Abstract
Background
Vertebrate limb development involves a reciprocal feedback loop between limb mesenchyme and the overlying apical ectodermal ridge (AER). Several gene pathways participate in this feedback loop, including Fgf signaling. In the forelimb lateral plate mesenchyme, Tbx5 activates Fgf10 expression, which in turn initiates and maintains the mesenchyme/AER Fgf signaling loop. Recent findings have revealed that Tbx5 transcriptional activity is regulated by dynamic nucleocytoplasmic shuttling and interaction with Pdlim7, a PDZ-LIM protein family member, along actin filaments. This Tbx5 regulation is critical in heart formation, but the coexpression of both proteins in other developing tissues suggests a broader functional role.
Results
Knock-down of Pdlim7 function leads to decreased pectoral fin cell proliferation resulting in a severely stunted fin phenotype. While early gene induction and patterning in the presumptive fin field appear normal, the pectoral fin precursor cells display compaction and migration defects between 18 and 24 hours post-fertilization (hpf). During fin growth fgf24 is sequentially expressed in the mesenchyme and then in the apical ectodermal ridge (AER). However, in pdlim7 antisense morpholino-treated embryos this switch of expression is prevented and fgf24 remains ectopically active in the mesenchymal cells. Along with the lack of fgf24 in the AER, other critical factors including fgf8 are reduced, suggesting signaling problems to the underlying mesenchyme. As a consequence of perturbed AER function in the absence of Pdlim7, pathway components in the fin mesenchyme are misregulated or absent, indicating a breakdown of the Fgf signaling feedback loop, which is ultimately responsible for the loss of fin outgrowth.
Conclusion
This work provides the first evidence for the involvement of Pdlim7 in pectoral fin development. Proper fin outgrowth requires fgf24 downregulation in the fin mesenchyme with subsequent activation in the AER, and Pdlim7 appears to regulate this transition, potentially through Tbx5 regulation. By controlling Tbx5 subcellular localization and transcriptional activity and possibly additional yet unknown means, Pdlim7 is required for proper development of the heart and the fins. These new regulatory mechanisms may have important implications how we interpret Tbx5 function in congenital hand/heart syndromes in humans.
Background
The basic morphological and genetic mechanisms underlying vertebrate limb formation are highly conserved, from pectoral and pelvic fins in fish to arms and legs in humans [1-3]. Along the flank of the embryo the primordial limb fields are established at specific sites in the lateral plate mesoderm (LPM) [1]. The limb first appears as an outgrowth of mesenchyme from the LPM, which is covered by a sheet of ectoderm. The distal ectoderm covering the limb mesenchyme specializes and thickens to form a transient structure called the apical ectodermal ridge (AER). Reciprocal communication between the AER and underlying mesenchyme promotes cell proliferation and limb outgrowth. Physical removal of the AER, for example in the chicken embryo, results in cessation of limb growth and truncation of distal elements [4,5].
In all vertebrates, one of the earliest determinants defining the forelimb field is the T-box transcription factor Tbx5 [6-10]. Expressed in the limb mesenchyme, Tbx5 is required for forelimb development and its functional disruption in zebrafish, chicken, and mouse results in a complete loss of the limb [11-13]. Tbx5 transcriptionally activates Fgf10 in the forelimb mesenchyme [12], and its secreted gene product then signals to the AER to induce the expression of ectodermal Fgfs such as Fgf4 and Fgf8 [14-16]. Fgf8 in turn signals back to the underlying mesenchyme to maintain Fgf10 expression, thereby creating a feedback loop needed to support limb outgrowth and establish the proximal-distal limb axis.
The Fgf signaling pathway is critical for limb initiation and outgrowth [17]. Genetic disruption of Fgf10, Fgf8, or Fgf4/Fgf8 results in severely malformed or truncated limbs [15,16,18-21]. The secreted Fgf ligands can bind to four Fgf receptors (Fgfr), with Fgfr1 and Fgfr2 being essential for limb development [17]. Fgfr1 is expressed in the limb mesenchyme and is required for distal limb and digit formation [22,23]. Fgfr2 in the mouse is alternatively spliced into two isoforms, Fgfr2b and Fgfr2c, which are expressed in the ectoderm and mesenchyme, respectively [24]. Knock-out of both Fgfr2 isoforms results in a failure of limb induction [25,26] and deletion of isoform Fgfr2b causes limb defects due to a loss of AER maintenance [27,28]. While critical for proximal-distal limb patterning, Fgf signaling is also an integral part of patterning the other limb axes. The expression of Shh, a central signal in anterior-posterior axis patterning, as well as Wnts and Bmps, which participate in dorso-ventral axis formation, are all dependent on Fgf signals from the AER (reviewed in [29]).
We previously identified a member of the PDZ-LIM protein family, Pdlim7, to be co-expressed with and bind to the transcription factors Tbx5 and Tbx4 [30]. PDZ-LIM proteins contain an N-terminal PDZ domain and one or three C-terminal LIM domains. PDZ and LIM domains are both protein interaction modules, providing this multi-domain protein family with diverse interaction opportunities [31]. Functional roles for PDZ-LIM proteins have been reported in signal transduction, cell migration, and differentiation [32-35,31]. In cell cultures and chicken and zebrafish embryos we have shown that Pdlim7 regulates Tbx5 nuclear/actin cytoskeleton-associated localization and activity during cardiac atrioventricular boundary and valve formation [36-38]. Work in zebrafish revealed that Pdlim7 is also required for proper skeletal muscle development and maintenance [38,31]. However, the common or distinct functional roles Pdlim7 and related PDZ-LIM proteins have in organ formation in the developing vertebrate embryo remain poorly understood.
In the zebrafish, pdlim7 is co-expressed with tbx5 during cardiac and pectoral fin development [38]. Loss of either Pdlim7 or Tbx5 function leads to a similar cardiac phenotype, a non-looped heart, although by opposing molecular mechanisms [38,11]. Elimination of Tbx5 results in a loss of Tbx5 responsive gene activation while reduction of Pdlim7 leads to an upregulation of Tbx5 target genes at the atrio-ventricular boundary. Along with the heart problems observed at later developmental stages, compromised Pdlim7 activity also results in pectoral fin defects early in embryogenesis [38]. However, detailed analysis of the fin phenotype has not been performed. Here we analyze the pectoral fin phenotype induced by morpholino knock-down and mRNA overexpression of pdlim7 and provide the first evidence of a critical role for PDZ-LIM proteins in vertebrate limb development.
Results
Pdlim7 is required for pectoral fin development
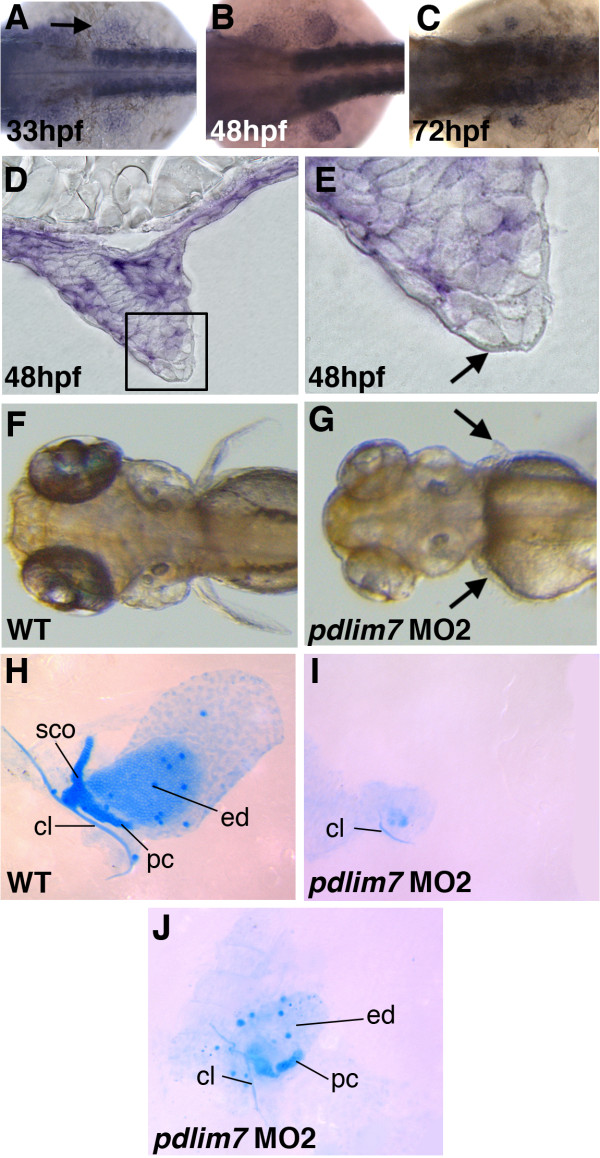
The PDZ-LIM protein, Pdlim7, was identified as a novel binding protein and regulator of the transcription factor Tbx5 [30,36,38]. Pdlim7 mRNA has been detected in several tissues of the vertebrate embryo including the limbs, heart, and skeletal muscle [30,36]. In the zebrafish embryo during fin development, using whole mount in situ hybridization, we first detected pdlim7 expression in the mesenchyme of the fin field at 33 hours post-fertilization (hpf) which was maintained in the fin up to 72 hpf (Fig. 1A-E; [38]). Expression of pdlim7 was not detected in the AER (Fig. 1D, E). Injection at the one-cell stage with 2 ng of pdlim7 antisense morpholino oligonucleotides, interfering with either protein translation (MO1) or RNA splicing (MO2), resulted in comparable defects in pectoral fin development (Fig. 1 and data not shown; see Methods). At 48 hpf, embryos injected with MO2 produced phenotypes with stunted fin buds as compared to control siblings, indicating a possible defect in forelimb outgrowth ([38]; data not shown). By four days of development in wild-type larvae the pectoral fins are clearly visible, while the pectoral fins of MO2 injected embryos were significantly smaller or absent (Fig. 1F-G). Both control and morphant larvae were stained with Alcian blue to visualize the extent of cartilage differentiation. In controls, all of the major cartilage elements were present in the pectoral fin (Fig. 1H). However, in pdlim7 MO2 injected larvae, cartilage development was severely impeded. In the majority of cases, only a fragment of the cleithrum bone along with limited unidentifiable cartilage condensation could be detected (Fig. 1I). Some phenotypic variability was observed among MO2 treated embryos and occasionally slight differences in phenotype were visualized between left and right pectoral fins within single embryos. Less severe pectoral fin phenotypes in morphant embryos resulted in larvae with elements of the cleithrum, postcoracoid process, and a greatly reduced endochondral disc (Fig. 1J). Based upon mRNA expression and gene knock-down data, Pdlim7 appears to be required for pectoral fin development.
