Abstract
Termination of murine rDNA transcription by RNA polymerase I (Pol I) requires pausing of Pol I by terminator-bound TTF-I (transcription termination factor for Pol I), followed by dissociation of the ternary complex by PTRF (Pol I and transcript release factor). To examine the functional correlation between transcription termination and initiation, we have compared transcription on terminator-containing and terminator-less rDNA templates. We demonstrate that terminated RNA molecules are more efficiently synthesized than run-off transcripts, indicating that termination facilitates reinitiation. Transcriptional enhancement is observed in multiple- but not single-round transcription assays measuring either promoter-dependent or promoter-independent Pol I transcription. Increased synthesis of terminated transcripts is observed in crude extracts but not in a PTRF-free reconstituted transcription system, indicating that PTRF-mediated release of pre-rRNA is responsible for transcriptional enhancement. Consistent with PTRF serving an important role in modulating the efficiency of rRNA synthesis, PTRF exhibits pronounced charge heterogeneity, is phosphorylated at multiple sites and fractionates into transcriptionally active and inactive forms. The results suggest that regulation of PTRF activity may be an as yet unrecognized means to control the efficiency of ribosomal RNA synthesis.
INTRODUCTION
Once a gene is activated, the amount of transcripts is determined primarily by the number of reinitiation events. Despite the remarkable progress that has been made in understanding the molecular mechanisms that govern basal and activated transcription in eukaryotes, relatively less effort has been directed towards understanding the mechanisms that mediate the recycling of RNA polymerases. For RNA polymerase II (Pol II), it has been suggested that the control over initiation and reinitiation can be uncoupled (1), raising the possibility that the two processes have different requirements. On the other hand, proper termination appears to be a prerequisite for recycling of Pol III to yield high transcription efficiency. Pol III was shown to be committed to reinitiate on the same gene, in a way that allows it to complete new cycles more rapidly than the initial one (2). These data imply that Pol III is not released from the template to be recruited to the promoter in order to start a new transcription cycle. Rather, a preferential termination pathway allows RNA release and reinitiation without release of Pol III.
Pol III-transcribed genes are small and, therefore, the facilitated reinitiation pathway could be brought about by a specific contact between terminating Pol III and a component of the preinitiation complex. However, a terminator-dependent reinitiation pathway has also been proposed to be responsible for the high transcription rate of Pol I transcription. The tandemly repeated mammalian rRNA genes are large, each ∼14 kb coding region being separated by ~30 kb of intergenic spacer sequences. Eukaryotic ribosomal transcription units are flanked both at their 5′- and 3′-side by one or more terminator elements. In the mouse, a repeated 18 bp sequence motif (AGGTCGACCAGA/TT/ANTCCG), termed ‘Sal box’, functions as the transcription terminator (3,4). In addition, a T-rich sequence upstream of the terminator has been shown to be involved in both release of terminated transcripts and 3′-terminal processing of pre-rRNA (5,6). The ‘Sal box’ is recognized by TTF-I (transcription termination factor), a specific DNA binding protein that stops elongating Pol I when bound to the terminator (7,8). DNA-bound TTF-I on its own is not sufficient for transcript release. In mammals, dissociation of the ternary transcription complex at the terminator requires PTRF (Pol I and transcript release factor), a 44 kDa protein that interacts with Pol I, TTF-I and the 3′ end of pre-rRNA (6,9). Substitution of 3′-terminal uridylates by guanine residues abolishes PTRF binding and impairs release activity. Thus, transcription termination in mammals requires two cis-acting DNA elements, the ‘Sal box’ and the T-rich element located upstream of the terminator, and two trans-acting factors, e.g. TTF-I that stops elongating Pol I and PTRF that dissociates TTF-I-paused transcription complexes.
The fact that binding sites for TTF-I are present both upstream and downstream of the rDNA transcription unit suggests a functional linkage between transcription termination and initiation. A model has been proposed in which each rDNA transcription unit forms a protein-mediated loop that juxtaposes the promoter and terminator element (10,11). According to this ‘ribomotor’ or ‘polymerase handover’ model, Pol I molecules that have terminated nascent pre-rRNA chains could be transferred directly to the gene promoter without being released from the template. TTF-I, which has binding sites near the promoter and the 3′ end of the rDNA transcription unit, would be a perfect candidate for mediating intramolecular interactions between the 5′ and 3′ end of rDNA. In support of this view, TTF-I has been shown to interact simultaneously with two separate DNA fragments bearing ‘Sal box’ elements (12) and, therefore, to be potentially capable of linking the promoter-proximal ‘Sal box’ with the distal terminators.
To test this model, we have compared transcription on templates that contain the upstream and downstream terminators or either of these elements, respectively. We found that both promoter-dependent and promoter-independent transcription was more efficient when termination was allowed to occur. However, TTF-I-mediated stimulation of Pol I transcription did not require the presence of the promoter-proximal ‘Sal box’, indicating that the increase in the amount of transcripts was not due to ‘handover’ of Pol I via DNA loops. We demonstrate that PTRF, by dissociating ternary complexes, stimulates recycling of Pol I. Moreover, we demonstrate that cellular PTRF is phosphorylated at multiple sites, a finding that suggests that 3′ end formation of pre-rRNA and/or recycling of Pol I may be regulated by changes in the phosphorylation pattern of PTRF.
MATERIALS AND METHODS
Plasmid constructs
pMrWT contains mouse rDNA sequences from –170 to +155. The minigene construct pMrT2 (8) contains the rDNA sequence from pMrWT fused to the murine terminator element T2 (nucleotides 604–685 downstream of the 3′ end of 28S rRNA). pMrT0mT2 is similar to pMrT2, but lacks three nucleotides within the upstream terminator that abolishes TTF-I binding. The plasmid pCAT-T6-T1 contains a 49 bp fragment from the 3′-terminal region of mouse rDNA (nucleotides 556–604 downstream of the 3′ end of 28S rRNA) including the terminator T1 and flanking sequences (8).
