Abstract
Type 1B diabetes (typically early onset; without islet autoantibodies) has been described in patients bearing small coding sequence mutations in the INS gene. Not all mutations in the INS gene cause the autosomal dominant Mutant INS-gene-induced Diabetes of Youth (MIDY) syndrome, but most missense mutations affecting proinsulin folding produce MIDY. MIDY patients are heterozygotes, with the expressed proinsulin mutants exerting dominant-negative (gain of toxic function) behavior in pancreatic beta cells. Herein, we focus primarily on proinsulin folding in the endoplasmic reticulum, providing insight into perturbations of this folding pathway in MIDY. Accumulated evidence indicates that in the molecular pathogenesis of the disease, misfolded proinsulin exerts dominant effects that initially inhibit insulin production, progressing to beta cell demise with diabetes.
Basics of proinsulin structure and risk of misfolding in the ER
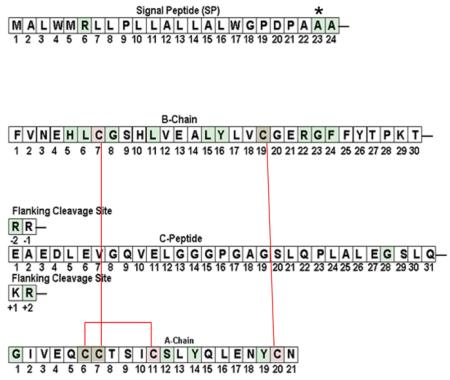
Despite a net weight in humans of only ~2 grams, pancreatic islets and their secreted peptide hormones are essential for normal body metabolism. This is especially true of glucose-regulated insulin production and insulin secretion from pancreatic beta cells. The protein production pathway begins when the INS gene product is translated as preproinsulin 1. From N- to C- terminus, the precursor is comprised sequentially of the signal peptide (SP, residues 1-24), insulin B-chain (residues 1-30), C-peptide (residues 1-31) plus its two sets of flanking dibasic cleavage sites, and the insulin A-chain (residues 1-21). The newly-synthesized preproinsulin signal peptide facilitates recruitment to the endoplasmic reticulum (ER), whereupon the growing nascent polypeptide is cotranslationally translocated across the ER membrane into the ER lumen.
Approximately half of daily insulin secretion accommodates ongoing anabolic needs under basal conditions between meals, while the other half is secreted in response to eating 2. Even with the substantial decrease in insulin expression and secretion under fasting conditions, proinsulin biosynthesis still dominates the beta cell proteome 3. Proinsulin monomer folding and dimerization is thought to occur within the ER lumen; traffic through the Golgi complex and into immature secretory granules brings proinsulin in contact with zinc facilitating its further assembly to hexamers 4. Ultimately, proteolytic excision of the C-peptide and its dibasic flanking residues, along with organelle remodeling, results in insulin storage in mature secretory granules 5, 6.
Much is known about proinsulin or insulin folding in vitro while relatively little is known about proinsulin folding in vivo 7. Indeed, experiments have not yet established whether the following processes are strictly sequential events: 1) completion of translocation, 2) excision of the signal peptide catalyzed by the ER signal peptidase, 3) initiation of B-chain folding, and 4) initiation of disulfide bond formation leading to the three evolutionary conserved disulfide bonds of proinsulin [destined to be ‘interchain’ within insulin: C(B7)-C(A7) and C(B19)-C(A20), as well as intrachain C(A6)-C(A11)]. In the ER, these events are estimated to occur within a time span of 20 seconds to perhaps a minute from the initiation of preproinsulin translocation. Given the competition between reaction rates in pancreatic beta cells and all eukaryotic cells, it is likely that within a cohort of nascent preproinsulins or other secretory proteins, molecules may engage in closely-spaced early events in a stochastic process 8, ie., with a sequence of events that are not precisely identical between major and minor subpopulations of preproinsulins.
With regard to initiation of B-chain folding, the secondary structure of proinsulin includes three alpha-helical domains. Based on proinsulin delivery into the ER lumen from N- to C-terminus, the B-chain has the earliest folding opportunity, and acquisition of the central B-chain alpha helix running from S(B9) - C(B19), including side-chain packing at residue V(B12)9, may be an early event to facilitating formation of the C(B19)-C(A20) disulfide bond and driving the folding pathway forwards 10. In conjunction with additional B-chain structural features 11, an N-terminal A-chain alpha-helix including V(A3) - A(A8) includes alignment of the C(B7)-C(A7) and C(A6)-C(A11) disulfide bonds while a C-terminal A-chain alpha helix including L(A13) - Y(A19) influences alignment of C(B19)-C(A20) 12, 13. Within the ER luminal environment, proinsulin disulfide bond formation is coupled to the reduction of one or more resident ER oxidoreductases 14, which are in turn catalytically recycled via their own reduction of Ero1beta and/or Ero1alpha 15, 16.
Formation of disulfide bonds within proinsulin that are destined to be ‘interchain’ within insulin, i.e., C(B7)-C(A7) and C(B19)-C(A20), are essential for stabilizing a three-dimensional structure suitable for anterograde export of the prohormone from the ER 17. Within the three-dimensional structure of proinsulin, hydrophobic core residues are buried in the interior of the molecule 12, whereas the C(B19)-C(A20) disulfide bond is not totally buried. and the C(B7)-C(A7) bond is nearly fully exposed on the surface of the folded polypeptide 18, so as to render proinsulin susceptible to destabilization by thiol attack.
As much as a third of all newly synthesized proteins may fail to fold properly, requiring either refolding or elimination 19; thus, the “substrate” for proinsulin misfolding already exists in the wild-type molecule 20. In the event of proinsulin misfolding, “sticky” residues from the hydrophobic core and unpaired or mispaired cysteine thiols create significant risk for non-native intermolecular interactions with nearest neighbors including the high concentration of bystander proinsulin molecules in the ER. Such interactions could confer ER retention on bystander proinsulin molecules, limiting their potential for insulin production.