Figure 1.
Pdlim7 is required for pectoral fin development. A-C: Whole-mount in situ hybridization using antisense RNA to show expression of pdlim7 in the developing pectoral fin mesenchyme at 33 hpf (A), 48 hpf (B), and 72 hpf (C). D-E: Sectioned embryos at 48 hpf of whole-mount in situ hybridization of pdlim7 show expression in fin mesenchyme. Boxed region in D is magnified in E to distinguish mesenchyme (purple color) and AER (arrow). F-G: Dorsal view of wild-type (F) and MO2 injected (G) embryos at 96 hpf. Arrows in G point to position of pectoral fin. H-J: Alcian blue stained cartilage preparations of dissected pectoral fins at 96 hpf. Wild-type (H), severe MO2 phenotype (I), and mild MO2 phenotype (J). I and J from same embryo. cl, cleithrum; pc, postcoracoid process; ed, endodermal disc; sco, scapulocoracoid.
Cell proliferation is decreased in pectoral fins after Pdlim7 knock-down
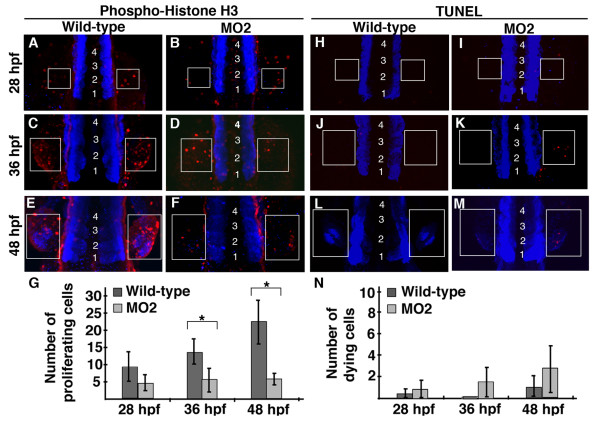
Knock-down of Pdlim7 results in the loss of, or severely truncated, pectoral fins. One possible cause for the fin phenotype could be due to alterations in cell proliferation or viability. To investigate this, embryos injected with pdlim7 MO2 were analyzed at stages of pectoral fin growth between 28 and 48 hpf for cell proliferation using an anti-phospho-histone H3 (p-H3) antibody or for cell death using TUNEL (Fig. 2; see Methods). Wild-type embryos displayed an increase in p-H3 antibody reactivity from 28, 36, to 48 hpf in the developing pectoral fin (Fig. 2A, C, E, boxed regions). At 28 hpf, in the pectoral fin field, MO2 injected embryos had a comparable number of dividing cells as wild-type (Fig. 2A, B). However, quantification of p-H3 positive cells revealed a steady increase of proliferating cells in wild-type pectoral fins as development progressed to 48 hpf, while cell proliferation remained at a low, constant level in pdlim7 knock-down embryos (Fig. 2D, F, G). Although the fin mesenchyme is smaller in morpholino injected embryos (see Figs. 3 and 4), the cells in the fin field including p-H3 positive cells appeared to be more scattered in the lateral plate mesoderm. Considering this, we used equal sized boxed regions in wild-type and MO2-treated embryos for analysis, which provides for a certain overestimation of dividing cells in morphant fins. Even with this conservative measure, we were able to detect a significant difference in p-H3 positive cells at 36 and 48 hpf between wild-type siblings and pdlim7 MO2 injected embryos (asterisks Fig. 2G; Additional file 1, Table S1).
Figure 2.
Pdlim7 knock-down pectoral fins have decreased cell proliferation. A-F: Dorsal view, anterior end of embryo is out of view to the bottom of image, of whole-mount anti-phospho-histone H3 (p-H3) antibody (red) staining on wild-type (A, C, E) and MO2 injected (B, D, F) embryos. Pectoral fins develop lateral to the 3rd somite, thus embryos were counterstained with MF20 (blue) to visualize somites, which are indicated by numbers (somite 1 refers to the most anterior somite). G: Quantification of p-H3 positive cells in pectoral fins of wild-type and MO2 injected embryos. 28 hpf wild-type n = 5, MO2 n = 5; p-value = 0.077. 36 hpf wild-type n = 4, MO2 n = 5; p-value = 0.036. 48 hpf wild-type n = 5, MO2 n = 5; p-value = 0.001. Experiment performed in triplicate, representative data from single replicate shown in G. Statistically significant p-values (<0.05) are denoted by asterisks. H-M: Whole-mount TUNEL assay on wild-type (H, J, L) and MO2 injected (I, K, M) embryos. Apoptotic cells in red with MF20 stained somites in blue, as described for p-H3 staining. N: Quantification of apoptotic cells in wild-type and MO2 injected embryos. 28 hpf wild-type n = 6, MO2 n = 4; p-value = 0.483. 36 hpf wild-type n = 6, MO2 n = 5; p-value = 0.05. 48 hpf wild-type n = 4, MO2 n = 6; p-value = 0.073. Experiment performed in triplicate, representative data from single replicate shown in N. White boxes indicate pectoral fin field at 28 hpf (A-B, H-I), 36 hpf (C-D, J-K), and 48 hpf (E-F, L-M).
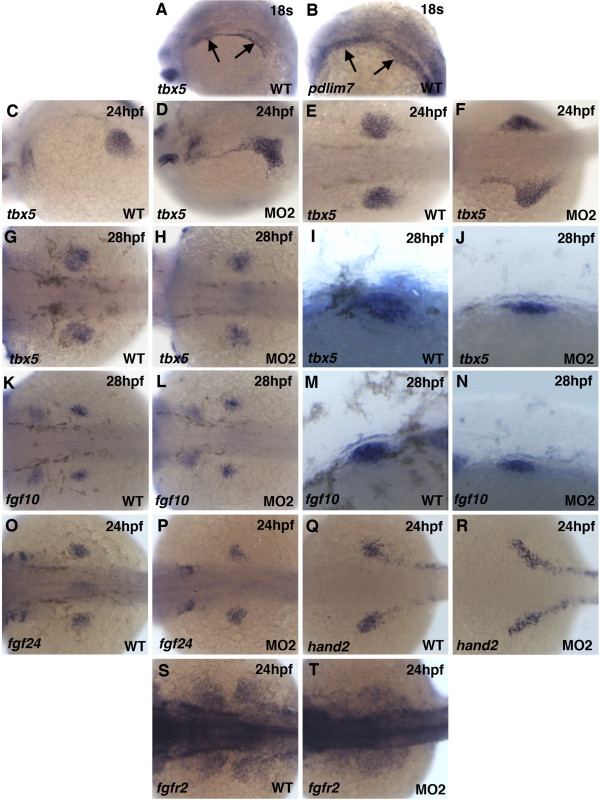
Figure 3.
Migration and compaction defects in pdlim7 MO2 injected embryos. A-R: Whole-mount antisense RNA in situ hybridization of wild-type and MO2 injected embryos. tbx5 (A-H) expression at 24 hpf (A-D) and 28 hpf (E-H) in wild-type (A, C, E, G) and MO2 injected (B, D, F, H) embryos. G and H magnified lateral view of pectoral fin. fgf10 (I-L) expression at 28 hpf in wild-type (I, K) and MO2 injected (J, L) embryos. K and L magnified lateral view of pectoral fin. Expression at 24 hpf in wild-type and MO2 injected embryos of fgf24 (M-N), hand2 (O-P), and fgfr2 (Q-R), respectively. Head is positioned to the left.
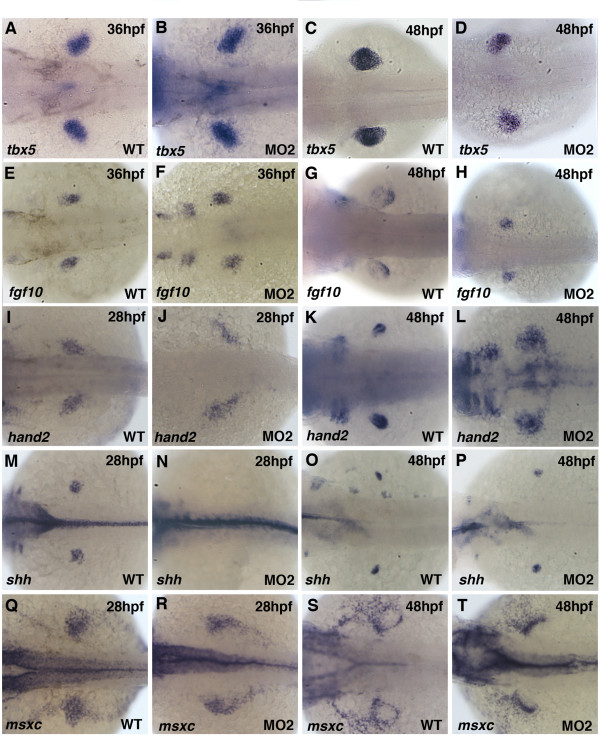
Figure 4.