In vitro transcription assays
Transcription in crude extracts or a reconstituted system has been described previously (8,13,14). Reactions (25 µl) contained 15–50 ng template DNA (pMrWT/NarI or pMrT2/EcoRI) and 100 µg S-100 extract proteins in 12 mM Tris–HCl pH 7.9, 0.1 mM EDTA, 0.5 mM DTE, 5 mM MgCl2, 80 mM KCl, 12% glycerol, 0.66 mM each of ATP, CTP, GTP, 0.01 mM UTP and 1 µCi [α-32P]UTP (3000 Ci/mmol). The reconstituted transcription system contained 2–4 µl Pol I [S-300 fraction (14)], 1 µl TIF-IA/TIF-IC (polylysine–agarose fraction), 3 µl TIF-IB (CM-400 fraction) and 5 ng UBF. After incubation for 60 min at 30°C, transcripts were isolated and analyzed on 4.5% polyacrylamide gels.
Transcription on bead-bound tailed templates
Transcription assays using immobilized tailed templates have been described (5,6,9). Briefly, the plasmids were cut with BglII and an oligonucleotide (3′-ACCAAAAAAACTAG-5′) was ligated to the cohesive ends to create a 10 nt 3′ overhang. The template was cut with HindIII, the free oligonucleotides were removed by precipitation with 7.5% polyethylene glycol 6000 in the presence of 0.9 M NaCl. For immobilization, biotin-14-dATP was incorporated into the HindIII restriction site using Klenow enzyme. Biotinylated template (10 µg) was bound to 500 µl streptavidin magnetic beads (Dynal) and incubated with bovine serum albumin and phosphatidylcholine (5 mg/ml each) to block non-specific binding sites as described (6). Transcription reactions (25 µl) containing 5 µl (100 ng) bead-bound template, 12 mM Tris–HCl pH 7.9, 5 mM MgCl2, 0.06 mM EDTA, 12% glycerol, 70 mM KCl and 0.5 mM UpG dinucleotide (Sigma) were pre-incubated for 10 min at 30°C with 0.5 U mouse Pol I and 30 ng TTF-I. Transcription was started by the addition of 0.6 mM each of ATP, UTP and CTP, 12.5 µM GTP and 8 µCi [α-32P]GTP. After incubation for 10 min in the presence or absence of PTRF, transcripts were separated into template-bound and released fractions. Then an equal volume of stop buffer (0.2 M ammonium acetate pH 5.2, 0.4% SDS, 1 mg/ml yeast tRNA) was added, RNA was extracted, precipitated with ethanol and resolved on 6% polyacrylamide/7 M urea gels.
Protein purification
FLAG-tagged UBF1 was immunopurified from baculovirus-infected Sf9 cells as described (14). Histidine-tagged mouse PTRF was expressed in Escherichia coli BL21(DE3)pLysS from pRSET-PTRF (9) and purified on Ni2+-chelate agarose and S-Sepharose (Pharmacia). Alternatively, pGEX-PTRF was used to express and purify a glutathione S-transferase (GST)–PTRF fusion protein. Histidine-tagged murine TTFΔN185 was expressed in baculovirus-infected Sf9 cells as described (15).
Western blots
Proteins were separated by SDS–PAGE and transferred to nitrocellulose. The membrane was first blocked in PBS containing 5% milk powder and 0.2% Tween-20 for 1 h, then incubated for 10 h at 4°C with affinity-purified chicken anti-PTRF antibodies (1:50), followed by incubation with anti-chicken IgY antibodies coupled with horseradish peroxidase (diluted 1:2000). Proteins were visualized by enhanced chemoluminescence (ECL, Amersham).
In vivo phosphorylation and tryptic phosphopeptide mapping
NIH 3T3 cells (1 × 105) were labeled for 8 h in phosphate-free DMEM containing 10% dialyzed FCS and 1 mCi/ml 32P-orthophosphate. Cells were lysed in RIPA buffer (20 mM Tris–HCl pH 8.0, 100 mM NaCl, 0.5% sodium deoxycholate, 0.5% NP-40, 0.5% SDS, 10 mM EGTA, 20 mM KF, 1 mM sodium orthovanadate, 10 mM K2HPO4, 2 µg/ml of each leupeptin, aprotinin and pepstatin) and incubated for 5 h at 4°C with rabbit α-PTRF antibodies coupled to protein A Sepharose CL-4B (Pharmacia). After extensive washing with RIPA buffer, immunoprecipitated proteins were separated by 10% SDS–PAGE, transferred to nitrocellulose and visualized by autoradiography. Labeled PTRF was cut out and processed for tryptic phosphopeptide mapping as described (16).
Two-dimensional (2D) gel electrophoresis
Proteins were precipitated, dissolved in 8 M urea, 0.5% (v/v) Pharmalyte (pH 4–7), 0.2% DTT, 0.5% Triton X-100, and subjected to isoelectric focusing on Immobiline Drystrips (Pharmacia), according to the instructions of the manufacturer. After isoelectric focusing, the strips were equilibrated for 10 min in 50 mM Tris–HCl pH 6.8, 30% glycerol, 6 M urea, 2% SDS and 0.2% DTT before being applied for electrophoresis on 10% SDS–polyacrylamide gels. Proteins were blotted onto nitrocellulose filters and PTRF was visualized by immunostaining.
