Abstract
Cancer cells commonly have a high rate of telomere loss, even when expressing telomerase, contributing to chromosome instability and tumor cell progression. This review addresses the hypothesis that this high rate of telomere loss results from a combination of four factors. The first factor is an increase in the frequency of double-strand breaks (DSBs) at fragile sites in cancer cells due to replication stress. The second factor is that telomeres are fragile sites. The third factor is that subtelomeric regions are highly sensitive to DSBs, so that DSBs near telomeres have an increased probability of resulting in chromosome instability. The fourth factor is that cancer cells may be deficient in chromosome healing, the de novo addition of telomeres to the sites of DSBs, a mechanism that prevents chromosome instability resulting from DSBs near telomeres. Understanding these factors and how they influence telomere loss will provide important insights into the mechanisms of chromosome instability and the development of novel approaches for anti-cancer therapy.
Introduction
Human telomeres are composed of a TTAGGG repeat sequence and associated proteins that together form a cap that keeps the ends of chromosomes from appearing as double-strand breaks (DSBs) and thereby prevents chromosome fusion (1, 2). In humans, telomeres are maintained in germ line cells, but shorten as somatic cells divide due to the down regulation of telomerase. Telomere shortening limits the replication of somatic cells, and as a result, cancer cells invariably maintain their telomeres, most often through the expression of telomerase, although approximately 10% of human tumors maintain telomeres through an alternative mechanism (3). Excessive telomere shortening prior to the expression of telomerase can lead to chromosome fusion, which has been proposed as a mechanism for chromosome instability (4). However, a high rate of telomere loss is common in a variety of different types of early passage cancer cells despite the expression of telomerase (5). This review will address the mechanisms responsible for this spontaneous telomere loss in human cancer cells and its importance in the chromosome instability associated with human cancer.
My laboratory has investigated the mechanisms and consequences of telomere loss using plasmid sequences integrated immediately adjacent to a telomere (6). These plasmids contain both positive and negative selectable marker genes, as well as a recognition site for the I-SceI endonuclease. Selection with ganciclovir for the loss of the herpes simplex virus thymidine kinase (HSV-tk) gene within the plasmid is used to identify cells in the population that have lost the marked telomere, either spontaneously or as a result of DSBs induced by the I-SceI endonuclease. This approach allows us to monitor both the rate of telomere loss and the sequence of events involved in the instability of an individual chromosome that has lost a telomere. Consistent with the results of Gisselsson et al. involving the analysis of a large number of early passage cancer cells (5), our more in-depth analysis demonstrated a high rate of spontaneous telomere loss in the human EJ-30 bladder cell carcinoma (7-9) and SCC-61 squamous cell carcinoma (unpublished observation) cell lines. A critical feature of the spontaneous telomere loss in these cancer cell lines is that it occurs at a low enough frequency so that the cells do not die, and therefore continue to divide and accumulate large numbers of chromosome rearrangements. With the EJ-30 human carcinoma cell line, the spontaneous rate of loss of an individual telomere is 10−4 events/cell/generation, which means that approximately one of the 92 telomeres in this near diploid cell line would be lost in every 100 cell divisions.