Pectoral fin mesenchymal gene expression in Pdlim7 knock-down embryos. Dorsal view of whole-mount antisense RNA in situ hybridization of wild-type (A, C, E, G, I, K, M, O, Q, S) and MO2 injected (B, D, F, H, J, L, N, P, R, T) embryos. A-D: tbx5 expression in wild-type and MO2 injected embryos at 36 hpf (A-B) and 48 hpf (C-D). E-H: fgf10 expression at 36 hpf (E-F) and 48 hpf (G-H). I-L: hand2 expression at 28 hpf (I-J) and 48 hpf (K-L). M-P: shh expression at 28 hpf (M-N) and 48 hpf (O-P). Q-T: msxc expression at 28 hpf (Q-R) and 48 hpf (S-T). Head is positioned to the left.
One possible explanation for lower numbers of p-H3 positive cells in the pectoral fins may be reduced cell survival after pdlim7 knock-down. Therefore, using a TUNEL assay, we investigated apoptosis in wild-type and pdlim7 MO2 injected pectoral fins at 28, 36, and 48 hpf (Fig. 2H-M). In wild-type embryos, at most only one to two apoptotic cells could be observed in the developing pectoral fin at any of the three time points tested (Fig. 2H, J, L). In MO2 injected embryos, a slight increase of apoptotic cells in pectoral fins was detected, especially at 48 hpf (Fig. 2I, K, M). However, quantification of the data revealed that the slight increase in apoptotic cells in pdlim7 morphants was suggestive but not statistically significant compared to wild-type pectoral fins (Fig. 2N; Additional file 1, Table S1). These findings indicate that loss of Pdlim7 has no significant effect on apoptosis; however, the protein appears necessary for normal cell proliferation in the pectoral fin field.
Pdlim7 knock-down embryos exhibit pectoral fin cell migration and compaction defects
Knock-down of Pdlim7 function leads to severe arrest in pectoral fin development and lower numbers of proliferating cells in the budding fin. We next sought to determine if the pectoral fin field was established correctly in the absence of pdlim7. In previous work we have demonstrated that Pdlim7 can regulate Tbx5 activity, one of the forelimb/pectoral fin field markers essential for limb outgrowth [36,38,8,10]. The cells of the fin field are derived from a population of cells in the LPM that initially comprises both heart and pectoral fin precursors [39]. The tbx5 expressing heart and pectoral fin progenitor cells remain indistinguishable until the 18-somite stage, when the pectoral fin precursor cells migrate posteriorly and separate from the adjacent anterior cardiac progenitors. The migratory behavior of these cells has been shown to be dependent upon Tbx5 activity [39]. Of note, in wild-type embryos both fin and heart precursor cells co-express tbx5 and pdlim7 (Fig. 3A, B), and by 24 hpf, tbx5 expressing cells are completely separated into the heart and pectoral fin primordia (Fig. 3C). In contrast, tbx5 expressing cells in 24 hpf pdlim7 morphants were detected in the LPM between the heart tube and pectoral fin field, connecting the two organ fields (Fig. 3D). Dorsal views of MO2 injected embryos also displayed a less compact pectoral fin field as visualized by tbx5 expression (Fig. 3E, F). In comparison to control embryos, at 28 hpf, the pectoral fin field was noticeably smaller in size and induction of limb outgrowth appeared defective (Fig. 3G-J). These results suggested aberrant or delayed cell migration of forelimb precursors into the fin field, although overall embryonic development did not appear significantly delayed in pdlim7 morphants as cardiac beating began as expected around 22 hpf.
The Tbx5 downstream target gene, fgf10, is expressed in the pectoral mesenchyme at 28 hpf; right at the time when the fin bud emerges from the LPM (Fig. 3K, M; [40]). Although fin bud growth was disrupted, fgf10 was induced normally in pdlim7 knock-down embryos (Fig. 3K-N), suggesting that the Pdlim7 mediated misregulation of Tbx5 transcriptional activity upon fgf10 may not be the cause of the compromised fin outgrowth.
To further investigate whether the pectoral fin primordium was fully established, we analyzed the expression of the early specification markers fgf24, hand2, and fgfr2. All of these genes are active in the LPM in the fin primordial cells [41-43]. Fgf24 is expressed in the pectoral fin precursor mesenchyme and is functionally required for cell migration and compaction to the presumptive fin field [41]. At 24 hpf in control embryos, fgf24 clearly delineated the future location of the pectoral fins (Fig. 3O). fgf24 expression was detected after pdlim7 knock-down, although the expression domain was slightly smaller and appeared closer to the midline of the embryo (Fig. 3P). hand2 was detected in the presumptive fin mesenchyme by 24 hpf in control and MO2 injected embryos (Fig. 3Q-R; [42]). However, in morphant embryos, hand2 expression failed to undergo mediolateral expansion in the LPM. Expression of fgfr2, which is thought to be downstream of Tbx5 [43], was detected in the compact primordial pectoral fin cells in wild-type embryos (Fig. 3S). In the morphants, fgfr2 expression was diffuse and did not display normal compaction (Fig. 3T), similar to the compaction defect of tbx5 expressing cells (Fig. 3F). In pdlim7 knock-down embryos, mesenchymal gene induction and pectoral fin cell specification appears to occur normally, however, early compaction of the primordial fin field is disrupted or delayed possibly due to incomplete precursor cell migration.
Knock-down of Pdlim7 does not disrupt pectoral fin mesenchyme patterning
Molecular markers for early pectoral fin specification were normal in pdlim7 compromised embryos, although precursor cell migration appeared delayed and the resulting fin field was smaller and less compact. In order to gain a better understanding of the cause of the pectoral fin phenotype, we examined the expression of several mesenchymal markers involved in limb patterning. We first analyzed tbx5, which is regulated by Pdlim7 in the zebrafish heart [38]. By 36 and 48 hpf, tbx5 expression could be detected in wild-type embryos throughout the developing pectoral fin mesenchyme (Fig. 4A, C; [11]). In pdlim7 knock-down embryos, tbx5 expression was maintained in the fin mesenchyme, however, the expression domain appeared less compact when compared to controls (Fig. 4A-D). Fgf10 is a direct downstream target of Tbx5 in the forelimb [12,13,40]. As expected, in wild-type and pdlim7 MO2 injected embryos at both 36 and 48 hpf, fgf10 was expressed in the limb mesenchyme; however, at the later time point the fgf10 expression domain appeared smaller in the morphants and less concentrated to the distal mesenchyme (Fig. 4E-H). The detection of fgf10 in the pectoral fin mesenchyme of MO2 treated embryos suggested that Tbx5 protein was transcriptionally functional and proximal-distal limb outgrowth had been initiated.
We next tested the expression of hand2 and shh, two genes required for limb development and involved in anterior-posterior patterning [44,42,46]. hand2 is normally expressed in the posterior mesenchyme of the developing pectoral fin (Fig. 4I, K; [42]). In pdlim7 MO2 injected embryos, hand2 expression was detected in the LPM between 28 and 48 hpf, though its expression was diffuse and covered a wider area as compared to the control embryos (Fig. 4I-L; data not shown). Similar to hand2, shh is asymmetrically expressed in a posterior mesenchymal domain of the developing fin (Fig. 4M, O; [47]). At 28 hpf, in wild-type embryos, shh revealed this posterior expression, however, in the MO2 injected embryos no expression could be detected in the pectoral fin (Fig. 4M, N). Of note, shh expression was observed in the floor plate of morphant embryos at the same 28 hpf time-point (Fig. 4N; data not shown). Expression of shh recovered in the pectoral fin by 48 hpf, suggesting the absence at 28 hpf may have been due to a delay in activation (Fig. 4O, P). Despite the significant reduction in fin size at 48 hpf, shh remained asymmetrically expressed in a posterior domain in the pdlim7 knock-down embryos (data not shown). Another gene involved in patterning all three limb axes is msxc [48]. At 28 hpf, msxc was expressed in the pectoral fin mesenchyme in both control and pdlim7 morphant embryos (Fig. 4Q-R; [49]). Similar to the other mesenchymal markers tested, msxc displayed a very diffuse pectoral fin expression at 36 and 48 hpf in MO2 injected embryos compared to the controls (Fig. 4S-T; data not shown). The localization of msxc in the mesenchyme adjacent to the AER appeared to recover by the later time-point, however, some msxc expressing cells remained in the LPM (Fig. 4T). Most of the key regulators tested in the pectoral fin mesenchyme remained expressed in pdlim7 knock-down embryos, however, the overall size and domain organization in the fin field appeared significantly affected.
Pdlim7 knock-down causes loss of AER gene expression
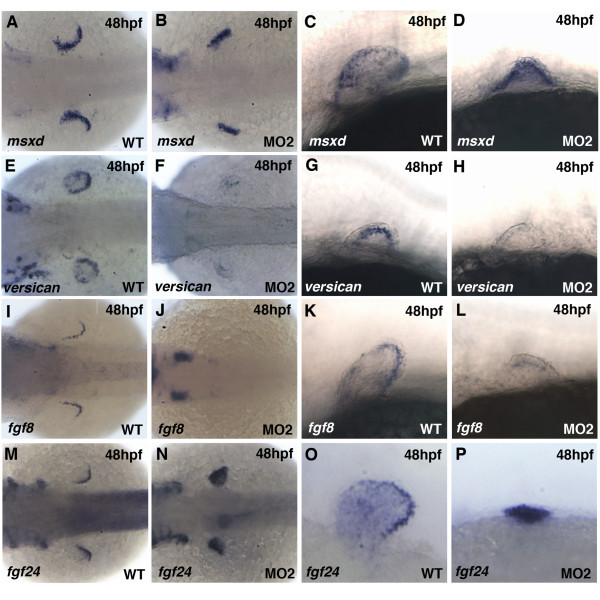
Knock-down of Pdlim7 results in a stunted fin phenotype (Fig. 1; [38]), suggesting a defect in limb outgrowth. Limb outgrowth requires continuous signaling between the distal AER and the underlying limb mesenchyme [1,2,29]. Since gene activities involved in patterning the pectoral fin mesenchyme were detected in the morphant embryos, we next asked whether gene expression in the AER was perturbed in Pdlim7 compromised embryos. At 48 hpf we detected expression of the fin AER marker msxd [49] in wild-type and pdlim7 MO2 injected embryos alike, indicating that the AER had formed (Fig. 5A-D). The proteoglycan versican was also detected in the AER of developing pectoral fins of wild-type embryos (Fig. 5E, G). We have found versican to be misregulated in the heart of pdlim7 MO2 injected embryos [38], and therefore wondered whether versican would be misexpressed in the pectoral fins as well. Indeed, versican was either greatly reduced or absent from the pectoral fins in morphant embryos (Fig. 5F, H). Another prominent AER marker gene is fgf8, whose function is required to maintain the signaling loop between the AER and underlying limb mesenchyme for appendage outgrowth [19,50,20]. Knock-down of Pdlim7 resulted in a significant reduction of fgf8, compared to sibling controls (Fig. 5I-L). The family member fgf24 is normally turned off in the fin mesenchyme by 48 hpf and subsequently expressed in the overlying AER (Fig. 5M, O; [41]). Importantly, unlike in the control embryos, we could not detect the switch from mesenchymal to AER expression for fgf24 after pdlim7 knock-down, but we rather observed ectopic expression in the fin mesenchyme (Fig. 5N, P). Therefore, the loss of fin outgrowth in pdlim7 morphant embryos appears to be due to a loss of proper AER function and disruption of the reciprocal signaling between AER and the adjacent mesenchyme.