RESULTS
Stimulation of the synthesis of terminated Pol I transcripts
To investigate a possible link between termination and initiation of Pol I transcription, we assayed transcriptional activity in S-100 extracts using templates that either contain or lack downstream or upstream terminator elements. pMrWT contains a murine rDNA fragment (from –170 to +155) including the promoter and the upstream terminator element T0. pMrT2 is an artificial minigene representing a fusion of the gene promoter with a 3′-terminal rDNA fragment including the second terminator T2 (Fig. 1A). On the terminator-less construct (pMrWT/NarI) recombinant TTF-I did not affect the amount of 378 nt run-off transcripts synthesized (Fig. 1B, lanes 1–3). On pMrT2, however, exogenous TTF-I not only efficiently terminated transcription, but also augmented the overall amount of transcripts synthesized. Thus, TTF-I enhances transcription on terminator-containing but not terminator-less constructs.
Figure 1.
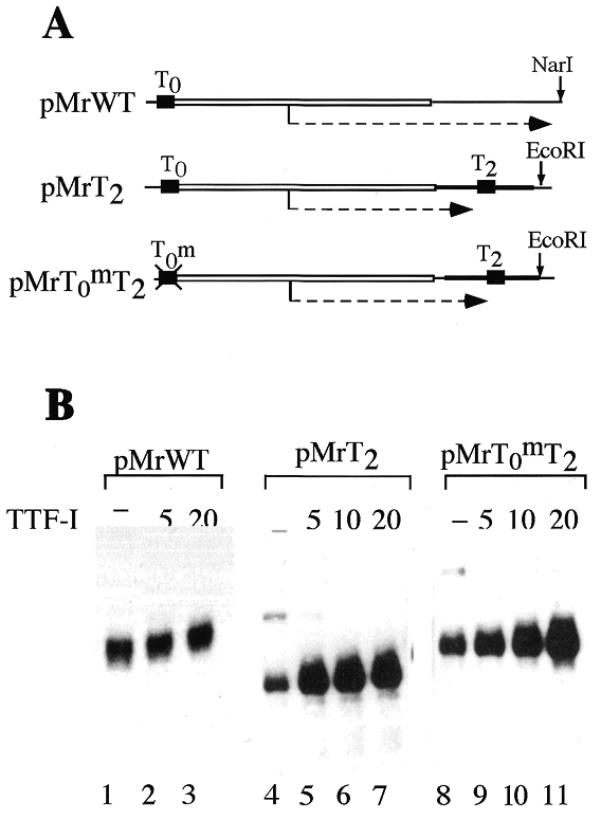
TTF-I stimulates transcription on terminator-containing templates. (A) Schematic representation of the templates used. The open bar marks 5′-terminal murine rDNA sequences, the black line 3′-terminal rDNA sequences and the black boxes indicate the position of the upstream and downstream terminator elements T0 and T2, respectively. Run-off and terminated transcripts are indicated by a dashed arrow. (B) Transcription assays. Reactions contained 20 ng of the indicated template DNA, 6 µl S-100 extract and either no TTF-I (lanes 1, 4 and 8), 5 fmol (lanes 2, 5 and 9), 10 fmol (lanes 6 and 10) or 20 fmol (lanes 3, 7 and 11) of recombinant TTFΔN185.
Previous results suggested a model in which simultaneous binding of TTF-I to the upstream and downstream terminator elements would facilitate ‘handover’ of Pol I from the terminator to the gene promoter, thereby increasing transcription efficiency (10–12). If this model was correct and the TTF-I-mediated transcriptional enhancement was due to a ‘handover’ of Pol I molecules from the terminator to the promoter, then transcription from a template containing no functional upstream terminator should not be stimulated by TTF-I. To test this, we used a template (pMrT0mT2) that is similar to pMrT2 but contains a mutated upstream terminator that is not recognized by TTF-I. Significantly, TTF-I stimulated the synthesis of terminated transcripts at the mutant template to the same degree as at pMrT2 (lanes 9–11), indicating that in the cell-free transcription system used transcriptional stimulation does not involve binding of TTF-I to the upstream terminator. Hence, TTF-I augments transcription by a mechanism that does not involve a cross-talk between the promoter-proximal terminator T0 and the downstream terminator T2.
Enhanced synthesis of terminated transcripts is due to elevated reinitiation
The more efficient transcription of templates containing a downstream terminator suggests that Pol I released at the terminator associates more efficiently with preinitiation complexes than Pol I that has been liberated from the linear template. To examine this, we measured the amount of run-off versus terminated transcripts at different incubation times. Quantitation of run-off and terminated transcripts by PhosphoImager analysis revealed a ratio of 1:1.3 at early time points (Fig. 2A, lanes 1 and 2), which increased to 1:3.6 at 90 min (lane 6), suggesting that proper termination augments reinitiation of Pol I.
Figure 2.
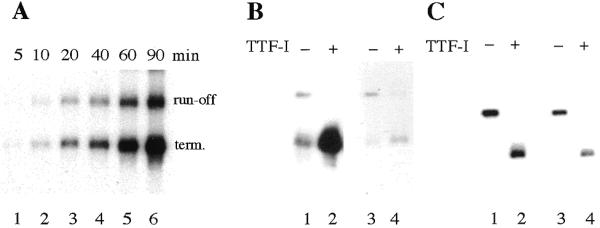
Termination facilitates reinitiation. (A) Time course of transcription. Assays contained 10 ng template DNA (pMrT2/EcoRI) and partially purified transcription factors. Reactions were started by addition of nucleotides and samples were taken at the time points indicated. (B) Sarkosyl sensitivity of TTF-I-mediated transcription stimulation. Transcription reactions contained 6 µl S-100 extracts and 30 ng pMrT2/EcoRI in the absence (lanes 1 and 3) or presence of 5 fmol recombinant TTFΔN185 (lanes 2 and 4). To prevent reinitiation, the reactions shown in lanes 3 and 4 contained 0.045% Sarkosyl. (C) TTF-I is not sufficient for the enhanced synthesis of terminated transcripts. Transcriptions were performed in a reconstituted system containing 20 ng pMrT2/EcoRI, 3 ng recombinant UBF, purified TIF-IA, TIF-IB, TIF-IC and two preparations of Pol I, i.e., H-400 (lanes 1 and 2) or S-300 (lanes 3 and 4). Where indicated, 5 fmol TTFΔN185 were added.