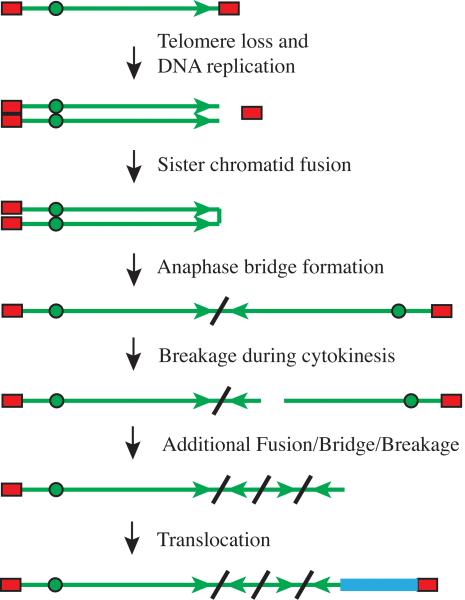
The analysis of the consequences of spontaneous telomere loss in individual gancilovir-resistant subclones of the EJ-30 human cancer cell line was conducted using Southern blot analysis, the cloning and sequencing of the DNA rearrangements involved, and cytogenetic analysis. Our results demonstrated that the most common events observed in HSV-tk-deficient subclones consisted of large deletions and gross chromosome rearrangements (7-10). All three of the HSV-tk-deficient subclones that were analyzed in more detail had undergone sister chromatid fusions that resulted in prolonged periods of chromosome instability involving breakage/fusion/bridge (B/F/B) cycles, demonstrating that this was a common consequence of spontaneous telomere loss. B/F/B cycles occur when the fused sister chromatids form a bridge during anaphase, which eventually breaks as the cell divides, resulting in inverted repeats on the end of the chromosome in one daughter cell and a terminal deletion on the chromosome in the other daughter cell (Fig. 1). Because these chromosomes still lack a telomere on one end, following DNA replication, the sister chromatids will fuse and break again in the next cell cycle. The repeated B/F/B cycles result in further amplification, generating arrays of inverted repeats that are typical of the amplified regions found in human cancer (6). The B/F/B cycles continue until the chromosome acquires a new telomere, most often by translocation of the end of another chromosome (9). However, the nonreciprocal translocations cause the loss of the telomere on the donor chromosome, resulting in the transfer of instability from one chromosome to another (9). Thus, even the loss of a single chromosome can result in prolonged chromosome instability involving multiple chromosomes. Importantly, the types of chromosome rearrangements resulting from telomere loss, including amplification, terminal deletions, isochromosomes, dicentrics, rings, and transloctions, are the same types of chromosome rearrangements typically associated with human cancer (6). The importance of chromosome instability resulting from telomere loss in promoting cancer has been demonstrated in mice with a knockout in the RNA component of telomerase. When crossed with p53 knockout mice, these telomerase-deficient knockout mice demonstrate a high frequency of carcinomas that are typical of human cancer (4). Moreover, the tumors of these mice show chromosome rearrangements involving amplified regions adjacent to translocations, identical to the rearrangements that we have reported for chromosome instability resulting from B/F/B cycles (8-10).
Figure 1.
Mechanism of chromosome instability involving breakage/fusion/bridge cycles. A double-strand break near a telomere before or during DNA replication results in telomere loss and sister chromatid fusion. Due to the presence of two centromeres, the fused sister chromatids break during anaphase, resulting in an inverted repeat on the end of one of the chromosomes in one daughter cell and a terminal deletion on the other chromosome in the other daughter cell. The absence of a telomere on the broken chromosomes leads to additional fusions, bridges, and breaks in subsequent cell cycles, resulting in further amplification. The acquisition of a telomere, most often by translocation from other chromosomes, eventually stabilizes the chromosome. Represented in the illustration are a chromosome (solid green line), the replicated sister chromatids (double green lines), telomeres (red rectangles), centromeres (green circles), the orientation of the subtelomeric region (arrows), sites of chromosome fusion (diagonal lines), and a translocation (heavy blue line).
Replication stress as a source of DSBs at fragile sites in human cancer cells
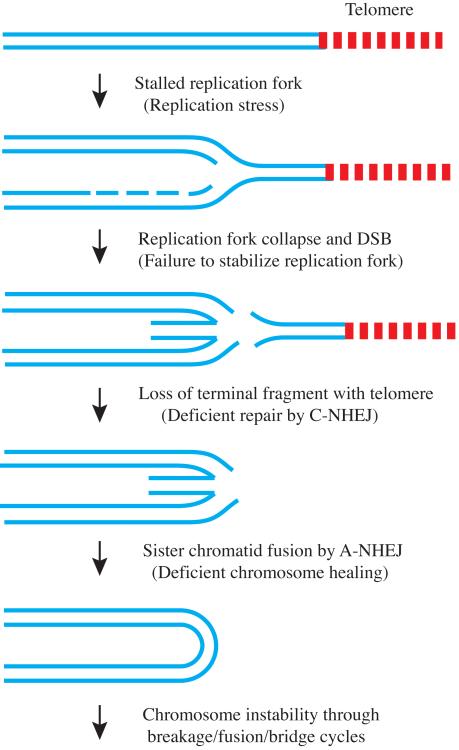
We have used I-SceI endonuclease to generate DSBs at the I-SceI site within the telomeric plasmid sequences to investigate how DSBs in subtelomeric regions affect telomere loss. These studies have been performed in both mouse embryonic stem (ES) cells (10-12) and in the EJ-30 human cancer cell line (13). The DSBs near telomeres in both cell types were found to produce GCRs that were similar to those resulting from spontaneous telomere loss in the EJ-30 tumor cell line. The I-SceI-induced GCRs in mouse ES cells primarily involved sister chromatid fusions (10-12), while additional GCRs, including fusions with other chromosomes, ring chromosomes, and translocations were also observed in EJ-30 (13). This high frequency of GCRs resulting from I-SceI-induced DSBs near telomeres suggests that the high rate of spontaneous telomere loss in human cancer cells might also result from DSBs near telomeres. How these DSBs might occur is not known, however, cancer cells have been demonstrated to have an increase in spontaneous DSBs as a result of oncogene-induced replication stress (14). Replication stress results when cancer cells that are continually traversing the cell cycle fail to adequately prepare the precursors required to replicate the genome. As a result, replication forks stall at fragile sites, which are regions that are difficult to replicate, leading to the formation of DSBs. A recent study using drug-induced replication stress has demonstrated that subtelomeric regions in mammalian cells are fragile sites, as shown by an increase in stalled replication forks near telomeres and telomere fragmentation (15). Based on these observations, it appears likely that subtelomeric regions in cancer cells experiencing replication stress would be sites of increased replication fork stalling and DSB formation (Fig. 2).