Figure 5.
Disruption of AER gene expression in pdlim7 MO2 injected embryos. A-D: Expression of msxd at 48 hpf in wild-type (A, C) and MO2 injected (B, D) embryos. E-H: Expression of versican in wild-type (E, G) and MO2 injected (F, H) embryos. I-L: Expression of fgf8 in wild-type (I, K) and MO2 injected (J, L) embryos. M-P: Expression of fgf24 in wild-type (M, O) and MO2 injected (N, P) embryos. Dorsal views (A, B, E, F, I, J, M, N) and lateral views (C, D, G, H, K, L, O, P). Head is positioned to the left.
Knock-down of pdlim7 affects Fgf signaling in the pectoral fin
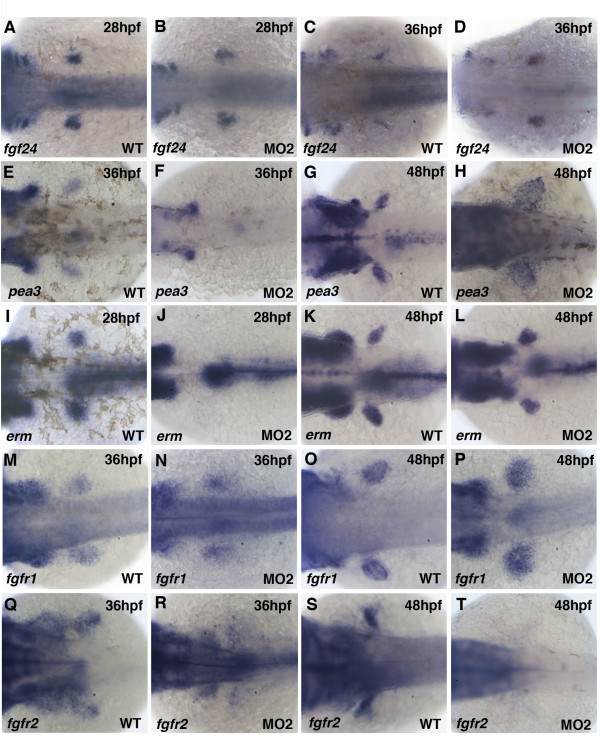
The disruption and loss of Fgf gene expression in the AER of pdlim7 MO2 injected embryos suggested a possible breakdown of the Fgf signaling loop between the mesenchyme and AER. To further explore this possibility, we tested additional Fgf signaling components in the pectoral fin after Pdlim7 protein reduction. Fgf24 is a zebrafish-specific factor that functions downstream of Tbx5 in pectoral fins and is required to activate fgf10 expression in the fin mesenchyme [41]. At 24 and 28 hpf, fgf24 was detected in the pectoral fin mesenchyme of wild-type embryos (Figs. 3O and 6A) and did not appear significantly different in embryos injected with pdlim7 MO2 (Figs. 3P and 6B) or earlier time-points (Fig. 5M, N). In contrast, in 36 hpf pdlim7 compromised embryos, fgf24 expression remained in the fin mesenchyme and did not switch to the AER (Fig. 6C-D). Ectopic expression of fgf24 in the pdlim7 morphant mesenchyme was maintained at 48 hpf with no detectable expression in the AER (Fig. 5M-P). We consequently tested two transcriptional targets of Fgf signaling in the fin mesenchyme, pea3 and erm [51,52]. We could detect pea3 expression in the pectoral fins at 36 and 48 hpf of control embryos (Fig. 6E, G). However, in 36 hpf morphant embryos, pea3 mRNA was absent from the LPM but appeared to recover by 48 hpf (Fig. 6F, H). Despite the presence of pea3 expression at 48 hpf, its localization was greatly diffused in the fin and did not show the normal restriction within the mesenchyme (Fig. 6G, H). A similar result was also observed for erm, a gene normally expressed in the budding pectoral fin (Fig. 6I, K). At 28 hpf in MO2 injected embryos, we could not detect erm mRNA in the early bud, but gene expression recovered by 48 hpf, although with a slightly smaller and irregular shaped domain than in controls (Fig. 6I-L).
Figure 6.
Fgf signaling pathway genes are disrupted after knock-down of Pdlim7. Dorsal view of whole-mount antisense RNA in situ hybridization of wild-type (A, C, E, G, I, K, M, O, Q, S) and MO2 injected (B, D, F, H, J, L, N, P, R, T) embryos. A-D: Expression of fgf24 at 28 hpf (A-B) and 36 hpf (C-D). E-H: Expression of pea3 at 36 hpf (E-F) and 48 hpf (G-H). I-L: Expression of erm at 28 hpf (I-J) and 48 hpf (K-L). M-P: Expression of fgfr1 at 36 hpf (M-N) and 48 hpf (O-P). Q-T: Expression of fgfr2 at 36 hpf (Q-R) and 48 hpf (S-T). Head is positioned to the left.
Sandwiched between the secreted Fgf signaling molecules and downstream target genes are the Fgf receptors, whose function is critical for signal transduction and limb formation [17]. We tested the expression of fgfr1 and fgfr2 in the developing pectoral fins of control and pdlim7 morphant embryos. At 36 and 48 hpf, fgfr1 was expressed in wild-type pectoral fins throughout the developing mesenchyme (Fig. 6M, O). In MO2 injected embryos; however, similar to other genes tested, fgfr1 displayed a loss of compaction (Fig. 6M-P). fgfr2 mRNA was also detected in the fin mesenchyme of control embryos at the two time-points, with expression restricted by 48 hpf to the proximal and anterior portion of the pectoral fin (Fig. 6Q, S). After knock-down of Pdlim7, fgfr2 levels were significantly reduced at 36 hpf and expression completely absent from the pectoral fin at 48 hpf (Fig. 6R, T). Thus, there appears to be a gradual loss of fgfr2 function between 24 and 48 hpf (Fig. 3 and Fig. 6). Of note, expression of fgfr2 in the head and brain was not affected by injection of pdlim7 MO2 (data not shown). Taken together, these results reveal that, after pdlim7 knock-down, the expression of several components of the Fgf signaling pathway known to be critical for limb outgrowth, including the AER/mesenchyme signaling loop, are misregulated or absent.
Pdim7 overexpression does not alter mesenchymal Fgf expression
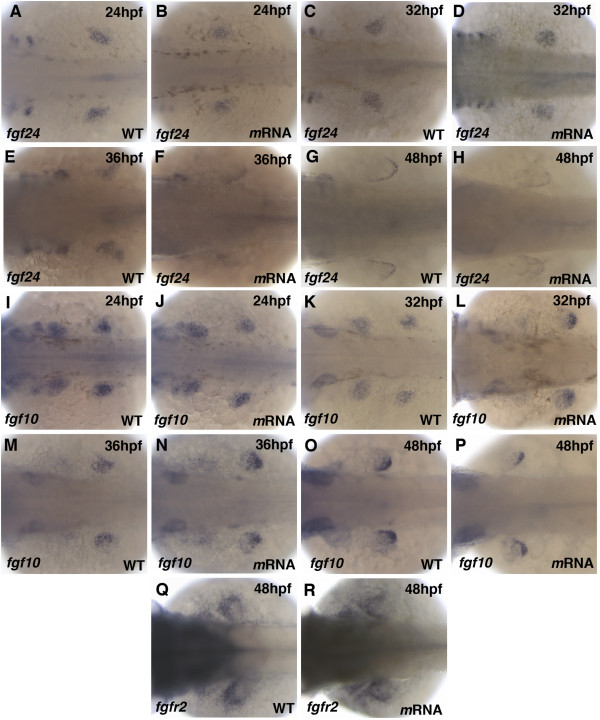
The ectopic expression of fgf24 in the fins of pdlim7 morpholino injected embryos suggested that fgf24 maybe a direct target of Tbx5 and subject to regulation by Pdlim7. In an effort to support this hypothesis and to complement the knock-down studies, we overexpressed pdlim7. Considering Pdlim7 induced shuttling of Tbx5, injection of 100 pg pdlim7 mRNA into one-cell stage embryos should sequester Tbx5 from the nucleus, resulting in the downregulation of target genes. Comparison of in situ hybridization for fgf24 between wild-type and mRNA injected embryos at 24, 32, 36, and 48 hpf displayed no significant changes in expression (Fig. 7A-H). Expression of fgf24 during the fin progenitor cell compaction appeared normal at 24 hpf (Fig. 7A, B) while the gene still transitioned from mesenchymal to strict AER localization between 32 and 48 hpf (Fig. 7C-H). In addition to fgf24, we monitored expression of fgf10 after pdlim7 overexpression. fgf10 did not reveal significant differences at 24, 32, 36, or 48 hpf between uninjected control siblings and pdlim7 mRNA injected embryos (Fig. 7I-P). fgf10 revealed its typical dynamic expression from broad mesenchymal (Fig. 7I-N) to concentrated location in the distal mesenchyme adjacent to the AER by 48 hpf (Fig. 7O, P). In several mRNA injected embryos we noticed a more pronounced fgf10 crescent shape expression domain adjacent to the AER, which is typical for a more mature limb and may indicate slightly accelerated differentiation caused by forced pdlim7 expression. Finally, since fgfr2 was greatly affected after pdlim7 knock-down and significantly downregulated by 48 hpf (Fig. 6), we determined the expression of fgfr2 after pdlim7 overexpression. Similar to the observations for fgf24 and fgf10, fgfr2 at 48 hpf did not display differential expression between wild-type versus the mRNA injected embryos and stained the proximal portion of the fin bud (Fig. 7Q, R). Control in situ hybridizations for the Tbx5 target gene tbx2b revealed, however, a reduction of expression at the heart AV boundary as previously published (data not shown and [38]), demonstrating that the injected synthetic mRNA was functional. The lack of change in fgf24 or fgf10 expression after pdlim7 overexpression does not rule out that fgf24 is directly activated by Tbx5 or regulated by Pdlim7, as maintenance of limb outgrowth and Fgf10 expression in the mouse is independent of Tbx5 [53].