This view is supported by single-round transcription assays (Fig. 2B). Initiation complexes were assembled at the rDNA promoter by preincubating the template pMrT2 with S-100 extract. After complex formation, TTF-I was added and transcription was initiated by addition of ribonucleotides. Reactions were performed in the absence (lanes 1 and 2) or presence of 0.045% Sarkosyl (lanes 4 and 5). At this concentration of Sarkosyl, preformed initiation complexes are not disrupted, but both the assembly of new complexes and reinitiation is prevented (17). Clearly, TTF-I mediated transcriptional stimulation was only observed in the absence of Sarkosyl, e.g., when reinitiation was allowed to occur. In single-round transcription assays, on the other hand, the overall level of transcripts remained the same irrespective of whether or not TTF-I was present. This indicates that transcription stimulation occurs at a step after initiation complex assembly.
For class III transcription, the presence of TFIIIB and TIFIIIC were sufficient to augment reinitiation by purified Pol III (2). To test whether the increased synthesis of terminated transcripts that occurs in crude extracts can also be observed with purified components, transcriptions were performed in a reconstituted transcription system. The system used contained template DNA, recombinant UBF and purified cellular Pol I, TIF-IB, TIF-IA and TIF-IC (14). Notably, in contrast to transcriptions in crude systems, TTF-I did not augment transcription in the reconstituted system (Fig. 2C). The overall amount of RNA was the same, regardless of whether run-off or terminated transcripts were synthesized. This result suggests that factor(s) being present in crude extracts but absent in the system containing purified transcription factors and Pol I is involved in TTF-I-mediated transcriptional enhancement.
PTRF stimulates transcription on tailed templates
The results presented so far suggest that the increase in transcriptional efficiency on templates containing downstream terminator(s) is due to facilitated recycling of Pol I after termination. Dissociation of the paused elongation complex by DNA-bound TTF-I requires PTRF, a factor that induces the release of Pol I and nascent transcripts from the template (9). We therefore reasoned that PTRF could be causally responsible for enhanced recycling of Pol I after transcription termination. To test this, transcription was performed in a ‘tailed’ template assay. This assay takes advantage of the fact that the presence of a 3′-terminal extension or ‘tail’ on a linear template allows specific initiation by Pol I in the absence of any other accessory factors. In the experiment shown in Figure 3, a template was used that contains 3′ terminal mouse rDNA sequences including the terminator T1 and natural flanking regions (pCAT-T6-T1). After linearization, an oligonucleotide was ligated to the 5′ end of the template to yield a single-stranded 3′ overhang that serves as an entry site for Pol I. To monitor dissociation of paused transcription complexes, a magnetic bead was attached to the downstream end of the template via a biotin–streptavidin linkage. This allows separation of template-bound ternary complexes containing nascent RNA molecules from transcripts released into the supernatant. In the presence of TTF-I, two transcripts were generated: a longer primary transcript (the length of which corresponds to the distance from the tail to just upstream of the terminator) and a smaller transcript, which is the product of a 3′-terminal processing or ‘backsliding’ reaction that removes four nucleotides from the primary transcript (5). In the absence of PTRF most transcripts remain associated with the template (Fig. 3A, lanes 1 and 2). Addition of increasing amounts of PTRF had two effects. First, the ratio of template-bound and free RNA molecules was altered indicating that transcripts were efficiently released (lanes 4, 6 and 8). Second, the amount of transcripts was augmented in the presence of an increasing amount of PTRF.
Figure 3.
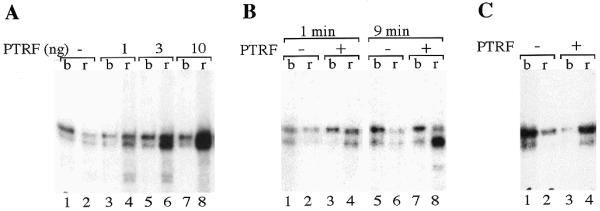
PTRF stimulates transcription on tailed templates. (A) PTRF-mediated transcript release and transcriptional stimulation. Reactions contained 0.5 U Pol I, immobilized tailed template (pCAT-T6-T1), 30 ng TTFΔN185 and 1, 3 or 10 ng histidine-tagged PTRF as indicated. Transcripts were separated into template-bound (b) and released (r) fractions by magnetic attraction. (B) Time-dependent transcript release and transcriptional stimulation. Transcription assays contained 100 ng of immobilized tailed template pCAT-T6-T1, 0.5 U mouse Pol I and 30 ng TTFΔN185. After incubation for 10 min at 30°C, 3 ng PTRF was added and incubation was continued for another 1 or 9 min. Transcripts were fractionated into template-bound (b) and released (r) molecules. (C) Transcript release on ternary transcription complexes. Transcription reactions contained bead-bound tailed template (pCAT-T6-T1), NTPs (including [α-32P]GTP), 0.5 U Pol I and 30 ng recombinant TTF-I. After incubation for 10 min, complexes paused at the terminator were isolated by magnetic attraction, washed with buffer AM-200 and incubated for another 10 min with 10 ng PTRF in the presence of cold NTPs.
These data indicate that, as a consequence of ternary transcription complex dissociation, PTRF augments reinitiation. If this assumption is correct, PTRF-mediated increase in transcriptional activity should be more pronounced at longer incubation times. Indeed, similar amounts of transcripts were synthesized within 1 min in the absence or presence of PTRF (Fig. 3B, lanes 1–4), whereas in the presence of PTRF ~10-fold more transcripts were synthesized after 9 min (lanes 5–8). This time dependence of transcription stimulation suggests that PTRF increases the ability of Pol I to carry out secondary initiation events subsequent to the initial round of transcription.