Evidence for proinsulin misfolding in the ER
The first indication of the possibility of such misfolding emerged upon biosynthetic expression of an insulin analog bearing the P(B28)K,K(B29)P double mutation (clinically known as ‘insulin lispro’), leading to low protein production of the precursor in the ER, with the expressed product containing aberrant disulfide isomers 21. This was surprising given that in vitro folding stability of some monomeric insulin analogs, including lispro, is thought to be comparable to that of native insulin 22, 23. One possible explanation was that foreshortening of the C-peptide in a bioengineered analog might impair chain alignment for disulfide bond formation 17; however, only extreme shortening triggers increased disulfide mismatch and loss of insulin bioactivity 24. In fact, aberrant proinsulin disulfide isomers are made even in pancreatic islets isolated from normal rodents in the absence of any agents that induce ER stress, although the abundance of such aberrant isomers is increased under conditions of increased proinsulin synthesis such as high glucose or genetic inability to maintain the phosphorylation state of Ser51 of eukaryotic initiation factor 2-alpha (eIF2alpha) 20, 25. To date, the precise disulfide mismatches in proinsulin have not been identified; however, artificially engineered insulin disulfide isomers bearing native-like structural features 26-28 might possibly correspond to proinsulin disulfide isomers that occur in the ER of pancreatic beta cells.
Accumulation of misfolded proteins in the ER is a cause of (and synonymous with) ER stress. Temporary or chronic imbalance in the proinsulin folding environment in beta cells leads to temporary or chronic ER stress. ER stress is not synonymous with the ER stress response (see Box 1); in normal physiology, the two are linked but in the pathophysiology of diabetes, imbalance between ER stress and the ER stress response may occur. For example, genetic 29, 30 or epigenetic 31 incompetence in the ER stress response only increases ER stress 25. This has been shown for misfolded secretory proteins other than proinsulin 32, 33, and may also occur in states of misfolded proinsulin accumulation such as upon dysregulation of proinsulin biosynthesis 34, 35 or degradation 36. Temporary and chronic ER stress response activation is linked to both pro-survival and pro-apoptotic pathways in pancreatic beta cells, as reviewed extensively by others 29-31, 37-39.
Box 1. ER stress response pathways and proinsulin biosynthesis and misfolding.
PERK (PKR-like Endoplasmic Reticulum Kinase) is an ER signaling protein responsible for phosphorylating eIF2alpha, and increased PERK activity facilitates the cellular response needed to coordinate the synthesis of proinsulin with its folding and trafficking 25, 76-81. Defects in PERK-mediated phosphorylation of eIF2alpha result in increased proinsulin misfolding and ER retention caused by impaired ER homeostasis 29; importantly, these phenotypes occur even in the absence of any INS gene mutations.
Ire1alpha is a conserved ER signaling protein that is also expressed in pancreatic beta cells. Knockdown of Ire1alpha in a pancreatic beta cell line markedly decreases proinsulin biosynthesis 34, while chronic activation of Ire1 can also result in decreased proinsulin biosynthesis through preproinsulin mRNA turnover 34, 82.
ATF6 is the third ER membrane signaling component of the tripartite ER stress response. ATF6 overexpression inhibits insulin gene expression 83 and can trigger beta cell apoptosis, but its physiological overexpression is held in check by facilitated degradation involving the Wolfram syndrome gene product, WFS1 84.
Insulinopathies
Most monogenic mutations in the coding sequence of insulin, including classical insulinopathies and MIDY mutants (the latter being the focus of this review), result in autosomal dominant disease with a spectrum of severity, ranging from late onset diabetes with hyperinsulinemia to early onset diabetes with insulin deficiency. Additionally, a smaller subset of patients with neonatal diabetes have recessive (loss-of-function) mutations in the INS gene, resulting in impaired insulin biosynthesis 40.
The classical insulinopathies (so named because the mutants have been known for many years), involve three molecular mechanisms: a) poor insulin binding to the insulin receptor with hyperinsulinemia secondary to poor clearance of mutant insulin-V(A3)L, -F(B24)S, or F(B25)L from the circulation 41; b) endoproteolytic cleavage site mutations 42; and c) mutant proinsulin-H(B10)D that exhibits altered protein storage in the secretory pathway43. Collectively, each of these mutations results in significant secretion of insulin or proinsulin to the bloodstream, and therefore none are likely to exert their effect exclusively within the secretory pathway of pancreatic beta cells.
Mutant INS-gene-induced Diabetes of Youth (MIDY)
Over the past three years, 26 new INS gene mutations have been associated with a monogenic diabetes syndrome 44-51 that we are calling Mutant Ins-gene-induced Diabetes of Youth (MIDY), defined as autosomal dominant diabetes with insulin deficiency in the absence of beta cell autoimmunity. Patients with MIDY were previously diagnosed most often as having permanent neonatal diabetes mellitus and less frequently as type 1b (autoantibody-negative) diabetes, MODY or type 2 diabetes. Genetic studies have shown that 80% of the mutations causing MIDY occur de novo with 20% transmitted from parent to child. MIDY is second in frequency only to mutations in the sulfonylurea receptor/Kir6.2 complex as a monogenic cause of permanent neonatal diabetes 52. Although MIDY mutations have been suggested to affect the earliest events of proinsulin biosynthesis in the ER 7, the precise molecular mechanisms of beta cell failure caused by these mutations are incompletely understood.
Mice have two functional, nonallelic insulin genes (with Ins2 corresponding to the human INS gene on chromosome 11). A heterozygous null genotype at the Ins2 locus — with or without a homozygous null genotype at the Ins1 locus — is insufficient to cause diabetes 53, 54, yet male Akita mice inheriting even one Ins2 allele encoding the MIDY mutant proinsulin-C(A7)Y develop insulin deficient-diabetes after weaning. This suggests that the MIDY mutant creates a gain of toxic function. Thus far, the Akita mouse, expressing proinsulin-C(A7)Y, is the most studied MIDY mutant. In the remainder of the review, we first provide an overview of human preproinsulin mutations causing MIDY, and then discuss potential molecular mechanisms of diabetes pathogenesis, relying heavily on data from the Akita mouse model.
MIDY clinical phenotypes linked to molecular phenotypes
Although most newly identified INS gene mutations cause severe insulin deficient diabetes (Table 1), the broad spectrum of clinical phenotypes suggests that different MIDY (pre)proinsulin mutations behave differently. Mutated amino acid residues causing MIDY have now been found within all domains of the preproinsulin molecule including the signal peptide, C-peptide and flanking cleavage sites. Over 70% of the mutations are contained with the insulin B- or A-chains 44-48, 55. Many of these mutations have implications for the maintenance of normal folding and bioactivity of mature (ie, processed) insulin 7, but it is highly likely that at least some conserved residues of proinsulin are involved primarily in the proinsulin folding pathway (ie, kinetics effects on achieving a final folded structure) rather than being important for structural stability (or even bioactivity) after normal disulfide pairing has been accomplished (ie, thermodynamic effects on the final folded structure) 56. Many of the MIDY mutants create or eliminate a cysteine residue at different positions along the polypeptide chain, resulting in an odd number of Cys residues with the potential to alter or disrupt normal proinsulin disulfide pairing. Of those mutations not altering preproinsulin cysteine content, most are located at evolutionarily conserved residues suggesting that they are indispensable for normal proinsulin folding. Ultimately, both cysteine and non-cysteine MIDY mutants highlight the importance of properly paired proinsulin disulfide bonds to prevent development of diabetes caused by misfolded proinsulin.