Figure 7.
Pdlim7 overexpression does not alter Fgf signaling genes. Dorsal view of whole-mount antisense RNA in situ hybridization of wild-type (A, C, E, G, I, K, M, O, Q) and 100 pg synthetic pdlim7 mRNA injected (B, D, F, H, J, L, N, P, R) embryos. A-H: Expression of fgf24 at 24 hpf (A-B), 32 hpf (C-D), 36 hpf (E-F), and 48 hpf (G-H). I-P: Expression of fgf10 at 24 hpf (I-J), 32 hpf (K-L), 36 hpf (M-N), and 48 hpf (O-P). Q-R: Expression of fgfr2 at 48 hpf.
Discussion
Pdlim7 functions to regulate Tbx5 transcriptional activity
T-box proteins including Tbx5 contain nuclear localization and nuclear export sequences, enabling these transcription factors to relocate between nuclear and cytoplasmic cell compartments [54-56]. We have previously shown that Pdlim7 is necessary for dynamic shuttling of Tbx5, sequestering the transcription factor to actin filaments outside the nucleus and thereby regulating Tbx5 target gene expression both in vitro and in vivo [30,36,38]. For example, tbx2b and nppa are downstream targets of Tbx5 during zebrafish heart valve development and can be indirectly regulated by Pdlim7 levels in cells of the atrio-ventricular (AV) boundary [38]. Loss of Pdlim7 function by morpholino knock-down results in increased tbx2b and nppa expression with excess valve tissue, while overexpression of Pdlim7 by synthetic mRNA injection into the embryo causes downregulation of these Tbx5 target genes and reduced valve tissue. These experimental results lead to a model in which a balance of Pdlim7 and Tbx5 within the cell regulates transcription factor activity. Does a similar molecular mechanism function during forelimb development? Fgf10 has been shown to be a transcriptional target of Tbx5 using in vitro reporter assays and in vivo during mouse forelimb development [12]. In addition, in cultured cells, Pdlim7 can regulate Tbx5 activation of an Fgf10-luciferase reporter construct [36]. Work from this study in the developing zebrafish pectoral fins, however, suggests that misregulation of Pdlim7 does not cause significant changes in fgf10 expression. Knock-down of pdlim7 did not result in an obvious upregulation or ectopic expression of fgf10 and respective overexpression of Pdlim7 did not lead to an apparent decrease in fgf10 expression (Fig. 3, 4, 7), however, the smaller fin size in the morphant embryos may obscure a correct assessment and contribute to this observation. Interestingly, the zebrafish specific Fgf24 has been placed in a pathway between Tbx5 and Fgf10 during pectoral fin induction (Fig. 8A; [41,43]). fgf24 is expressed in the presumptive pectoral fin cells in the LPM at 18 hpf and is maintained in the fin mesenchyme until 28 hpf, before the gene is downregulated and then activated in the AER by 32 hpf (Fig. 6 and 7; [41]). Of note, we could detect induction of pdlim7 mRNA by 32 hpf in the budding fin, the time point when fgf24 mesenchymal expression is turned off. The switch between mesenchymal to AER expression of fgf24 does not occur in pdlim7 knock-down embryos; however, we could demonstrate ectopic expression of fgf24 in the fin mesenchyme up to 48 hpf (Fig. 5 and Fig. 6). The fact that Pdlim7 function can influence fgf24 expression in the fin mesenchyme, coupled with previous findings that loss of Tbx5 results in a loss of fgf24 expression [41], suggests that Fgf24 may be the critical Fgf target of Tbx5 in the zebrafish fin.
Figure 8.
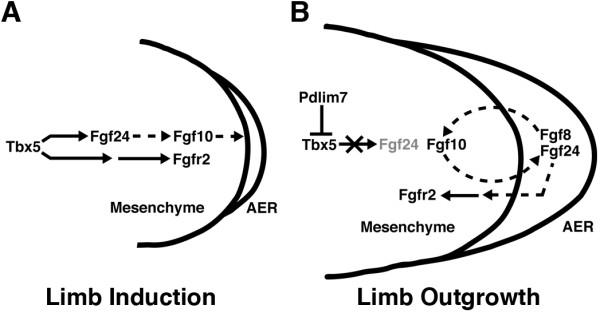
Model for vertebrate limb induction and outgrowth. During zebrafish fin induction, Tbx5 activates parallel pathways in the lateral plate mesoderm. In one pathway, Tbx5 transcriptionally activiates fgf24, which then initiates a signaling cascade leading to the activation of fgf10. In a second pathway, Tbx5 activity indirectly regulates the expression of fgfr2 (A). During fin outgrowth, Tbx5 transcriptional activity is negatively regulated by Pdlim7-mediated nucleocytoplasmic shuttling. Relocation and retention of Tbx5 at the actin cytoskeleton results in transcriptional restriction of fgf24 in the limb mesenchyme. The mesenchymal downregulation is required for initiating Fgf24 expression in the AER, which is a critical step to establish the reciprocal signaling loop with the mesenchyme to maintain fgf10 and fgfr2 expression (B). Solid arrows denote transcriptional regulation. Dashed arrows indicate signal transduction regulation.
Based upon this suggestion we asked whether overexpression of pdlim7 would inhibit mesenchymal fgf24 expression before the normal appearance of pdlim7. Pdlim7 mRNA injection, however, did not yield an obvious reduction of fgf24 or fgf10 expression levels at 24 hpf in the fin mesenchyme, a time window before endogenous pdlim7 appears (Fig. 7A, B, I, J). This may be because expression of Pdlim7 and Tbx5 in a cell does not automatically result in binding and sequestration of the transcription factor. This notion is supported by co-expression of Pdlim7 and Tbx5 in undifferentiated cultured chicken epicardial cells that have strict nuclear localization of Tbx5, suggesting that the interaction of both proteins is regulated by yet unknown mechanisms, possibly posttranslational modifications [37]. Thus, early ectopic expression of pdlim7 may not be sufficient to result in failure of fgf24 expression. During the later stages (32-48 hpf) of limb outgrowth, pdlim7 overexpression also did not result in significantly perturbed fgf24 or fgf10 expression. The reason for this may lie in the possibility that Tbx5 is not required for maintenance of fgf24 and fgf10 expression. Recent experiments in the mouse have shown that Tbx5 is only required for induction of Fgf10 during limb initiation but not for maintenance during limb outgrowth [53]. If a similar regulation operates in the zebrafish, overexpression of Pdlim7 in the fin mesenchyme, during stages after 32 hpf when Tbx5 nuclear activity may not be critical, will have little to no affect on Tbx5 target gene expression. This could explain why fgf24 and fgf10 are maintained after pdlim7 mRNA injection. In contrast, loss of Pdlim7 may act as a gain-of-function with regard to Tbx5 activity resulting in ectopic fgf24 in the fin mesenchyme. Consistent with this hypothesis, the genomic sequence just upstream of the fgf24 gene contains a putative consensus Tbx5 binding element ([57], data not shown), which would provide a mechanism for fgf24 regulation by Pdlim7/Tbx5 interactions. Additional experiments at the genomic DNA level will be needed to fully resolve this point.
Knock-down of pdlim7 causes persistent high levels of nuclear Tbx5 and continued ectopic expression of fgf24 in mesenchymal fin cells, which likely accounts for fgf10 maintenance despite the loss of AER genes such as fgf8. The expression of fgfr2, which appears to be in a parallel pathway downstream of Tbx5 during fin induction [43], is induced normally in pdlim7 MO2 injected embryos but becomes lost at post-induction stages possibly due to the absence of reciprocal AER signaling. We hypothesize that the signal required in the AER to maintain fgfr2 expression during fin outgrowth is Fgf24. fgf24 is never activated in the AER of pdlim7 morphants and the downregulation of fgfr2 appears to begin when fgf24 is supposed to switch from mesenchymal to ectodermal expression. This new information adds to our understanding of vertebrate limb induction and outgrowth and allows the extension of current models (Fig. 8A, B). During limb induction Tbx5 operates in at least two feed-forward parallel pathways. In one branch of the pathway, Tbx5 activates transcription of fgf24, which then leads to the expression of fgf10 and signaling to the AER. The other branch regulates via a genetic cascade leading to fgfr2 activation (Fig. 8A). During limb outgrowth after 32 hpf, Pdlim7 in the fin mesenchymal cells control Tbx5 activity by removing the transcription factor from the nucleus (Fig 8B). As a consequence, fgf24 becomes down-regulated, allowing a switch from mesenchymal to AER expression. Fgf signaling molecules, including Fgf24 and Fgf8, emanating from the AER maintain fgf10 and fgfr2 expression in the underlying mesenchyme. The reciprocal signaling between the AER and mesenchyme becomes independent of Tbx5 activity during limb outgrowth (Fig. 8B; [53]). Although the new data presented here clearly demonstrate that appropriate regulation of Tbx5 via Pdlim7 is necessary for establishment of a functional AER through Fgf24 expression, future investigations confirming this hypothesis may reveal the mechanistic details for Pdlim7 regulating Tbx5 balance and transcriptional activity in the developing forelimbs.