This view is supported by another experimental approach that measures dissociation of stalled ternary transcription complexes but not reinitiation. In the experiment shown in Figure 3C, reactions containing bead-bound tailed template, Pol I and TTF-I were briefly incubated with nucleotides to allow the formation of ternary transcription complexes that have paused at the terminator. Then, paused ternary complexes were isolated by magnetic attraction, washed and incubated further in transcription buffer in the presence or absence of PTRF. Consistent with previous results (6,9), PTRF induced the release of both the primary and processed transcript (Fig. 3C). In this case, however, the overall amount of transcripts remained the same.
Separation of transcriptionally active and inactive forms of PTRF
The finding that PTRF stimulates recycling of Pol I suggests that this factor may serve a regulatory role in rDNA transcription. To investigate whether the activity of cellular PTRF is regulated, we tried to isolate functionally different forms of PTRF. For this, nuclear extract proteins were separated on a phosphocellulose column followed by chromatography on S-Sepharose using a linear salt gradient from 100 to 500 mM KCl. Individual fractions were assayed in the tailed template assay for transcript release activity and on immunoblots to monitor PTRF protein. The western blot in Figure 4A demonstrates that >50% of cellular PTRF eluted at 200 mM KCl, the rest eluted at between 400 and 450 mM KCl. Notably, there was no correlation between transcript release activity and the amount of PTRF present in the fraction. The fractions eluting at 200 mM KCl which contain the majority of PTRF were significantly less active than those eluting between 400 and 450 mM KCl (Fig. 4B). Moreover, PTRF-containing fractions with low release activity failed to augment overall transcription. This finding indicates that PTRF activity and transcriptional stimulation are intimately coupled. Separation of active and inactive forms of PTRF was also observed on other chromatographic resins tested (data not shown). Thus, cellular PTRF appears to exist in functionally distinct forms that differ in their capability to liberate terminated transcripts from the template and augment transcription.
Figure 4.
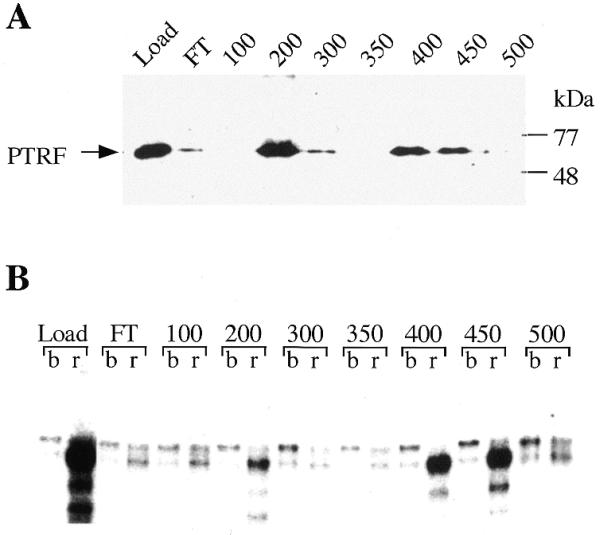
Chromatographic separation of active and inactive forms of PTRF. (A) Western blot. PTRF-containing fractions from a phosphocellulose column were pooled (Load) and fractionated on S-Sepharose. The flow-through (FT) and individual fractions eluting at the salt concentrations indicated were analyzed on western blots using chicken anti-PTRF antibodies. (B) Transcript release assay. The reactions contained bead-bound 3′-end tailed pCAT-T6-T1, 0.5 U Pol I, 30 ng TTF-I and 5 µl of the respective fractions shown at the top of the gel. After incubation, the assays were separated into template-bound (b) and released (r) fractions.
PTRF is phosphorylated at multiple sites
The fact that native PTRF fractionates into transcriptionally active and inactive forms suggests that the activity of PTRF may be modified by phosphorylation. As a first step to address this issue, we analyzed the migration of cellular PTRF by 2D gel electrophoresis. In the first dimension (isoelectric focusing), the proteins are separated by charge, whereas in the second dimension (SDS–PAGE) the proteins are separated by size. On 2D gels, two populations of cellular PTRF were observed with isoelectric points (IEP) of about pH 4.2 and 5.6 (Fig. 5A). The pronounced charge heterogeneity suggests that PTRF is modified at multiple sites.
Figure 5.
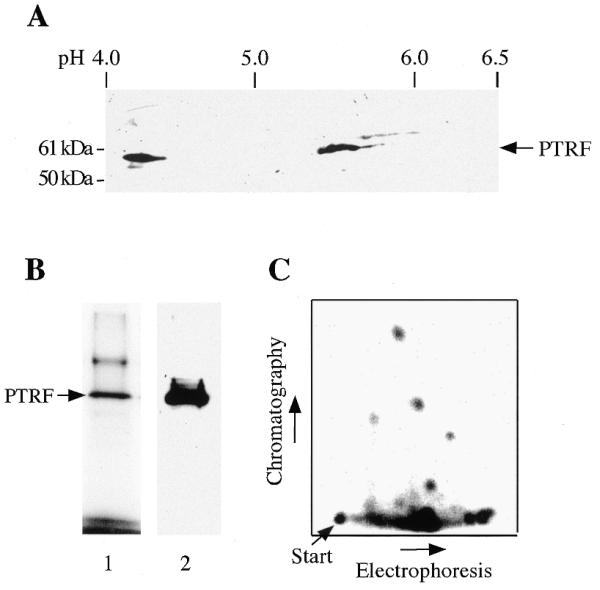
PTRF is phosphorylated at multiple sites. (A) 2D gel electrophoresis of PTRF. Nuclear extract proteins were separated by 2D electrophoresis, and PTRF was detected on western blots using anti-PTRF antibodies. (B) Tryptic phosphopeptide map of PTRF labeled in vivo. NIH 3T3 cells were metabolically labeled with 32P-orthophosphate, PTRF was immunoprecipitated, resolved by 10% SDS–PAGE and detected by autoradiography (lane 1). In parallel, PTRF was immunoprecipitated from unlabeled cells and visualized on immunoblots with anti-PTRF antibodies (lane 2). (C) Radiolabeled PTRF was digested with trypsin and subjected to 2D peptide mapping (right panel).