Figure 2.
Mechanisms proposed for spontaneous telomere loss in cancer cells. During DNA replication, the two DNA strands (blue lines) are converted into four strands as the replication fork progresses. Telomere loss in cancer cells occurs because replication forks stall at fragile sites, which includes telomeres, as a result of replication stress caused by continuous cell division. The failure to stabilize stalled replication forks in the proximity of a telomere would result in replication fork collapse and the formation of a double-strand break (DSBs), causing the loss of the terminal fragment containing the telomere. Similarly, replication forks encountering an I-SceI-induced DSB near a telomere would also result in telomere loss. Alternatively, the increased frequency of loss of the terminal fragment containing the telomere could result from a deficiency in DSB repair near telomeres. Finally, a failure to stabilize the chromosome after the loss of the telomere due to a deficiency in chromosome healing in cancer cells would further increase the likelihood of sister chromatid fusion or other gross chromosome rearrangements, which would then initiate chromosome instability.
Increased sensitivity of subtelomeric regions to DSBs
Oncogene-induced replication stress would affect DNA replication at numerous fragile sites within the cancer cell genome. However, subtelomeric regions are highly sensitive to DSBs, and therefore would be much more likely to experience gross chromosome rearrangements (GCRs) and chromosome instability in response to stalled replication forks than most fragile sites. The analysis of the consequences of I-SceI-induced DSBs at different locations along a chromosome in yeast demonstrated that DSBs in subtelomeric regions were much more likely to result in GCRs than DSBs at interstitial sites (16). Consistent with this observation, we have demonstrated that in the EJ-30 human tumor cell line, the frequency of large deletions, terminal deletions, and GCRs that result from I-SceI-induced DSBs is much greater at subtelomeric sites than at interstitial sites (13). This is not due to a difference in the frequency of DSBs, since the frequency of small deletions, which we (13) and others (17, 18) have found to be the most common type of event at interstitial I-SceI-induced DSBs, is the same at both locations. Our results therefore demonstrate that DSBs in subtelomeric regions in mammalian cells have a much greater probability of resulting in GCRs and chromosome instability than DSBs at interstitial sites.
The mechanism responsible for the increased sensitivity of subtelomeric regions to DSBs could also be responsible for the high rate of spontaneous telomere loss in cancer cells (Fig. 2). One possible mechanism would be a deficiency in repair of DSBs in subtelomeric regions by either nonhomologous end joining (NHEJ) or homologous recombination repair, since as mentioned earlier, replication stress in cancer cells is likely to result in DSBs near telomeres. Homologous recombination repair utilizes the sister chromatid as a template, while NHEJ involves the rejoining of the broken ends, often after processing (19). There are two forms of NHEJ, classical NHEJ (C-NHEJ) and alternative NHEJ (A-NHEJ) (20). C-NHEJ is a well-defined pathway and is relatively efficient in the rejoining DSBs, most often resulting in the restoration of I-SceI sites during repair of I-SceI-induced DSBs (21). In contrast, much less is known about A-NHEJ, which is responsible for most small deletions at I-SceI-induced DSBs (20, 21), and is commonly involved in the formation of GCRs (20). The similarity in the frequency of small deletions that we observed at interstitial and subtelomeric DSBs in the EJ-30 tumor cell line suggests that the efficiency of repair by A-NHEJ is not affected by the proximity of a telomere. This deficiency in repair of DSBs could result from the inhibition of ATM by the telomeric protein TRF2, as part of its role in protecting chromosome ends and preventing chromosome fusion (22). ATM is required for the repair of DSBs in heterochromatin (19), and subtelomeric regions have been demonstrated to consist of heterochromatin (23). Studies in yeast demonstrated that the relative frequency of NHEJ is decreased near telomeres (16), which was found to result from an increase in the repair of DSBs through unconventional mechanisms that resulted in the loss of the terminal fragment.