Table 1.
Sequence of preproinsulin and site of MIDY mutations
|
MIDY mutations |
Ages of onset |
Original diagnosis |
References |
|||
|---|---|---|---|---|---|---|
| Nomenclature by peptide chain |
Nomenclature by preproinsulin a.a. |
|||||
| Signal Peptide |
R(SP6)C or H | R6C or H | 15-65yr1 | MODY1 | 50,56,60 |
|
| A(SP23)S | A23S | 6 yr | Type 1 | 66 | ||
| A(SP24)D | A24D | most <6 mo2 | Neonatal DM2 | 49,50,62 | ||
|
| ||||||
| Insulin B Chain |
H(B5)D | H29D | <6 mo | Neonatal DM | 50 | |
| L(B6)M. V. or P | L30M, V or P | <6 mo or 17-38 yr3 | Neonatal DM or MODY3 | 53,60 | ||
| G(B8)S or R | G32S or R | most <6 mo4 | Neonatal DM or Type 14 | 49-50,54,66 | ||
| L(B11)P | L35P | <6 mo | Neonatal DM | 50,53 | ||
| LY(B15,16)H | L39Y delinsH | <6 mo | Neonatal DM | 53 | ||
| C(B19)G | C43G | most <6 mo5 | Neonatal DM5 | 49-50 | ||
| R(B22)Q | R46Q | 17-24 yr | MODY | 51,56 | ||
| G(B23)V | G47V | <6 mo | Neonatal DM | 49-50 | ||
| F(B24)C | F48C | <6 mo | Neonatal DM | 49-50 | ||
|
| ||||||
| B-C junction |
R(Cpep -2)C | R55C | <6 mo or 8-37 yr | Type 1 or MODY | 51,60 | |
| C-Peptide | G(Cpep28)R | G84R | <6 mo | Neonatal DM | 50 | |
| C-A junction |
R(Cpep+2)C | R89C | most <6 mo6 | Neonatal DM6 | 49-54 | |
|
| ||||||
| Insulin A Chain |
G(A1)C | G90C | <6 mo | Neonatal DM | 49-50 | |
| C(A6)Y | C95Y | <6 mo | Neonatal DM | 53 | ||
| C(A7)Y or S | C36Y or S | <6 mo | Neonatal DM | 49-50,62,55 | ||
| S(A12)C | S101C | <6 mo | Neonatal DM | 50 | ||
| Y(A14)C | Y103C | <6 mo | Neonatal DM | 14 | ||
| Y(A19)C or Stop | YlOSC or stop | <6 mo | Neonatal DM | 49-50,53 | ||
Age of onset 5 wk - 7 y in 3 members of the same family (50)
Proband’s mother, age 68, carries mutation without DM (60)
Age of onset 5 mo - 22 y in 4 members of the same family(50)
Proband’s father developed diet-controlled type 2 DM beginning at age 30 (49)
Three family members developed DM age 2 - 4 y (51).
The native SP23 of chimpanzee preproinsulin is Ser. In the cartoon on the right, the normal position of Cys residues is overlaid in pink; disulfide pairs shown with red lines
MIDY mutation sites overlaid in green.
Four preproinsulin mutations located either near the N or C-terminal end of the signal peptide are likely to affect one or more of the four earliest events of proinsulin biosynthesis: 1) completion of translocation, 2) excision of the signal peptide catalyzed by the ER signal peptidase, 3) initiation of B-chain folding, and 4) initiation of disulfide bond formation. Preproinsulin-R(SP6)C and - R(SP6)H mutations result in deficiency within the signal peptide of an N-terminal positive charge, which has been shown to play an important role for targeting newly synthesized secretory proteins to the ER 57. Both of these mutations resulted in an ER stress response when the recombinant preproinsulin mutants were expressed in transfected cell lines, yet mutant preproinsulin molecules were properly delivered to and through the secretory pathway 58. The translocation efficiency of these mutants, as well as the origin of the ER stress, needs further exploration. Preproinsulin-A(SP24)D mutation disrupts normal signal peptide cleavage 11, which is likely to interfere with downstream proinsulin folding and trafficking. Expression in a transfected beta cell line demonstrates proper ER targeting of preproinsulin-A(SP24)D but > 95% of the mutant molecules were retained intracellularly as proteins of increased molecular mass (consistent with uncleaved preproinsulin) 59. By contrast, another study reported that a meaningful fraction of mutant proinsulin reaches secretory granules, and neither expression of the mutant nor a demonstrably associated ER stress response attenuates secretion of co-expressed wild-type insulin or C-peptide 60. Thus, the biological mechanisms by which this mutant brings about diabetes require further clarification. Finally, whereas preproinsulin-A(SP23)S mutation was reported recently in a patient originally diagnosed with type 1 diabetes 61, Ser is the native residue in this position of chimpanzee preproinsulin, leaving open the question of whether this substitution is a real contributor to diabetes.
Several new proinsulin mutations have been reported in the C-peptide and its flanking dibasic cleavage sites. The proinsulin-G(C28)R mutant was originally identified in a subject diagnosed with type 1 diabetes 45. However, a new study reports that the proinsulin-G(C28)R variant can efficiently exit the ER and be released from transfected cells 59, raising questions about whether this mutation is diabetogenic. Of the two MIDY mutations reported at flanking cleavage sites, R(Cpep −2)C and R(Cpep +2)C, the latter is more commonly reported, encoded at a CpG hot spot for mammalian mutations 44. Unlike other mutations occurring at this same amino acid residue, R(Cpep +2)P, R(Cpep +2)L and R(Cpep +2)H, which cause hyperproinsulinemia diagnosed in adulthood 42, 43, most patients with MIDY mutations at flanking cleavage sites have early onset insulin deficient diabetes. Direct side-by-side comparison of the recombinant proinsulin-R(Cpep +2)C and proinsulin-R(Cpep +2)L mutants showed a profound inhibition of secretion linked to the mutant Cys residue with no inhibition linked to the mutant Leu residue 48. This finding underscores the potency of unpaired proinsulin cysteines as a perturbant.and highlights the range of clinical severity and age of onset of diabetes associated with different mutant alleles even when they encode distinct amino acids at the same position of the preproinsulin polypeptide chain.