To test this, NIH 3T3 cells were metabolically labeled with 32P-orthophosphate, PTRF was immunoprecipitated and radiolabeled proteins were resolved by SDS–PAGE. As shown in Figure 5B, PTRF represented the major phosphorylated protein in the immunoprecipitate. After digestion with trypsin, peptides were subjected to 2D fingerprint analysis (Fig. 5C). Consistent with multiple phosphorylations being responsible for the charge heterogeneity of PTRF, a complex pattern of tryptic phosphopeptides was observed. Although we still do not know about both the kinases that modify PTRF and the sites that are phosphorylated, the finding that PTRF is phosphorylated at multiple sites suggests that changes in the phosphorylation pattern may alter PTRF activity.
As a first step to find out whether phosphorylation may alter PTRF activity, fractions containing active and inactive forms of PTRF were analyzed by 2D gel electrophoresis. For this, PTRF was purified by a series of chromatographic steps (Fig. 6A). Fractions from the final purification step (MQII) were assayed for their capability to dissociate ternary transcription complexes. Again, the amount of PTRF did not correlate with transcript release activity. Early eluting fractions (#18) failed to dissociate ternary transcription complexes (Fig. 6B, lanes 1–4), whereas equal amounts of PTRF eluting at higher salt concentrations (#24) were highly active (lanes 5–8). On 2D gels, both fractions showed a similar heterogeneity, the only difference being an additional spot at pH 4.5 in the active fraction #24 that was not present in the inactive fraction #18 (Fig. 6C). The spot at pH 4.5, on the other hand, was reproducibly found in release-competent fractions. We believe that this highly modified subpopulation of PTRF may represent the enzyme entity that catalyzes the dissociation of ternary transcription complexes. Whether or not the other forms of PTRF serve different cellular functions remains to be investigated.
Figure 6.
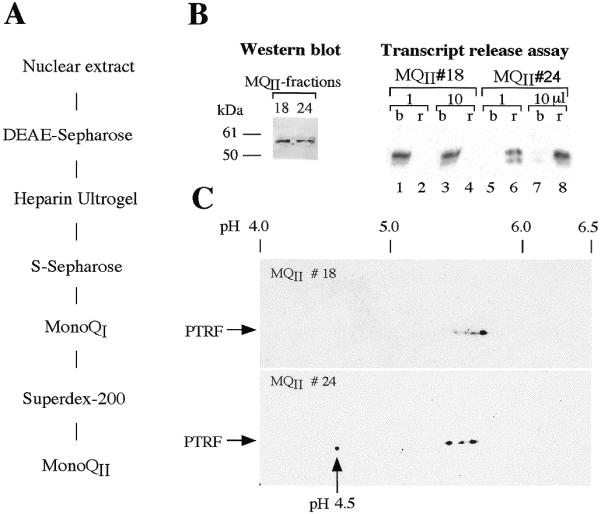
Charge heterogeneity of active and inactive forms of PTRF. (A) Diagram showing the chromatographic steps used to purify PTRF from nuclear extracts. (B) Chromatographic separation of release-competent and -incompetent forms of PTRF. Individual fractions from the last purification step, e.g., Mono QII, were analyzed on western blots or assayed for their capability to dissociate ternary transcription complexes. Ternary complexes were formed by pre-incubating bead-bound tailed template (pCAT-T6-T1) for 5 min with Pol I, TTF-I and nucleotides to allow Pol I to reach the terminator. Paused complexes were removed by magnetic attraction, washed with buffer AM-200, and then incubated for another 5 min with cold nucleotides in the presence of fractions MQII #18 and #24, respectively. Transcripts were separated into template-bound (b) and released (r) fractions. 2D gel electrophoresis of fractions MQII #18 and #24. The two fractions were subjected to 2D gel electrophoresis, blotted onto nitrocellulose filters, and PTRF was immunostained with anti-PTRF antibodies.
DISCUSSION
Based on the initial observation that in crude transcription systems terminated transcripts are more efficiently synthesized than run-off transcripts, we have investigated the functional linkage between transcription termination and initiation. We were intrigued by the ‘ribomotor’ model (10), which could account for the marked efficiency of cellular Pol I transcription and may provide a molecular explanation of how the process of transcription initiation and termination could be mechanistically coupled. According to this model, each rDNA transcription unit forms a loop which may channel RNA polymerases directly to the promoter after termination, thus bypassing the pool of free Pol I and ensuring efficient recycling of Pol I. This model is supported by the finding that in yeast and mammals the rDNA transcription units are flanked at both ends by terminator elements that are recognized by the respective transcription termination factor, e.g. TTF-I in mammals and Reb1p in yeast (4,5,8,18,19). TTF-I has been shown to form oligomers in solution and to be capable of interacting with two ‘Sal box’-containing DNA fragments in trans (12). By simultaneously binding to both the promoter-proximal and downstream terminator elements, TTF-I could mediate interactions between the 5′ and 3′ end of rDNA, thereby facilitating ‘hand over’ of Pol I from the terminator to the transcription start site.