A second mechanism that could account for both the increased sensitivity of subtelomeric regions to DSBs and the increased rate of telomere loss in cancer cells is an inability to stabilize stalled replication forks near telomeres (Fig. 2). The stabilization of stalled replication forks is required to prevent replication fork collapse and the formation of DSBs (24). Stalled replication forks resulting from either replication stress or as a result of a replication fork encountering an I-SceI-induced DSB might be more likely to collapse near telomeres due to a deficiency in ATR, which like ATM, is inhibited at telomeres to prevent chromosome fusion (22). ATR is important in the stabilization of stalled replication forks, and cells deficient in ATR are highly sensitive to replication stress and DNA lesions (24). An additional factor that could further promote telomere loss would be the absence of origins of replication within the terminal fragment distal to the DSB, since dormant origins of replication can fire under conditions of replication stress as a way of ensuring complete DNA replication (25).
Chromosome healing compensates for deficient repair of DSBs near telomeres
Chromosome healing is the de novo addition of telomeric repeat sequences at the sites of DSBs, which in yeast has been demonstrated to involve telomerase. Chromosome healing in yeast is inhibited by the PIF1 helicase, which has been proposed as a mechanism to prevent chromosome healing from interfering with DSB repair (26). Consistent with this conclusion, the inhibition of chromosome healing by PIF1 at interstitial sites in yeast requires it to be phosphorylated by a MEC1/RAD53-dependent pathway in response to DSBs (27). Chromosomal healing also occurs in human germ line cells, as demonstrated by the role of terminal deletions resulting from the addition of telomeres to the ends of broken chromosomes in human genetic disease (28). However, chromosome healing has not been observed at interstitial DSBs generated by I-SceI in human cell lines that express telomerase (17, 29), and the expression of telomerase has little effect on the response of mouse cells to ionizing radiation (30). Therefore, as in yeast, chromosome healing appears to be closely regulated in mammalian cells.
Although chromosome healing is not observed at interstitial DSBs in mammalian cells, we have demonstrated that chromosome healing is a common event at DSBs near telomeres in mouse ES cells, where it accounts for approximately one-third of the rearrangements (11, 12). The absence of chromosome healing at DSBs near telomeres in ES cell lines with a knockout of the catalytic subunit of telomerase, mTERT (11), and the restoration of chromosome healing upon expression of mTERT in these cells (unpublished observation), demonstrates that chromosome healing involves telomerase in these ES cell lines, as it does in yeast. However, we have also observed that chromosome healing is a frequent event at DSBs near telomeres in a mouse ES cell line that had acquired the ability to maintain telomeres through a telomerase-independent pathway (11). Thus, human tumor cells that maintain telomeres through a telomerase-independent pathway might also be capable of performing chromosome healing. Regardless of the mechanism of chromosome healing, our studies in mouse ES cells demonstrated that unlike sister chromatid fusions, which typically occur following degradation, chromosome healing always occurred at or near the site of the DSB (11). Therefore, chromosome healing precedes and prevents degradation, GCRs, and chromosome instability resulting from DSBs near telomeres. Based on this observation and the fact that chromosome healing rarely occurs at DSBs at interstitial sites, we have hypothesized that chromosome healing serves as an alternative mechanism for dealing with DSBs near telomeres to compensate for the increased sensitivity to DSBs in these regions (13, 29). Although chromosome healing results in terminal deletions, little DNA would be lost with DSBs near telomeres, and therefore terminal deletions would be preferable to the alternative, which involves GCRs and chromosome instability.