Before proceeding on to a detailed consideration of MIDY mutations located within the B- and A-chains that create or delete a Cys residue (represented by proinsulin-C(A7)Y, which is discussed below), we wish to mention a final group of mutants involving missense mutations that neither create nor delete a cysteine yet nevertheless cause MIDY 7. Intriguingly, no such mutations have yet been found in the A-chain. By contrast, multiple mutations have been reported in the B-chain, affecting the highly conserved residues 5, 6, 8, 11, 22, or 23. Residue H(B5) facilitates alignment of the proinsulin C(B7)-C(A7) disulfide partners 62 and recent evidence suggests that sequence variation at this site is a primary reason for the formation of aberrant disulfide isomers involving C(B7) of IGF-1, a proinsulin homolog 11. Invariant residue L(B6), which contributes strongly to the structure of bioactive insulin, has been proposed to play a role in maintaining local structure in the vicinity of the C(B7)-C(A7) disulfide and allows flexibility of the N-terminal region to either join the central B-chain alpha helix or turn from it 63. Similarly substitution of residue G(B8)S mutagenizes a site critical to formation of the C(B7)-C(A7) disulfide bond 64. Perturbations around residue L(B11) also impair insulin structure 9, and computational biology studies suggest L(B11) and R(B22) as two of the 5 most important residues for the stability of individual insulin monomers 65, including hydrogen bonding 46 critical to B-chain/A-chain alignment. Finally, substitution at residue G(B23)V may implicate this residue in rotational flexibility of the proinsulin polypeptide backbone 66,which might facilitate C(B19)-C(A20) pairing; this is a hypothesis that deserves testing. To summarize, the evidence from this group of MIDY mutants points towards the idea that proper B-chain structure and flexibility are essential elements for proper cysteine alignments for disulfide bonding in the proinsulin folding pathway. Failure to achieve these alignments sets off the molecular pathogenesis of the autosomal dominant diabetes known as MIDY.
Molecular mechanisms of beta cell failure caused by misfolded preproinsulins: lessons from the Akita mouse
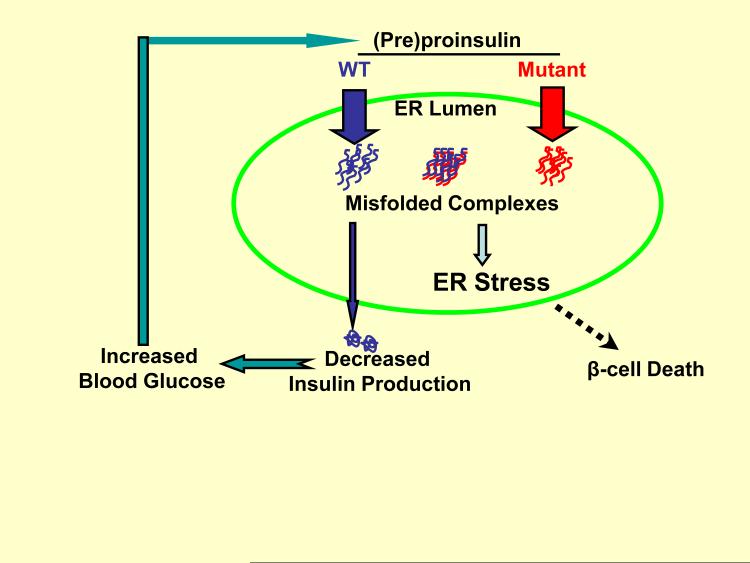
Although not fully characterized, progress is being made to advance our understanding of the pathogenesis of diabetes caused by misfolded (pre)proinsulin, using MIDY as the most clear-cut example. In brief, we suggest that the disease may be initiated when trafficking of normal proinsulin is adversely affected by co-expressed misfolded mutant proinsulin, resulting in a cascade of additional, overlapping defects that culminate in beta cell demise (Fig. 1). The most detailed results to date come from data using Akita mice in which a heterozygous proinsulin-C(A7)Y mutation is identical to a heterozygous mutation causing human MIDY. Immunohistological analysis of the pancreas of male Akita mice after diabetes onset excludes lymphocytic infiltration or islet inflammation as a cause of diabetes, yet there is a decrease in insulin immunostaining 67.
Figure 1.
Proposed molecular mechanisms of beta cell failure caused by misfolded (pre)proinsulin. When WT (blue) and mutant (red) proinsulin are co-expressed in beta cells, the mutant proinsulin promotes WT proinsulin retention in the ER, decreasing insulin production, and forcing beta cells to use up insulin secretory granules from their progressively depleted insulin storage pool. The insufficiency of insulin increases blood glucose that initially stimulates even more production of wild-type and mutant preproinsulins, exacerbating ER stress induced by misfolded proinsulin-containing protein complexes. Inexorable progression of insulin insufficiency leads to frank diabetes, further ER stress, and beta cell demise.
From immunohistological analysis in a population of Akita males and females at postnatal day 1, there is a normal insulin positive area and islet mass 68, suggesting that individuals bearing MIDY mutations are not born with a deficiency of beta cells. And yet, at the electron microscope level, the beta cells of these animals at postnatal day 1 exhibit expansion of compartments of the early secretory pathway including the ER, which is accompanied by an increase in mitochondria (suggesting a need for increased energy production) and a decrease in insulin secretory granules 68. Normally, beta cells use their biosynthetic machinery to generate a large reserve of accumulated secretory granules that represent the major pool of pancreatic insulin. Thus, given that Akita mice have normal insulin sensitivity (and thus no need to secrete more insulin than normal), a morphological decrease in the number of insulin granules on postnatal day 1 suggests that there is already a significant block in insulin production. Indeed, in such animals, the pancreatic content of insulin, while not yet low enough yet to trigger detectable diabetes, has already decreased to less than one-third of that of wild-type controls 68.