To test this model, transcriptions were performed on templates that contain various combinations of the promoter-proximal and downstream ‘Sal box’ elements. We consistently observed TTF-I-mediated transcriptional enhancement on terminator-containing templates. However, this enhancement was not due to communication between the upstream and downstream TTF-I binding sites, because it was also observed on constructs that lack the upstream terminator, indicating that transcriptional activation was brought about by the downstream terminator alone. Consequently, stimulation of RNA synthesis on terminator-containing templates appears to be due to enhanced reinitiation frequency of Pol I rather than a ‘hand over’ of Pol I from the terminator to the promoter.
Evidence is accumulating that terminators may play a basic role in transcription. For Pol III, templates that lack a consensus terminator neither assemble transcription complexes in vitro nor function efficiently in vivo (20,21). This linkage between termination and initiation is most easily explained in the study by Dieci and Sentenac (2), which demonstrated that Pol III preferentially recycles on the same template. After the first initiation event on a given preinitiation complex, Pol III becomes committed to more rapidly transcribing the same gene in a way that is termination dependent. This optimization of transcription after a single gene activation event allows RNA release and efficient transcription reinitiation without release of RNA polymerase III.
The results presented in this study suggest that transcription termination also facilitates reinitiation of Pol I. Thus, a terminator-dependent reinitiation pathway appears to be responsible for the high transcriptional efficiency of both class I and III genes. However, whereas Pol III transcription reinitiation occurs without release of the transcribing enzyme from the template, Pol I needs to be liberated at the terminator in order to be recruited to the promoter and start a new transcription cycle. This mechanistic difference is due to the fact that Pol I genes are large and separated by intergenic spacer sequences, which in some organisms are considerably larger than the pre-rRNA coding region. When reaching the terminator, Pol I has to leave the densely packed template and, irrespective of whether or not TTF-I is involved in maintaining a loop structure of the rDNA transcription unit in vivo, is recruited to a preformed preinitiation complex.
Not surprisingly, the central player in termination-dependent transcriptional enhancement is PTRF, a factor that dissociates transcription complexes paused by DNA-bound TTF-I, thereby releasing both Pol I and nascent transcripts from the template (7,9). By inducing dissociation of Pol I from the template, PTRF can convert transcription elongation complexes that are arrested at the terminator to ones that reinitiate at the promoter. Consistent with its function in transcription termination, PTRF has been demonstrated to associate with both Pol I and TTF-I (9). It is not yet known whether PTRF remains bound to the RNA polymerase after being released from the template. Moreover, we still do not know whether PTRF is capable of interacting with the preinitiation complex, thereby directing Pol I to the promoter.
PTRF has striking functional similarities to the autoimmune antigen La, a 50 kDa phosphoprotein that is transiently associated with the precursors for tRNAs, 5S RNA and other transcripts synthesized by Pol III (22). La protein binds preferentially to RNAs ending in a run of uridine residues and has been shown to be involved in 3′ end formation of Pol III transcripts (23–26). La is not required for basal levels of termination by Pol III, but appears to increase the termination efficiency (27) and stimulate reinitiation (28,29). La fractionates into transcriptionally active and inactive forms, depending on phosphorylation of serine 366 by casein kinase II (30). The phosphorylated form of La is transcriptionally inactive and can be reactivated by dephosphorylation.
Although there is no significant sequence homology between PTRF and La, the functional homology between both proteins is intriguing. Both proteins resolve into several bands in isoelectric focusing (IEF) gels, indicating an intrinsic propensity for charge heterogeneity. The cDNA of PTRF encodes a 44 kDa polypeptide which migrates on SDS–polyacrylamide gels like a 52 kDa protein. Moreover, PTRF is phosphorylated at many sites. In contrast to La, where phosphorylation of serine 366 by casein kinase II appears to inhibit activity (30), the functional relevance of PTRF phosphorylation is not yet known. Up to now we had failed to correlate PTRF activity with a specific phosphorylation pattern. Both recombinant and cellular PTRF are active in promoting transcript release and activation of transcription (9) and, therefore, specific phosphorylation may inhibit PTRF activity. Inhibition of PTRF activity would suppress dissociation of ternary Pol I transcription complexes. Consequently, the movement of tightly packed Pol I molecules along the rDNA transcription unit would be blocked, thereby providing a most effective means to ‘freeze’ transcription elongation complexes and silence rRNA synthesis. In this scenario, external signals that affect the phosphorylation pattern of PTRF could alter the activity of PTRF, thus enabling the cell to regulate rRNA synthesis without having to disassemble and reassemble the entire transcription initiation complex.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Steve Mason, Renate Voit and Joachim Klein for help and advice. This work was supported in part by the Deutsche Forschungsgemeinschaft and the Fond der Chemischen Industrie.
