [Google Scholar]
- 25.Collin L, Schlessinger K, Hall A. APC nuclear membrane association and microtubule polarity. Biol. Cell. 2008;100:243–252. doi: 10.1042/BC20070123. [DOI] [PubMed] [Google Scholar]
- 26.Kim J, et al. Functional genomic screen for modulators of ciliogenesis and cilium length. Nature. 464:1048–1051. doi: 10.1038/nature08895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Konig R, et al. Human host factors required for influenza virus replication. Nature. 463:813–817. doi: 10.1038/nature08699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Konig R, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gunsalus KC, et al. Predictive models of molecular machines involved in Caenorhabditis elegans early embryogenesis. Nature. 2005;436:861–865. doi: 10.1038/nature03876. [DOI] [PubMed] [Google Scholar]
- 30.Lunardi A, et al. A genome-scale protein interaction profile of Drosophila p53 uncovers additional nodes of the human p53 network. Proc. Natl. Acad. Sci. USA. 107:6322–6327. doi: 10.1073/pnas.1002447107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Denker SP, Huang DC, Orlowski J, Furthmayr H, Barber DL. Direct binding of the Na--H exchanger NHE1 to ERM proteins regulates the cortical cytoskeleton and cell shape independently of H(+) translocation. Mol. Cell. 2000;6:1425–1436. doi: 10.1016/s1097-2765(00)00139-8. [DOI] [PubMed] [Google Scholar]
- 32.Chanda SK, et al. Genome-scale functional profiling of the mammalian AP-1 signaling pathway. Proc. Natl. Acad. Sci. USA. 2003;100:12153–12158. doi: 10.1073/pnas.1934839100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.LaCount DJ, et al. A protein interaction network of the malaria parasite Plasmodium falciparum. Nature. 2005;438:103–107. doi: 10.1038/nature04104. [DOI] [PubMed] [Google Scholar]
- 34.Qi M, Elion EA. MAP kinase pathways. J. Cell Sci. 2005;118:3569–3572. doi: 10.1242/jcs.02470. [DOI] [PubMed] [Google Scholar]
- 35.Remm M, Storm CE, Sonnhammer EL. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- 36.Konig R, et al. A probability-based approach for the analysis of large-scale RNAi screens. Nat. Methods. 2007;4:847–849. doi: 10.1038/nmeth1089. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.