Pdlim7 in cell migration and compaction
Tbx5 has been shown to be required for proper pectoral fin cell migration during zebrafish development [39]. We also found evidence for defective cell migration in pdlim7 MO2 injected embryos (Fig. 3). tbx5 expressing pectoral fin cells display delayed migration and disrupted compaction of the fin field. This defect does not appear to be due to an overall developmental delay in pdlim7 morphants, as the heart begins to beat in a comparable time window as uninjected controls. A similar phenotype is also seen in ikarus (ika) zebrafish mutants, which lack fgf24 function [41]. In these mutant embryos, tbx5 expressing cells fail to undergo appropriate compaction at the pectoral fin field. The primordial fin cells at 18-somites express tbx5, pdlim7 as well as fgf24 (Fig. 3, [10,41]). Considering fgf24 is a target of Tbx5, it is possible that Pdlim7/Tbx5 protein interactions regulate fgf24 expression already in the 18-somite-stage embryo. Increased nuclear Tbx5, by loss of Pdlim7, would result in misregulation of fgf24. This idea is supported by the similar compaction defects observed after pdlim7 knock-down and in fgf24 ika mutants (Fig. 3; [41]). Overexpression as well as loss of Tbx5 causes defects in cell migration in cultured chicken proepicardial cells [58]. It is plausible that Pdlim7 functions at an initial stage of migration, in part by regulating Tbx5 balance in the nucleus and cytoplasm.
An alternative view would be that Pdlim7 itself, by its nature as an actin-associated protein, may modulate actin cytoskeleton dynamics and in this way directly influence cell migration [30,36]. In this context it is of interest that members of the PDZ-LIM protein family have been reported to be involved in cell migration [59-61]. However, more direct experimentation is required to fully elucidate this intriguing possibility.
Pdlim7 may be involved in Fgf signal transduction
In zebrafish, knock-down of pdlim7 by morpholino injection results in severely stunted pectoral fins, decreased cell proliferation in the fin region, and a loss or altered expression of Fgf pathway genes. Fgf signal transduction from the AER is required for limb outgrowth and proliferation of the undifferentiated limb mesenchyme [17,62]. How might Pdlim7 be involved in Fgf signaling in the developing pectoral fin? The delayed migration of cells into the pectoral fin field, coupled with the reduced ability of the cells in the limb field to compact after loss of Pdlim7 function, may contribute to decreased Fgf signaling. The lower proliferation rate in the budding fins further equates to fewer cells secreting and responding to signaling molecules, lowering the signaling potential. Thus, it is possible that a certain minimum threshold needed for signal propagation is not reached and the mesenchyme/epithelial reciprocal interactions required for limb outgrowth are not established or maintained. This scenario may account for the maintenance of mesenchymal Fgfs but loss of Fgf expression in the AER.
A second possibility could be that Pdlim7 plays a more direct role in Fgf signal transduction. Pdlim7 can interact with transmembrane receptor tyrosine kinases [31,63,64] as well as with protein kinase C (PKC; [65]), a component downstream of the Fgf receptor signal transduction pathway [17]. Pdlim7 may be necessary as an adapter, bringing together transmembrane proteins and intracellular signal transducers. Without Pdlim7 function, the fin mesenchyme may not be competent to respond to either paracrine signals from the AER or autocrine signals, despite the expression of fgf10 and fgfr1 in the fin mesenchyme. The eventual loss of fgfr2 and misexpression of pea3 and erm, coupled with the lack of fgf8 expression, suggest a breakdown of Fgf response in the fin mesenchyme.
However, the Fgf pathway genes that remain active in the fin, such as fgf10 and fgf24, may be a consequence of Tbx5 misregulation caused by knock-down of Pdlim7. The ectopic activity of Tbx5 could account for the loss of specific mesenchymal and AER genes. Work by others has shown Tbx5 to function upstream of Fgf24, which may be a direct target, and Fgf10 during pectoral fin development (Fig. 8; [11,41]). fgf10 expression may be maintained in the fin mesenchyme by ectopic expression of fgf24 as a result of increased nuclear Tbx5 levels, despite the loss of AER signaling.
Conclusion
Here we provide the first evidence for a role of a PDZ-LIM family member in vertebrate limb development. Pdlim7 is expressed in developing zebrafish pectoral fins and is required for normal outgrowth. In line with the model that Pdlim7 regulates Tbx5 transcriptional activity by altering its subcellular location, during fin development this regulation appears to have direct consequences on fgf24. Loss of Pdlim7 function results in ectopic fgf24 expression in the fin mesenchyme but lack of fgf24 induction in the AER, resulting in a breakdown of the Fgf signaling loop required for fin mesenchyme proliferation and outgrowth.
Methods
Zebrafish
Wild-type (TU) stocks were maintained at 28.5°C. Embryos were staged according to Kimmel et al. [66]. Embryos were cultured in 0.0045% phenylthiourea in Danieau buffer beginning at 24 hpf to inhibit pigmentation.
Morpholino and mRNA injection
Antisense morpholino (MO) oligonucleotides were obtained from Gene Tools (Gene Tools, LLC, Philomath, OR). Pdlim7 MO2 was targeted to the splice donor site of exon 2 and detailed MO controls are described in Camarata et al. [38]. Embryos were injected at the one-cell stage and fixed at appropriate time points with 4% formaldehyde prepared from paraformaldehyde. Uninjected sibling embryos were fixed along with morpholino injected embryos as controls.
Zebrafish pdlim7 mRNA was synthesized using the mMessage mMachine kit (Ambion), as described [38]. Embryos were injected with 100 pg of pdlim7 mRNA at the one-cell stage and fixed at appropriate time points with 4% formaldehyde prepared from paraformaldehyde. Uninjected sibling embryos were fixed along with mRNA injected embryos as controls.
In situ hybridization
Whole mount in situ hybridization was performed as previously described [38] using an Intavis Insitu Pro VSi (Koeln, Germany). Antisense RNA probes used were pdlim7, tbx5 [38], hand2 [42], shh, msxc, msxd [49], fgf24 [41], pea3, erm [67], fgfr1, fgfr2 [68], versican [69], and fgf8. fgf10 cDNA was cloned into Bluescript KS+ using primers previously described [18]. Embryos were imaged on a Leica MZ16 stereomicroscope fitted with a Leica DFC490 color camera using ImagePro MC (MediaCybernetics) software. Images were processed using Adobe Photoshop CS3.
Embryo sectioning
Whole mount in situ hybridization using pdlim7 probe was performed on embryos fixed at 48 hpf. Embryos were allowed to sink in 30% sucrose, embedded in O.C.T. Compound (Tissue-Tek), and 10 micron sections obtained using a Leica CM3050 S cryostat. Sections were imaged on a Leica DMR upright microscope fitted with a QImaging Retiga-4000R Fast 1394 color camera using OpenLab software. Images were processed using Adobe Photoshop CS3.
Immunohistochemistry and TUNEL
For detecting proliferating cells and developing somites simultaneously, 28-48 hpf fixed embryos were incubated in a block solution consisting of 10% sheep serum, 2 mg/mL BSA, and 0.2% saponin in PBS with Tween-20 (PBT) for 1 hour at room temperature. Next, the Anti-phospho-histone-h3 antibody (Anti-p-H3) (Millipore) was used at a 1:20 dilution, while the MF20 antibody was used at a 1:40 dilution in PBT with 0.2% saponin. Embryos were incubated in the primary antibody solution for 1 hour at room temperature or at 4 degrees Celcius overnight. Embryos were then washed in PBT multiple times before incubation for 1 hour at room temperature in secondary antibodies diluted at 1:100 in PBT with 0.2% saponin. For detection of apoptotic cells in whole mount embryos TUNEL staining was performed essentially as previously described [70]. Briefly, 28-48 hpf fixed embryos were incubated in proteinase K for 5 - 10 minutes, subjected to several washes of PBT, and subsequently incubated at 37°C in a TUNEL cell death detection reagent for 1 hour (in situ cell death Detection Kit-TMR Red, Roche Diagnostics). Embryos were then washed multiple times in PBT and then incubated for 1 hour at RT in the block solution prior to incubation with the MF20 antibody (developed by D. A. Fischman), which was obtained from the Developmental Studies Hybridoma Bank (University of Iowa), to visualize somites. Antibody block was made as previously described and MF20 was diluted to 1:40 in PBT with 0.2% saponin as described above. Confocal microscopy was performed on a Zeiss LSM510. Statistical significance between wild-type and MO2 injected embryos was determined using two-tailed student's t-test.
Alcian blue staining
For cartilage analysis, zebrafish larvae were fixed at 96 hpf and treated with Alcian blue solution dissolved in 80% ethanol/20% glacial acetic acid (acid alcohol) for several hours or overnight. Larvae were destained in several washes of acid alcohol before being transferred to a 1% KOH:3% hydrogen peroxide solution for further clearing of pigment cells. Larval tissue was then digested in trypsin, followed by dissection of pectoral fin cartilages, which were then flat mounted and visualized on a Leica MZ16F.
Authors' contributions
TC and HGS conceived and designed the experiments. TC, DS, TS, JK, and BH, performed the experiments. TC, DS, TS, JK, BH, and HGS wrote and edited the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Table S1. Supporting quantitative data for p-H3 and TUNEL staining shown in Fig. 2.
Contributor Information
Troy Camarata, Email: tcamarata@partners.org.
Diana Snyder, Email: DSnyder@childrensmemorial.org.