References
- 1.Jiang Y. and Gralla,J.D. (1993) Uncoupling of initiation and reinitiation rates during HeLa RNA polymerase II transcription in vitro. Mol. Cell. Biol., 13, 4572–4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dieci G. and Sentenac,A. (1996) Facilitated recycling pathway for RNA polymerase III. Cell, 84, 245–252. [DOI] [PubMed] [Google Scholar]
- 3.Grummt I., Maier,U., Öhrlein,A., Hassouna,N. and Bachellelerie,J.-P. (1985) Transcription of mouse rDNA terminates downstream of the 3′ end of 28S RNA and involves interaction of factors with repeated sequences in the 3′ spacer. Cell, 43, 801–810. [DOI] [PubMed] [Google Scholar]
- 4.Bartsch I., Schoneberg,C. and Grummt,I. (1987) Evolutionary changes of sequences and factors that direct transcription termination of human and mouse ribosomal genes. Mol. Cell. Biol., 7, 2521–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuhn A., Bartsch,I. and Grummt,I. (1990) Specific interaction of the murine transcription termination factor TTF I with class-I RNA polymerases. Nature, 344, 559–562. [DOI] [PubMed] [Google Scholar]
- 6.Mason S.W., Sander,E.E. and Grummt,I. (1997) Identification of a transcript release activity acting on ternary transcription complexes containing murine RNA polymerase I. EMBO J., 16, 163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evers R., Smid,A., Rudloff,U., Lottspeich,F. and Grummt,I. (1995) Different domains of the murine RNA polymerase I-specific termination factor mTTF-I serve distinct functions in transcription termination. EMBO J., 14, 1248–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grummt I., Rosenbauer,H., Niedermeyer,I., Maier,U. and Öhrlein,A. (1986) A repeated 18 bp sequence motif in the mouse rDNA spacer mediates binding of a nuclear factor and transcription termination. Cell, 45, 837–846. [DOI] [PubMed] [Google Scholar]
- 9.Jansa P., Mason,S.W., Hoffmann-Rohrer,U. and Grummt,I. (1998) Cloning and functional characterization of PTRF, a novel protein which induces dissociation of paused ternary transcription complexes. EMBO J., 17, 2855–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kempers-Veenstra A.E., Oliemans,J., Offenberg,H., Dekker,A.F., Piper,P.W., Planta,R.J. and Klootwijk,J. (1986) 3′-End formation of transcripts from the yeast rRNA operon. EMBO J., 5, 2703–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kulkens T., van der Sande,C.A., Dekker,A.F., van Heerikhuisen,H. and Planta,R.J. (1992) A system to study transcription by yeast RNA polymerase I within the chromosomal context: functional analysis of the ribosomal DNA enhancer and the RBP1/REB1 binding sites. EMBO J., 11, 4665–4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sander E.E. and Grummt,I. (1997) Oligomerization of the transcription termination factor TTF-I: Implications for the structural organization of ribosomal transcription units. Nucleic Acids Res., 25, 1142–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smid A., Finsterer,M. and Grummt,I. (1992) Limited proteolysis unmasks specific DNA-binding of the murine RNA polymerase I-specific transcription termination factor TTF-I. J. Mol. Biol., 227, 635–647. [DOI] [PubMed] [Google Scholar]
- 14.Schnapp A. and Grummt,I. (1996) Purification, assay and properties of RNA polymerase I and class I- specific transcription factors in mouse. Methods Enzymol., 273, 233–248. [DOI] [PubMed] [Google Scholar]
- 15.Sander E.E., Mason,S.W., Munz,C. and Grummt,I. (1996) The amino-terminal domain of the transcription termination factor TTF-I causes protein oligomerization and inhibition of DNA binding. Nucleic Acids Res., 24, 3677–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voit R., Schäfer,K. and Grummt,I. (1997) Mechanism of repression of RNA polymerase I transcription by the retinoblastoma protein. Mol. Cell. Biol., 17, 4230–4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schnapp A., Pfleiderer,C., Rosenbauer,H. and Grummt,I. (1990) A growth-dependent transcription initiation factor (TIF-IA) interacting with RNA polymerase I regulates mouse ribosomal RNA synthesis. EMBO J., 9, 2857–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ju Q.D., Morrow,B.E. and Warner,J.R. (1990) REB1, a yeast DNA-binding protein with many targets, is essential for growth and bears some resemblance to the oncogene myb. Mol. Cell. Biol., 10, 5226–5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lang W.H. and Reeder,R.H. (1993) The REB1 site is an essential component of a terminator for RNA polymerase I in Saccharomyces cerevisiae. Mol. Cell. Biol., 13, 649–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allison D.S. and Hall,B.D. (1985) Effects of alterations in the 3′ flanking sequence on in vivo and in vitro expression of the yeast SUP4-o tRNATyr gene. EMBO J., 4, 2657–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chu W.M., Liu,W.M. and Schmid,C.W. (1995) RNA polymerase III promoter and terminator elements affect Alu RNA expression. Nucleic Acids Res., 23, 1750–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lerner M.R., Boyle,J.A., Hardin,J.A. and Steitz,J.A. (1981) Two novel classes of small ribonucleoproteins detected by antibodies associated with lupus erythematosus. Science, 211, 400–402. [DOI] [PubMed] [Google Scholar]
- 23.Stefano J.E. (1984) Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell, 36, 145–154. [DOI] [PubMed] [Google Scholar]
- 24.Gottlieb E. and Steitz,J.A. (1989) The RNA binding protein La influences both the accuracy and the efficiency of RNA polymerase III transcription in vitro. EMBO J., 8, 841–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gottlieb E. and Steitz,J.A. (1989) Function of the mammalian La protein: Evidence for its action in transcription termination by RNA polymerase III. EMBO J., 8, 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bachmann M., Pfeifer,K., Schroder,H.C. and Muller,W.E. (1990) Characterization of the autoantigen La as a nucleic acid-dependent ATPase/dATPase with melting properties. Cell, 60, 85–93. [DOI] [PubMed] [Google Scholar]
- 27.Cozzarelli N.R., Gerrard,S.P., Schlissel,M., Brown,D.D. and Bogenhagen,D.F. (1983) Purified RNA polymerase III accurately and efficiently terminates transcription of 5S RNA genes. Cell, 34, 829–835. [DOI] [PubMed] [Google Scholar]
- 28.Maraia R.J., Kenan,D.J. and Keene,J.D. (1994) Eukaryotic transcription termination factor La mediates transcript release and facilitates reinitiation by RNA polymerase III. Mol. Cell. Biol., 14, 2147–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maraia R.J. (1996) Transcription termination factor La is also initiation factor for RNA polymerase III. Proc. Natl Acad. Sci. USA, 93, 3383–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan H., Sakulich,A.L., Goodier,J.L., Zhang,X., Qin,J. and Maraia,R.J. (1997) Phosphorylation of the human La antigen on serine 366 can regulate recycling of RNA polymerase III transcription complexes. Cell, 88, 707–715. [DOI] [PubMed] [Google Scholar]