An interesting observation made in the course of our studies is that the frequency of chromosome healing at DSBs near telomeres in the EJ-30 human cancer cell line is much lower than the frequency of chromosome healing in mouse ES cells. Unlike mouse ES cells, where chromosome healing is observed in approximately one-third of the HSV-tk-deficient subclones resulting from I-SceI-induced DSBs (11), chromosome healing in EJ-30 is observed in less than one percent of the HSV-tk-deficient subclones resulting from I-SceI-induced DSBs (13). Although this could be a result of differences between human and mouse cells, it may also be a result of a deficiency in chromosome healing near telomeres in human tumor cells. A deficiency in chromosome healing would promote chromosome instability by allowing sister chromatid fusions or other GCRs (Fig. 2). Thus, a deficiency in chromosome healing could be selected for in human cancer cells, where chromosome instability provides an advantage by increasing the frequency of chromosome rearrangements leading to tumor cell progression. Presumably, these changes in the efficiency of chromosome healing could involve alterations in the modification of PIF1 or other proteins that regulate telomerase, leading to the same type of suppression of chromosome healing near telomeres that normally occurs at interstitial DSBs. Understanding the regulation of chromosome healing by PIF1 or other proteins may therefore lead to new approaches for promoting chromosome healing in human cancer cells, and thereby limiting the extent of chromosome instability resulting from spontaneous telomere loss. This could lead to new anti-cancer therapies for use in combination with chemotherapeutic drugs to prevent the amplification of genes involved in tumor cell resistance. Moreover, this approach might also selectively sensitize cancer cells to ionizing radiation or chemotherapeutic drugs that kill cancer cells by producing DSBs. Interfering with the regulation of chromosome healing would lead to de novo telomere addition at interstitial DSBs and therefore interfere with DSB repair. While this would have little effect on normal human somatic cells that do not express telomerase, it would could have a dramatic effect on most human cancer cells that typically over-express telomerase.
Acknowledgements
This work was supported by National Institutes of Health grant CA12025.
Abbreviations
- (B/F/B)
breakage/fusion/bridge
- (DSBs)
double-strand breaks
- (GCRs)
gross chromosome rearrangements
- (HSV-tk)
herpes simplex thymidine kinase
- (HRR)
homologous recombination repair
- (NHEJ)
nonhomologous end joining
Footnotes
Disclosure of Potential Conflicts of Interest No conflicts of interest to disclose.
References
- 1.Chan SR, Blackburn EH. Telomeres and telomerase. Philos Trans R Soc Lond B Biol Sci. 2004;359:109–21. doi: 10.1098/rstb.2003.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–34. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 3.Reddel RR, Bryan TM. Alternative lengthening of telomeres: dangerous road less travelled. Lancet. 2003;361:1840–1. doi: 10.1016/S0140-6736(03)13538-6. [DOI] [PubMed] [Google Scholar]
- 4.Maser RS, DePinho RA. Connecting chromosomes, crisis, and cancer. Science. 2002;297:565–9. doi: 10.1126/science.297.5581.565. [DOI] [PubMed] [Google Scholar]
- 5.Gisselsson D, Jonson T, Petersen A, et al. Telomere dysfunction triggers extensive DNA fragmentation and evolution of complex chromosome abnormalities in human malignant tumors. Proc Natl Acad Sci USA. 2001;98:12683–8. doi: 10.1073/pnas.211357798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murnane JP. Telomeres and chromosome instability. DNA Repair (Amst) 2006;5:1082–92. doi: 10.1016/j.dnarep.2006.05.030. [DOI] [PubMed] [Google Scholar]
- 7.Fouladi B, Miller D, Sabatier L, Murnane JP. The relationship between spontaneous telomere loss and chromosome instability in a human tumor cell line. Neoplasia. 2000;2:540–54. doi: 10.1038/sj.neo.7900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lo AWI, Sabatier L, Fouladi B, Pottier G, Ricoul M, Murnane JP. DNA amplification by breakage/fusion/bridge cycles initiated by spontaneous telomere loss in a human cancer cell line. Neoplasia. 2002;6:531–8. doi: 10.1038/sj.neo.7900267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sabatier L, Ricoul M, Pottier G, Murnane JP. The loss of a single telomere can result in genomic instability involving multiple chromosomes in a human tumor cell line. Mol. Cancer Res. 2005;3:139–50. doi: 10.1158/1541-7786.MCR-04-0194. [DOI] [PubMed] [Google Scholar]