When Akita animals begin to show overt signs of lower body weight and higher blood glucose indicating insulin deficiency after a few weeks of postnatal life, a trend towards a decrease in insulin positive area of the islets first becomes noticeable 68. However even within the remaining beta cells there is a decrease in ‘steady-state’ insulin content that is much greater than can be accounted for by loss of one wild-type allele 69. Thus, although age-related progression of diabetes leads ultimately to an obvious loss of beta cell mass 70, and the underlying genetic background of the individual clearly influences the severity and progression of the diabetes, it appears that in this representative example of MIDY, a block in insulin production is a primary defect, preceding the decline of beta cell mass 71. At a stage of overt diabetes but prior to massive loss of islets, a loss of insulin production is detected despite supranormal levels of proinsulin biosynthesis from the remaining wild-type alleles, indicating a defect of wild-type proinsulin trafficking to secretory granules 72. It is quite possible that secondary effects from the hyperglycemia of diabetes further exacerbate the underlying proinsulin trafficking defects and concomitant ER stress 69.
In the case of homozygosity of the Akita mutation, animals still have two functional Ins1 alleles, but the higher level of misfolded proinsulin results in fulminant diabetes within two weeks of life, associated with a dramatic loss of total islet area and a reduction in the islet content of beta cells 68. Given the presence of the remaining wild-type mouse Ins1 alleles, the likelihood is that animals bearing the heterozygous Akita mutation probably synthesize less MIDY proinsulin than their human counterparts (who have only one INS gene) whereas homozygous animals probably synthesize an equal or greater fraction of MIDY proinsulin than their human counterparts. A critical lesson from the comparison of heterozygous and homozygous animals is that the actual fraction of total newly-synthesized proinsulin comprised of misfolded proinsulin will have a great impact on the acceleration/progression of diabetes.
If two copies of Ins1 are sufficient to avoid diabetes 53, 54, how can we account for the insulin deficiency before any loss of beta cell mass? In islets isolated from Akita mice, the mutant proinsulin inhibits insulin production from co-expressed wild-type proinsulin alleles 72. Further, species-specific or epitope-tagged versions of proinsulin allowed clear distinction between wild-type ‘innocent proinsulin bystanders’ and mutant gene products. Using a construct known as hProCpepGFP (human proinsulin with green fluorescent protein contained within the C-peptide), human insulin and CpepGFP are generated in the secretory granules of rodent pancreatic beta cells 72. Transgenic mice expressing hProCpepGFP in pancreatic beta cells are normal, but when mated to Akita mice to develop MIDY, the bystander hProCpepGFP precursor is blocked in the secretory pathway of islet beta cells; in parallel, production of the CpepGFP product also becomes blocked 70. Interestingly, an hProCpepGFP bearing the C(A7)Y MIDY mutation inhibited the export of co-expressed wild-type proinsulin (resulting in high molecular weight protein complexes that included both mutant and nonmutant gene products), while an untagged version of mouse Akita protein inhibited wild-type human insulin production in pancreatic beta cells 72. Initially, the trafficking blockade seems to involve specific recruitment of bystander proinsulin into aberrant disulfide-linked proinsulin-containing complexes while other secretory proteins are normally trafficked and secreted 69, 72. More work is needed to precisely characterize the aberrant disulfide-linked proinsulin-containing protein complexes, but recent studies suggest that improperly oxidized, high molecular weight species of wild-type proinsulin are recovered in increased amounts when the oxidizing environment of the ER is perturbed 15. Alternatives that need to be tested (which are not mutually exclusive) are that proinsulin mutants causing MIDY might directly engage wild-type proinsulin in aberrant disulfide-linked protein complexes, or might indirectly recruit wild-type proinsulin into such complexes through changes in the oxidizing environment of the ER. Through either of these mechanisms, the expression of proinsulin mutants that cause MIDY blocks wild-type proinsulin in the ER, causing insulin deficiency.
Findings consistent with this basic paradigm are now being replicated with many of the newly-described MIDY mutants. For example, recent examinations of human MIDY mutants typically exploit species-specific antibodies and constructs, or distinct, epitope-tagged versions 58-60. To greater or lesser degrees, each of these most recent studies supports the notion of dominant-negative inhibition of trafficking of co-expressed proinsulin, which occurs at a stage before beta cell death; indeed, demonstration of these bystander effects requires ongoing secretory protein synthesis.
In later stages of MIDY, as beta cell dysfunction in diabetic individuals worsens, other co-expressed secretory proteins may become blocked in the secretory pathway 73. Moreover, affected individuals convert from having normal beta cell mass to having a loss of beta cell mass 70. Indeed, the higher the expressed fraction of misfolded proinsulin, the more beta cell demise is accelerated, and the greater is the activation of ER stress responses that progress even further after diabetes onset 69. Among other things, chronic activation of ER stress response pathways leads to induction of the Chop protein that is strongly implicated in beta cell apoptosis 74, 75. While the present review has not focused specifically on the links between ER stress response pathways and beta cell death, we do not exclude the fact that cell death pathways are highly likely to participate in the natural progression of MIDY, as a final common pathway leading ultimately to a decrease in beta cell mass.
Conclusion
We are beginning to understand that MIDY links the earliest steps of proinsulin folding and misfolding to the cell biology of the secretory pathway in pancreatic beta cells. In MIDY, evidence indicates that dominant-negative diabetes is directly linked to the quantity of misfolded proinsulin expressed. Yet other genetic and epigenetic modifiers may contribute to MIDY phenotypes. Patients with several MIDY mutations, including proinsulin-A(SP24)D, G(B8)S, G(B8)R, C(B19)G, or R(Cpep -2)C, have a wide range of clinical severity even among family members bearing the same mutation — including the possibility of absence of diabetes throughout life. Developing therapeutics that may bring about such an outcome may have relevance far beyond MIDY. Specifically, the work described in this review raises new questions about beta cell failure in more common forms of type 2 diabetes, in which augmented absolute levels of misfolded proinsulin might bring about similar dominant inhibition of insulin production, ER stress, and beta cell demise.