Tyler Schwend, Email: tschwend@ksu.edu.
Julian Klosowiak, Email: j-klosowiak@fsm.northwestern.edu.
Brandon Holtrup, Email: BHoltrup@childrensmemorial.org.
Hans-Georg Simon, Email: hgsimon@northwestern.edu.
Acknowledgements
We are grateful to Drs. D. Yelon, C. Neumann, and K. Poss for sharing in situ hybridization probes. We would like to thank J. Krcmery for expert technical advice and critical reading of the manuscript. This work is supported by NIH grant R01HL085834 (to H.-G. Simon).
References
- Capdevila J, Izpisua Belmonte JC. Patterning mechanisms controlling vertebrate limb development. Annu Rev Cell Dev Biol. 2001;17:87–132. doi: 10.1146/annurev.cellbio.17.1.87. [DOI] [PubMed] [Google Scholar]
- Logan M. Finger or toe: the molecular basis of limb identity. Development. 2003;130:6401–10. doi: 10.1242/dev.00956. [DOI] [PubMed] [Google Scholar]
- Mercader N. Early steps of paired fin development in zebrafish compared with tetrapod limb development. Dev Growth Differ. 2007;49:421–37. doi: 10.1111/j.1440-169X.2007.00942.x. [DOI] [PubMed] [Google Scholar]
- Saunders JW Jr. The proximo-distal sequence of origin of the parts of the chick wing and the role of the ectoderm. J Exp Zool. 1948;108:363–403. doi: 10.1002/jez.1401080304. [DOI] [PubMed] [Google Scholar]
- Summerbell D. A quantitative analysis of the effect of excision of the AER from the chick limb-bud. J Embryol Exp Morphol. 1974;32:651–60. [PubMed] [Google Scholar]
- Gibson-Brown JJ, Agulnik SI, Chapman DL, Alexiou M, Garvey N, Silver LM, Papaioannou VE. Evidence of a role for T-box genes in the evolution of limb morphogenesis and the specification of forelimb/hindlimb identity. Mech Dev. 1996;56:93–101. doi: 10.1016/0925-4773(96)00514-X. [DOI] [PubMed] [Google Scholar]
- Isaac A, Rodriguez-Esteban C, Ryan A, Altabef M, Tsukui T, Patel K, Tickle C, Izpisua-Belmonte JC. Tbx genes and limb identity in chick embryo development. Development. 1998;125:1867–75. doi: 10.1242/dev.125.10.1867. [DOI] [PubMed] [Google Scholar]
- Logan M, Simon HG, Tabin C. Differential regulation of T-box and homeobox transcription factors suggests roles in controlling chick limb-type identity. Development. 1998;125:2825–35. doi: 10.1242/dev.125.15.2825. [DOI] [PubMed] [Google Scholar]
- Ohuchi H, Takeuchi J, Yoshioka H, Ishimaru Y, Ogura K, Takahashi N, Ogura T, Noji S. Correlation of wing-leg identity in ectopic FGF-induced chimeric limbs with the differential expression of chick Tbx5 and Tbx4. Development. 1998;125:51–60. doi: 10.1242/dev.125.1.51. [DOI] [PubMed] [Google Scholar]
- Begemann G, Ingham PW. Developmental regulation of Tbx5 in zebrafish embryogenesis. Mech Dev. 2000;90:299–304. doi: 10.1016/S0925-4773(99)00246-4. [DOI] [PubMed] [Google Scholar]
- Garrity DM, Childs S, Fishman MC. The heartstrings mutation in zebrafish causes heart/fin Tbx5 deficiency syndrome. Development. 2002;129:4635–45. doi: 10.1242/dev.129.19.4635. [DOI] [PubMed] [Google Scholar]
- Agarwal P, Wylie JN, Galceran J, Arkhitko O, Li C, Deng C, Grosschedl R, Bruneau BG. Tbx5 is essential for forelimb bud initiation following patterning of the limb field in the mouse embryo. Development. 2003;130:623–33. doi: 10.1242/dev.00191. [DOI] [PubMed] [Google Scholar]
- Rallis C, Bruneau BG, Del Buono J, Seidman CE, Seidman JG, Nissim S, Tabin CJ, Logan MP. Tbx5 is required for forelimb bud formation and continued outgrowth. Development. 2003;130:2741–51. doi: 10.1242/dev.00473. [DOI] [PubMed] [Google Scholar]
- Ohuchi H, Nakagawa T, Yamamoto A, Araga A, Ohata T, Ishimaru Y, Yoshioka H, Kuwana T, Nohno T, Yamasaki M. et al. The mesenchymal factor, FGF10, initiates and maintains the outgrowth of the chick limb bud through interaction with FGF8, an apical ectodermal factor. Development. 1997;124:2235–44. doi: 10.1242/dev.124.11.2235. [DOI] [PubMed] [Google Scholar]
- Min H, Danilenko DM, Scully SA, Bolon B, Ring BD, Tarpley JE, DeRose M, Simonet WS. Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes Dev. 1998;12:3156–61. doi: 10.1101/gad.12.20.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine K, Ohuchi H, Fujiwara M, Yamasaki M, Yoshizawa T, Sato T, Yagishita N, Matsui D, Koga Y, Itoh N. et al. Fgf10 is essential for limb and lung formation. Nat Genet. 1999;21:138–41. doi: 10.1038/5096. [DOI] [PubMed] [Google Scholar]
- Coumoul X, Deng CX. Roles of FGF receptors in mammalian development and congenital diseases. Birth Defects Res C Embryo Today. 2003;69:286–304. doi: 10.1002/bdrc.10025. [DOI] [PubMed] [Google Scholar]
- Norton WH, Ledin J, Grandel H, Neumann CJ. HSPG synthesis by zebrafish Ext2 and Extl3 is required for Fgf10 signalling during limb development. Development. 2005;132:4963–73. doi: 10.1242/dev.02084. [DOI] [PubMed] [Google Scholar]
- Lewandoski M, Sun X, Martin GR. Fgf8 signalling from the AER is essential for normal limb development. Nat Genet. 2000;26:460–3. doi: 10.1038/82609. [DOI] [PubMed] [Google Scholar]
- Sun X, Mariani FV, Martin GR. Functions of FGF signalling from the apical ectodermal ridge in limb development. Nature. 2002;418:501–8. doi: 10.1038/nature00902. [DOI] [PubMed] [Google Scholar]
- Boulet AM, Moon AM, Arenkiel BR, Capecchi MR. The roles of Fgf4 and Fgf8 in limb bud initiation and outgrowth. Dev Biol. 2004;273:361–72. doi: 10.1016/j.ydbio.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Xu X, Weinstein M, Li C, Deng C. Fibroblast growth factor receptors (FGFRs) and their roles in limb development. Cell Tissue Res. 1999;296:33–43. doi: 10.1007/s004410051264. [DOI] [PubMed] [Google Scholar]
- Li C, Xu X, Nelson DK, Williams T, Kuehn MR, Deng CX. FGFR1 function at the earliest stages of mouse limb development plays an indispensable role in subsequent autopod morphogenesis. Development. 2005;132:4755–64. doi: 10.1242/dev.02065. [DOI] [PubMed] [Google Scholar]
- Orr-Urtreger A, Bedford MT, Burakova T, Arman E, Zimmer Y, Yayon A, Givol D, Lonai P. Developmental localization of the splicing alternatives of fibroblast growth factor receptor-2 (FGFR2) Dev Biol. 1993;158:475–86. doi: 10.1006/dbio.1993.1205. [DOI] [PubMed] [Google Scholar]
- Xu X, Weinstein M, Li C, Naski M, Cohen RI, Ornitz DM, Leder P, Deng C. Fibroblast growth factor receptor 2 (FGFR2)-mediated reciprocal regulation loop between FGF8 and FGF10 is essential for limb induction. Development. 1998;125:753–65. doi: 10.1242/dev.125.4.753. [DOI] [PubMed] [Google Scholar]
- Arman E, Haffner-Krausz R, Chen Y, Heath JK, Lonai P. Targeted disruption of fibroblast growth factor (FGF) receptor 2 suggests a role for FGF signaling in pregastrulation mammalian development. Proc Natl Acad Sci USA. 1998;95:5082–7. doi: 10.1073/pnas.95.9.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Moerlooze L, Spencer-Dene B, Revest JM, Hajihosseini M, Rosewell I, Dickson C. An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development. 2000;127:483–92. doi: 10.1242/dev.127.3.483. [DOI] [PubMed] [Google Scholar]
- Revest JM, Spencer-Dene B, Kerr K, De Moerlooze L, Rosewell I, Dickson C. Fibroblast growth factor receptor 2-IIIb acts upstream of Shh and Fgf4 and is required for limb bud maintenance but not for the induction of Fgf8, Fgf10, Msx1, or Bmp4. Dev Biol. 2001;231:47–62. doi: 10.1006/dbio.2000.0144. [DOI] [PubMed] [Google Scholar]
- Niswander L. Pattern formation: old models out on a limb. Nat Rev Genet. 2003;4:133–43. doi: 10.1038/nrg1001. [DOI] [PubMed] [Google Scholar]
- Krause A, Zacharias W, Camarata T, Linkhart B, Law E, Lischke A, Miljan E, Simon HG. Tbx5 and Tbx4 transcription factors interact with a new chicken PDZ-LIM protein in limb and heart development. Dev Biol. 2004;273:106–20. doi: 10.1016/j.ydbio.2004.05.024. [DOI] [PubMed] [Google Scholar]
- Krcmery J, Camarata T, Kulisz A, Simon HG. Nucleocytoplasmic functions of the PDZ-LIM protein family: new insights into organ development. BioEssays. 2010;32:100–8. doi: 10.1002/bies.200900148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawid IB, Breen JJ, Toyama R. LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet. 1998;14:156–62. doi: 10.1016/S0168-9525(98)01424-3. [DOI] [PubMed] [Google Scholar]
- Fanning AS, Anderson JM. PDZ domains: fundamental building blocks in the organization of protein complexes at the plasma membrane. J Clin Invest. 1999;103:767–72. doi: 10.1172/JCI6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach I. The LIM domain: regulation by association. Mech Dev. 2000;91:5–17. doi: 10.1016/S0925-4773(99)00314-7. [DOI] [PubMed] [Google Scholar]