- 10.Lo AWI, Sprung CN, Fouladi B, et al. Chromosome instability as a result of double-strand breaks near telomeres in mouse embryonic stem cells. Mol Cell Biol. 2002;22:4836–50. doi: 10.1128/MCB.22.13.4836-4850.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao Q, Reynolds GE, Wilcox A, et al. Telomerase-dependent and -independent chromosome healing in mouse embryonic stem cells. DNA Repair. 2008;7:1233–49. doi: 10.1016/j.dnarep.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sprung CN, Reynolds GE, Jasin M, Murnane JP. Chromosome healing in mouse embryonic stem cells. Proc Natl Acad Sci USA. 1999;96:6781–6. doi: 10.1073/pnas.96.12.6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zschenker O, Kulkarni A, Miller D, et al. Increased sensitivity of subtelomeric regions to DNA double-strand breaks in a human tumor cell line. DNA Repair. 2009;8:886–900. doi: 10.1016/j.dnarep.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsantoulis PK, Kotsinas A, Sfikakis PP, et al. Oncogene-induced replication stress preferentially targets common fragile sites in preneoplastic lesions. A genome-wide study. Oncogene. 2008;27:3256–64. doi: 10.1038/sj.onc.1210989. [DOI] [PubMed] [Google Scholar]
- 15.Sfeir A, Kosiyatrakul ST, Hockemeyer D, et al. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ricchetti M, Dujon B, Fairhead C. Distance from the chromosome end determines the efficiency of double-strand break repair in subtelomeres of haploid yeast. J Mol Biol. 2003;328:847–62. doi: 10.1016/s0022-2836(03)00315-2. [DOI] [PubMed] [Google Scholar]
- 17.Varga T, Aplan PD. Chromosomal aberrations induced by double strand DNA breaks. DNA Repair (Amst) 2005;4:1038–46. doi: 10.1016/j.dnarep.2005.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Honma M, Sakuraba M, Koizumi T, Takashima Y, Sakamoto H, Hayashi M. Non-homologous end-joining for repairing I-SceI-induced DNA double strand breaks in human cells. DNA Repair (Amst) 2007;6:781–8. doi: 10.1016/j.dnarep.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 19.Goodarzi AA, Noon AT, Jeggo PA. The impact of heterochromatin on DSB repair. Biochem Soc Trans. 2009;37:569–76. doi: 10.1042/BST0370569. [DOI] [PubMed] [Google Scholar]
- 20.Zha S, Boboila C, Alt FW. Mre11: roles in DNA repair beyond homologous recombination. Nat Struct Mol Biol. 2009;16:798–800. doi: 10.1038/nsmb0809-798. [DOI] [PubMed] [Google Scholar]
- 21.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–71. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 23.Benetti R, Garcia-Cao M, Blasco MA. Telomere length regulates the epigenetic status of mammalian telomeres and subtelomeres. Nat Genet. 2007;39:243–50. doi: 10.1038/ng1952. [DOI] [PubMed] [Google Scholar]
- 24.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–27. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ge XQ, Jackson DA, Blow JJ. Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev. 2007;21:3331–41. doi: 10.1101/gad.457807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou J-Q, Monson EK, Teng S-C, Schultz VP, Zakian VA. Pif1p helicase, a catalytic inhibitor of telomerase in yeast. Science. 2000;289:771–4. doi: 10.1126/science.289.5480.771. [DOI] [PubMed] [Google Scholar]
- 27.Makovets S, Blackburn EH. DNA damage signalling prevents deleterious telomere addition at DNA breaks. Nat Cell Biol. 2009;11:1383–6. doi: 10.1038/ncb1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varley H, Di S, Scherer SW, Royle NJ. Characterization of terminal deletions at 7q32 and 22q13.3 healed by de novo telomere addition. Am J Hum Genet. 2000;67:610–22. doi: 10.1086/303050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kulkarni A, Zschenker O, Reynolds G, Miller D, Murnane JP. The effect of telomere proximity on telomere position effect, chromosome healing and sensitivity to DNA double-strand breaks in a human tumor cell line. Mol Cell Biol. 2010 doi: 10.1128/MCB.01137-09. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Latre L, Genesca A, Martin M, et al. Repair of DNA broken ends is similar in embryonic fibroblasts with and without telomerase. Radiat Res. 2004;162:136–42. doi: 10.1667/rr3203. [DOI] [PubMed] [Google Scholar]