Acknowledgements
This review was supported primarily by National Institutes of Health Grant R01 DK 48280 (to P.A.), and also to R00 DK077441A (to I.H.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Welsh M, et al. Translational control of insulin biosynthesis. Evidence for regulation of elongation, initiation and signal-recognition-particle-mediated translational arrest by glucose. Biochem J. 1986;235:459–467. doi: 10.1042/bj2350459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rendell MS, Jovanovic L. Targeting postprandial hyperglycemia. Metabolism. 2006;55:1263–1281. doi: 10.1016/j.metabol.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 3.Permutt MA. Biosynthesis of insulin. In: Cooperstein SJ, Watkins D, editors. The Islets of Langerhans. Academic Press; 1981. pp. 75–95. [Google Scholar]
- 4.Huang XF, Arvan P. Intracellular transport of proinsulin in pancreatic b-cells: structural maturation probed by disulfide accessibility. J. Biol. Chem. 1995;270:20417–20423. doi: 10.1074/jbc.270.35.20417. [DOI] [PubMed] [Google Scholar]
- 5.Kuliawat R, et al. Differential sorting of lysosomal enzymes out of the regulated secretory pathway in pancreatic ß-cells. J. Cell Biol. 1997;137:595–608. doi: 10.1083/jcb.137.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuliawat R, Arvan P. Protein targeting via the “constitutive-like” secretory pathway in isolated pancreatic islets: passive sorting in the immature granule compartment. J. Cell Biol. 1992;118:521–529. doi: 10.1083/jcb.118.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiss MA. Proinsulin and the genetics of diabetes mellitus. J. Biol. Chem. 2009;284:19159–19163. doi: 10.1074/jbc.R109.009936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnsson N, Varshavsky A. Ubiquitin-assisted dissection of protein transport across membranes. EMBO J. 1994;13:2686–2698. doi: 10.1002/j.1460-2075.1994.tb06559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu SQ, et al. Steric requirements at position B12 for high biological activity in insulin. Biochemistry (Mosc) 1993;32:2631–2635. doi: 10.1021/bi00061a022. [DOI] [PubMed] [Google Scholar]
- 10.Hua QX, et al. The folding nucleus of the insulin superfamily: a flexible peptide model foreshadows the native state. J. Biol. Chem. 2006;281:28131–28142. doi: 10.1074/jbc.M602616200. [DOI] [PubMed] [Google Scholar]
- 11.Sohma Y, et al. Contribution of residue B5 to the folding and function of insulin and IGF-I: constraints and fine-tuning in the evolution of a protein family. J. Biol. Chem. 2010;285:5040–5055. doi: 10.1074/jbc.M109.062992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker EN, et al. The structure of 2Zn pig insulin crystals at 1.5 A resolution. Phil. Trans. R. Soc. Lond. - Biol. Sci. 1988;319:369–456. doi: 10.1098/rstb.1988.0058. [DOI] [PubMed] [Google Scholar]
- 13.Hua QX, et al. Mechanism of insulin chain combination. Asymmetric roles of A-chain alpha-helices in disulfide pairing. J. Biol. Chem. 2002;277:43443–43453. doi: 10.1074/jbc.M206107200. [DOI] [PubMed] [Google Scholar]
- 14.Appenzeller-Herzog C, Ellgaard L. The human PDI family: versatility packed into a single fold. Biochim. Biophys. Acta. 2008;1783:535–548. doi: 10.1016/j.bbamcr.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 15.Zito E, et al. ERO1-beta, a pancreas-specific disulfide oxidase, promotes insulin biogenesis and glucose homeostasis. J. Cell Biol. 2010;188:821–832. doi: 10.1083/jcb.200911086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sevier CS, Kaiser CA. Ero1 and redox homeostasis in the endoplasmic reticulum. Biochim. Biophys. Acta. 2008;1783:549–556. doi: 10.1016/j.bbamcr.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 17.Liu M, et al. Role of the connecting peptide in insulin biosynthesis. J. Biol. Chem. 2003;278:14798–14805. doi: 10.1074/jbc.M212070200. [DOI] [PubMed] [Google Scholar]
- 18.Guo ZY, et al. The in vitro oxidative folding of the insulin superfamily. Antioxidants & redox signaling. 2008;10:127–139. doi: 10.1089/ars.2007.1860. [DOI] [PubMed] [Google Scholar]
- 19.Schubert U, et al. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- 20.Liu M, et al. Proinsulin disulfide maturation and misfolding in the endoplasmic reticulum. J. Biol. Chem. 2005;280:13209–13212. doi: 10.1074/jbc.C400475200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang B.-y, et al. Behavior in the eukaryotic secretory pathway of insulin-containing fusion proteins and single-chain insulins bearing various B-chain mutations. J. Biol. Chem. 2003;278:3687–3693. doi: 10.1074/jbc.M209474200. [DOI] [PubMed] [Google Scholar]
- 22.Kaarsholm NC, et al. Comparison of solution structural flexibility and zinc binding domains for insulin, proinsulin, and miniproinsulin. Biochemistry (Mosc) 1989;28:4427–4435. doi: 10.1021/bi00436a046. [DOI] [PubMed] [Google Scholar]
- 23.Sohma Y, Kent SB. Biomimetic synthesis of lispro insulin via a chemically synthesized “mini-proinsulin” prepared by oxime-forming ligation. J. Am. Chem. Soc. 2009;131:16313–16318. doi: 10.1021/ja9052398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajpal G, et al. Single-chain insulins as receptor agonists. Mol. Endocrinol. 2009;23:679–688. doi: 10.1210/me.2008-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheuner D, et al. Control of mRNA translation preserves reticulum function in beta cells and maintains glucose homeostasis. Nat. Med. 2005;11:757–764. doi: 10.1038/nm1259. [DOI] [PubMed] [Google Scholar]
- 26.Sieber P, et al. Synthesis and biological activity of two disulphide bond isomers of human insulin: [A7-A11,A6-B7-cystine]- and [A6-A7,A11-B7-cystine]insulin (human) Hoppe Seyler’s Z. Physiol. Chem. 1978;359:113–123. doi: 10.1515/bchm.1978.359.1.113. [DOI] [PubMed] [Google Scholar]
- 27.Hua QX, et al. A protein caught in a kinetic trap: structures and stabilities of insulin disulfide isomers. Biochemistry (Mosc) 2002;41:14700–14715. doi: 10.1021/bi0202981. [DOI] [PubMed] [Google Scholar]
- 28.Hua QX, et al. Structure of a protein in a kinetic trap. Nat. Struct. Biol. 1995;2:129–138. doi: 10.1038/nsb0295-129. [DOI] [PubMed] [Google Scholar]