- Kadrmas JL, Beckerle MC. The LIM domain: from the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol. 2004;5:920–31. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- Camarata T, Bimber B, Kulisz A, Chew TL, Yeung J, Simon HG. LMP4 regulates Tbx5 protein subcellular localization and activity. J Cell Biol. 2006;174:339–48. doi: 10.1083/jcb.200511109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bimber B, Dettman RW, Simon HG. Differential regulation of Tbx5 protein expression and sub-cellular localization during heart development. Dev Biol. 2007;302:230–42. doi: 10.1016/j.ydbio.2006.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camarata T, Krcmery J, Snyder D, Park S, Topczewski J, Simon HG. Pdlim7 (LMP4) regulation of Tbx5 specifies zebrafish heart atrio-ventricular boundary and valve formation. Dev Biol. 2010;337:233–45. doi: 10.1016/j.ydbio.2009.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn DG, Kourakis MJ, Rohde LA, Silver LM, Ho RK. T-box gene tbx5 is essential for formation of the pectoral limb bud. Nature. 2002;417:754–8. doi: 10.1038/nature00814. [DOI] [PubMed] [Google Scholar]
- Ng JK, Kawakami Y, Buscher D, Raya A, Itoh T, Koth CM, Rodriguez Esteban C, Rodriguez-Leon J, Garrity DM, Fishman MC. et al. The limb identity gene Tbx5 promotes limb initiation by interacting with Wnt2b and Fgf10. Development. 2002;129:5161–70. doi: 10.1242/dev.129.22.5161. [DOI] [PubMed] [Google Scholar]
- Fischer S, Draper BW, Neumann CJ. The zebrafish fgf24 mutant identifies an additional level of Fgf signaling involved in vertebrate forelimb initiation. Development. 2003;130:3515–24. doi: 10.1242/dev.00537. [DOI] [PubMed] [Google Scholar]
- Yelon D, Ticho B, Halpern ME, Ruvinsky I, Ho RK, Silver LM, Stainier DY. The bHLH transcription factor hand2 plays parallel roles in zebrafish heart and pectoral fin development. Development. 2000;127:2573–82. doi: 10.1242/dev.127.12.2573. [DOI] [PubMed] [Google Scholar]
- Harvey SA, Logan MP. sall4 acts downstream of tbx5 and is required for pectoral fin outgrowth. Development. 2006;133:1165–73. doi: 10.1242/dev.02259. [DOI] [PubMed] [Google Scholar]
- Charite J, McFadden DG, Olson EN. The bHLH transcription factor dHAND controls Sonic hedgehog expression and establishment of the zone of polarizing activity during limb development. Development. 2000;127:2461–70. doi: 10.1242/dev.127.11.2461. [DOI] [PubMed] [Google Scholar]
- Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, Beachy PA. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–13. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- Neumann CJ, Grandel H, Gaffield W, Schulte-Merker S, Nusslein-Volhard C. Transient establishment of anteroposterior polarity in the zebrafish pectoral fin bud in the absence of sonic hedgehog activity. Development. 1999;126:4817–26. doi: 10.1242/dev.126.21.4817. [DOI] [PubMed] [Google Scholar]
- Krauss S, Concordet JP, Ingham PW. A functionally conserved homolog of the Drosophila segment polarity gene hh is expressed in tissues with polarizing activity in zebrafish embryos. Cell. 1993;75:1431–44. doi: 10.1016/0092-8674(93)90628-4. [DOI] [PubMed] [Google Scholar]
- Lallemand Y, Nicola MA, Ramos C, Bach A, Cloment CS, Robert B. Analysis of Msx1; Msx2 double mutants reveals multiple roles for Msx genes in limb development. Development. 2005;132:3003–14. doi: 10.1242/dev.01877. [DOI] [PubMed] [Google Scholar]
- Akimenko MA, Johnson SL, Westerfield M, Ekker M. Differential induction of four msx homeobox genes during fin development and regeneration in zebrafish. Development. 1995;121:347–57. doi: 10.1242/dev.121.2.347. [DOI] [PubMed] [Google Scholar]
- Moon AM, Capecchi MR. Fgf8 is required for outgrowth and patterning of the limbs. Nat Genet. 2000;26:455–9. doi: 10.1038/82601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehl H, Nusslein-Volhard C. Zebrafish pea3 and erm are general targets of FGF8 signaling. Curr Biol. 2001;11:503–7. doi: 10.1016/S0960-9822(01)00143-9. [DOI] [PubMed] [Google Scholar]
- Raible F, Brand M. Tight transcriptional control of the ETS domain factors Erm and Pea3 by Fgf signaling during early zebrafish development. Mech Dev. 2001;107:105–17. doi: 10.1016/S0925-4773(01)00456-7. [DOI] [PubMed] [Google Scholar]
- Hasson P, Del Buono J, Logan MP. Tbx5 is dispensable for forelimb outgrowth. Development. 2007;134:85–92. doi: 10.1242/dev.02622. [DOI] [PubMed] [Google Scholar]
- Collavoli A, Hatcher CJ, He J, Okin D, Deo R, Basson CT. TBX5 nuclear localization is mediated by dual cooperative intramolecular signals. J Mol Cell Cardiol. 2003;35:1191–5. doi: 10.1016/S0022-2828(03)00231-1. [DOI] [PubMed] [Google Scholar]
- Zaragoza MV, Lewis LE, Sun G, Wang E, Li L, Said-Salman I, Feucht L, Huang T. Identification of the TBX5 transactivating domain and the nuclear localization signal. Gene. 2004;330:9–18. doi: 10.1016/j.gene.2004.01.017. [DOI] [PubMed] [Google Scholar]
- Kulisz A, Simon HG. An evolutionarily conserved nuclear export signal facilitates cytoplasmic localization of the Tbx5 transcription factor. Mol Cell Biol. 2008;28:1553–64. doi: 10.1128/MCB.00935-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh TK, Packham EA, Bonser AJ, Robinson TE, Cross SJ, Brook JD. Characterization of the TBX5 binding site and analysis of mutations that cause Holt-Oram syndrome. Hum Mol Genet. 2001;10:1983–94. doi: 10.1093/hmg/10.18.1983. [DOI] [PubMed] [Google Scholar]
- Hatcher CJ, Diman NY, Kim MS, Pennisi D, Song Y, Goldstein MM, Mikawa T, Basson CT. A role for Tbx5 in proepicardial cell migration during cardiogenesis. Physiol Genomics. 2004;18:129–40. doi: 10.1152/physiolgenomics.00060.2004. [DOI] [PubMed] [Google Scholar]
- Healy NC, O'Connor R. Sequestration of PDLIM2 in the cytoplasm of monocytic/macrophage cells is associated with adhesion and increased nuclear activity of NF-kappaB. J Leukoc Biol. 2009;85:481–90. doi: 10.1189/jlb.0408238. [DOI] [PubMed] [Google Scholar]
- Loughran G, Healy NC, Kiely PA, Huigsloot M, Kedersha NL, O'Connor R. Mystique is a new insulin-like growth factor-I-regulated PDZ-LIM domain protein that promotes cell attachment and migration and suppresses Anchorage-independent growth. Mol Biol Cell. 2005;16:1811–22. doi: 10.1091/mbc.E04-12-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura N, Ohno K, Katayama T, Kanayama N, Sato K. The PDZ-LIM protein CLP36 is required for actin stress fiber formation and focal adhesion assembly in BeWo cells. Biochem Biophys Res Commun. 2007;364:589–94. doi: 10.1016/j.bbrc.2007.10.064. [DOI] [PubMed] [Google Scholar]
- Prykhozhij SV, Neumann CJ. Distinct roles of Shh and Fgf signaling in regulating cell proliferation during zebrafish pectoral fin development. BMC Dev Biol. 2008;8:91. doi: 10.1186/1471-213X-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durick K, Wu RY, Gill GN, Taylor SS. Mitogenic signaling by Ret/ptc2 requires association with enigma via a LIM domain. J Biol Chem. 1996;271:12691–4. doi: 10.1074/jbc.271.27.15934. [DOI] [PubMed] [Google Scholar]
- Wu RY, Gill GN. LIM domain recognition of a tyrosine-containing tight turn. J Biol Chem. 1994;269:25085–90. [PubMed] [Google Scholar]
- Kuroda S, Tokunaga C, Kiyohara Y, Higuchi O, Konishi H, Mizuno K, Gill GN, Kikkawa U. Protein-protein interaction of zinc finger LIM domains with protein kinase C. J Biol Chem. 1996;271:31029–32. doi: 10.1074/jbc.271.49.31029. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Munchberg SR, Ober EA, Steinbeisser H. Expression of the Ets transcription factors erm and pea3 in early zebrafish development. Mech Dev. 1999;88:233–6. doi: 10.1016/S0925-4773(99)00179-3. [DOI] [PubMed] [Google Scholar]
- Poss KD, Shen J, Nechiporuk A, McMahon G, Thisse B, Thisse C, Keating MT. Roles for Fgf signaling during zebrafish fin regeneration. Dev Biol. 2000;222:347–58. doi: 10.1006/dbio.2000.9722. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B. Antivin, a novel and divergent member of the TGFbeta superfamily, negatively regulates mesoderm induction. Development. 1999;126:229–40. doi: 10.1242/dev.126.2.229. [DOI] [PubMed] [Google Scholar]
- Schwend T, Ahlgren SC. Zebrafish con/disp1 reveals multiple spatiotemporal requirements for Hedgehog-signaling in craniofacial development. BMC Dev Biol. 2009;9:59. doi: 10.1186/1471-213X-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Supporting quantitative data for p-H3 and TUNEL staining shown in Fig. 2.