- 29.Scheuner D, Kaufman RJ. The unfolded protein response: a pathway that links insulin demand with beta-cell failure and diabetes. Endocr. Rev. 2008;29:317–333. doi: 10.1210/er.2007-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fonseca SG, et al. Endoplasmic reticulum stress in beta-cells and development of diabetes. Curr Opin Pharmacol. 2009;9:763–770. doi: 10.1016/j.coph.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eizirik DL, et al. The role for endoplasmic reticulum stress in diabetes mellitus. Endocr. Rev. 2008;29:42–61. doi: 10.1210/er.2007-0015. [DOI] [PubMed] [Google Scholar]
- 32.Hidvegi T, et al. Accumulation of mutant alpha1-antitrypsin Z in the endoplasmic reticulum activates caspases-4 and -12, NFkappaB, and BAP31 but not the unfolded protein response. J. Biol. Chem. 2005;280:39002–39015. doi: 10.1074/jbc.M508652200. [DOI] [PubMed] [Google Scholar]
- 33.Hidvegi T, et al. Regulator of G Signaling 16 is a marker for the distinct endoplasmic reticulum stress state associated with aggregated mutant alpha1-antitrypsin Z in the classical form of alpha1-antitrypsin deficiency. J. Biol. Chem. 2007;282:27769–27780. doi: 10.1074/jbc.M704330200. [DOI] [PubMed] [Google Scholar]
- 34.Lipson KL, et al. Regulation of insulin biosynthesis in pancreatic beta cells by an endoplasmic reticulum-resident protein kinase IRE1. Cell Metab. 2006;4:245–254. doi: 10.1016/j.cmet.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 35.Lipson KL, et al. The role of IRE1alpha in the degradation of insulin mRNA in pancreatic beta-cells. PLoS ONE. 2008;3:e1648. doi: 10.1371/journal.pone.0001648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allen JR, et al. High ER stress in ß-cells stimulates intracellular degradation of misfolded insulin. Biochem. Biophys. Res. Commun. 2004;324:166–170. doi: 10.1016/j.bbrc.2004.09.035. [DOI] [PubMed] [Google Scholar]
- 37.Oyadomari S, et al. Endoplasmic reticulum stress-mediated apoptosis in pancreatic beta-cells. Apoptosis. 2002;7:335–345. doi: 10.1023/a:1016175429877. [DOI] [PubMed] [Google Scholar]
- 38.Marciniak SJ, Ron D. Endoplasmic reticulum stress signaling in disease. Physiol. Rev. 2006;86:1133–1149. doi: 10.1152/physrev.00015.2006. [DOI] [PubMed] [Google Scholar]
- 39.Lai E, et al. Endoplasmic reticulum stress: signaling the unfolded protein response. Physiology (Bethesda) 2007;22:193–201. doi: 10.1152/physiol.00050.2006. [DOI] [PubMed] [Google Scholar]
- 40.Garin I, et al. Recessive mutations in the INS gene result in neonatal diabetes through reduced insulin biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 2010;107:3105–3110. doi: 10.1073/pnas.0910533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vinik A, Bell G. Mutant insulin syndromes. Horm. Metab. Res. 1988;20:1–10. doi: 10.1055/s-2007-1010736. [DOI] [PubMed] [Google Scholar]
- 42.Nakashima N, Umeda F. Etiology and molecular biology of hyperproinsulinemia. Nippon Rinsho. 1994;52:2556–2561. [PubMed] [Google Scholar]
- 43.Steiner DF, et al. Lessons learned from molecular biology of insulin-gene mutations. Diabetes Care. 1990;13:600–609. doi: 10.2337/diacare.13.6.600. [DOI] [PubMed] [Google Scholar]
- 44.Stoy J, et al. Insulin gene mutations as a cause of permanent neonatal diabetes. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15040–15044. doi: 10.1073/pnas.0707291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edghill EL, et al. Insulin mutation screening in 1,044 patients with diabetes: mutations in the INS gene are a common cause of neonatal diabetes but a rare cause of diabetes diagnosed in childhood or adulthood. Diabetes. 2008;57:1034–1042. doi: 10.2337/db07-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Molven A, et al. Mutations in the insulin gene can cause MODY and autoantibody-negative type 1 diabetes. Diabetes. 2008;57:1131–1135. doi: 10.2337/db07-1467. [DOI] [PubMed] [Google Scholar]
- 47.Polak M, et al. Heterozygous missense mutations in the insulin gene are linked to permanent diabetes appearing in the neonatal period or in early infancy: a report from the French ND (Neonatal Diabetes) Study Group. Diabetes. 2008;57:1115–1119. doi: 10.2337/db07-1358. [DOI] [PubMed] [Google Scholar]
- 48.Colombo C, et al. Seven mutations in the human insulin gene linked to permanent neonatal/infancy-onset diabetes mellitus. J. Clin. Invest. 2008;118:2148–2156. doi: 10.1172/JCI33777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rubio-Cabezas O, et al. Testing for monogenic diabetes among children and adolescents with antibody-negative clinically defined Type 1 diabetes. Diabetic Medicine. 2009;26:1070–1074. doi: 10.1111/j.1464-5491.2009.02812.x. [DOI] [PubMed] [Google Scholar]
- 50.Ahamed A, et al. Permanent neonatal diabetes mellitus due to a C96Y heterozygous mutation in the insulin gene. A case report. JOP. 2008;9:715–718. [PubMed] [Google Scholar]
- 51.Boesgaard TW, et al. Further evidence that mutations in INS can be a rare cause of Maturity-Onset Diabetes of the Young (MODY) BMC Med Genet. 2010;11:42. doi: 10.1186/1471-2350-11-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Flechtner I, et al. Diabetes in very young children and mutations in the insulinsecreting cell potassium channel genes: therapeutic consequences. Endocr Dev. 2007;12:86–98. doi: 10.1159/000109636. [DOI] [PubMed] [Google Scholar]
- 53.Leroux L, et al. Compensatory responses in mice carrying a null mutation for Ins1 or Ins2. Diabetes. 2001;50(Suppl):S150–S153. doi: 10.2337/diabetes.50.2007.s150. [DOI] [PubMed] [Google Scholar]
- 54.Duvillie B, et al. Increased islet cell proliferation, decreased apoptosis, and greater vascularization leading to beta-cell hyperplasia in mutant mice lacking insulin. Endocrinology. 2002;143:1530–1537. doi: 10.1210/endo.143.4.8753. [DOI] [PubMed] [Google Scholar]
- 55.Rubio-Cabezas O, et al. Testing for monogenic diabetes among children and adolescents with antibody-negative clinically defined Type 1 diabetes. Diabet. Med. 2009;26:1070–1074. doi: 10.1111/j.1464-5491.2009.02812.x. [DOI] [PubMed] [Google Scholar]
- 56.Liu M, et al. Crystal structure of a “nonfoldable” insulin: impaired folding efficiency despite native activity. J. Biol. Chem. 2009;284:35259–35272. doi: 10.1074/jbc.M109.046888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.von Heijne G. Transcending the impenetrable: how proteins come to terms with membranes. Biochim. Biophys. Acta. 1988;947:307–333. doi: 10.1016/0304-4157(88)90013-5. [DOI] [PubMed] [Google Scholar]
- 58.Meur G, et al. Insulin gene mutations resulting in early-onset diabetes: marked differences in clinical presentation, metabolic status, and pathogenic effect through endoplasmic reticulum retention. Diabetes. 2010;59:653–661. doi: 10.2337/db09-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park SY, et al. Mutant proinsulin proteins associated with neonatal diabetes are retained in the endoplasmic reticulum and not efficiently secreted. Biochem. Biophys. Res. Commun. 2010;391:1449–1454. doi: 10.1016/j.bbrc.2009.12.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rajan S, et al. In vitro processing and secretion of mutant insulin proteins that cause permanent neonatal diabetes. Am J Physiol Endocrinol Metab. 2010;298:E403–410. doi: 10.1152/ajpendo.00592.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bonfanti R, et al. Insulin gene mutations as cause of diabetes in children negative for five type 1 diabetes autoantibodies. Diabetes Care. 2009;32:123–125. doi: 10.2337/dc08-0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hua QX, et al. A conserved histidine in insulin is required for the foldability of human proinsulin: structure and function of an ALAB5 analog. J. Biol. Chem. 2006;281:24889–24899. doi: 10.1074/jbc.M602617200. [DOI] [PubMed] [Google Scholar]
- 63.Nakagawa SH, Tager HS. Implications of invariant residue LeuB6 in insulin-receptor interactions. J. Biol. Chem. 1991;266:11502–11509. [PubMed] [Google Scholar]
- 64.Steiner DF, et al. A brief perspective on insulin production. Diabetes Obes Metab. 2009;11(Suppl 4):189–196. doi: 10.1111/j.1463-1326.2009.01106.x. [DOI] [PubMed] [Google Scholar]
- 65.Zoete V, Meuwly M. Importance of individual side chains for the stability of a protein fold: computational alanine scanning of the insulin monomer. J Comput Chem. 2006;27:1843–1857. doi: 10.1002/jcc.20512. [DOI] [PubMed] [Google Scholar]
- 66.Nakagawa SH, et al. Chiral mutagenesis of insulin. Contribution of the B20-B23 beta-turn to activity and stability. J. Biol. Chem. 2006;281:22386–22396. doi: 10.1074/jbc.M603547200. [DOI] [PubMed] [Google Scholar]
- 67.Yoshioka M, et al. A novel locus, Mody4, distal to D7Mit189 on chromosome 7 determines early-onset NIDDM in nonobese C57BL/6 (Akita) mutant mice. Diabetes. 1997;46:887–894. doi: 10.2337/diab.46.5.887. [DOI] [PubMed] [Google Scholar]
- 68.Kayo T, Koizumi A. Mapping of murine diabetogenic gene mody on chromosome 7 at D7Mit258 and its involvement in pancreatic islet and beta cell development during the perinatal period. J. Clin. Invest. 1998;101:2112–2118. doi: 10.1172/JCI1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang J, et al. A mutation in the insulin 2 gene induces diabetes with severe pancreatic beta-cell dysfunction in the Mody mouse. J. Clin. Invest. 1999;103:27–37. doi: 10.1172/JCI4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hodish I, et al. Misfolded proinsulin affects bystander proinsulin in neonatal diabetes. J. Biol. Chem. 2009 doi: 10.1074/jbc.M109.038042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gupta S, et al. PERK (EIF2AK3) regulates proinsulin trafficking and quality control in the secretory pathway. Diabetes. 2010 doi: 10.2337/db09-1064. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu M, et al. Proinsulin maturation, misfolding, and proteotoxicity. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15841–15846. doi: 10.1073/pnas.0702697104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Izumi T, et al. Dominant negative pathogenesis by mutant proinsulin in the Akita diabetic mouse. Diabetes. 2003;52:409–416. doi: 10.2337/diabetes.52.2.409. [DOI] [PubMed] [Google Scholar]
- 74.Oyadomari S, et al. Targeted disruption of the CHOP gene delays endoplasmic reticulum stress-mediated diabetes. J. Clin. Invest. 2002;109:525–532. doi: 10.1172/JCI14550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Song B, et al. Chop deletion reduces oxidative stress, improves beta cell function, and promotes cell survival in multiple mouse models of diabetes. J. Clin. Invest. 2008;118:3378–3389. doi: 10.1172/JCI34587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lu PD, et al. Cytoprotection by pre-emptive conditional phosphorylation of translation initiation factor 2. EMBO J. 2004;23:169–179. doi: 10.1038/sj.emboj.7600030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yamaguchi Y, et al. Endoplasmic reticulum (ER) chaperone regulation and survival of cells compensating for deficiency in the ER stress response kinase, PERK. J. Biol. Chem. 2008;283:17020–17029. doi: 10.1074/jbc.M802466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Harding HP, et al. Diabetes mellitus and exocrine pancreatic dysfunction in perk-/-mice reveals a role for translational control in secretory cell survival. Mol. Cell. 2001;7:1153–1163. doi: 10.1016/s1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- 79.Back SH, et al. Translation attenuation through eIF2alpha phosphorylation prevents oxidative stress and maintains the differentiated state in beta cells. Cell Metab. 2009;10:13–26. doi: 10.1016/j.cmet.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang W, et al. PERK EIF2AK3 control of pancreatic beta cell differentiation and proliferation is required for postnatal glucose homeostasis. Cell Metab. 2006;4:491–497. doi: 10.1016/j.cmet.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 81.Feng D, et al. Acute ablation of PERK results in ER dysfunctions followed by reduced insulin secretion and cell proliferation. BMC Cell Biol. 2009;10:61. doi: 10.1186/1471-2121-10-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Han D, et al. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009;138:562–575. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Seo HY, et al. Endoplasmic reticulum stress-induced activation of activating transcription factor 6 decreases insulin gene expression via up-regulation of orphan nuclear receptor small heterodimer partner. Endocrinology. 2008;149:3832–3841. doi: 10.1210/en.2008-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fonseca SG, et al. Wolfram syndrome 1 gene negatively regulates ER stress signaling in rodent and human cells. J. Clin. Invest. 2010;120:744–755. doi: 10.1172/JCI39678. [DOI] [PMC free article] [PubMed] [Google Scholar]