Abstract
The Vif protein of HIV is essential for the effective propagation of this pathogenic retrovirus in vivo. Vif acts by preventing virion encapsidation of two potent antiviral factors, the APOBEC3G and APOBEC3F cytidine deaminases. Decreased encapsidation in part involves Vif-mediated recruitment of a ubiquitin E3 ligase complex that promotes polyubiquitylation and proteasome-mediated degradation of APOBEC3G/F. The resultant decline in intracellular levels of these enzymes leads to decreased encapsidation of APOBECG/F into budding virions. This review discusses recent advances in our understanding of the dynamic interplay of Vif with the antiviral APOBEC3 enzymes.
Keywords: HIV-1, Vif, APOBEC3G, APOBEC3F, non-structural protein
1) Introduction
Human immunodeficiency virus-1 (HIV-1) encodes four accessory proteins—Vif, Vpu, Vpr, and Nef. Early studies indicated that these accessory proteins are not always required for viral replication in cell cultures, but each is important for the success of natural infections. These accessory proteins often function by modulating host immune responses, including countering the intrinsic antiviral effects of host restriction factors (Bieniasz, 2004; Malim and Emerman, 2008). Vif, a cytoplasmic, 23-kDa basic phosphoprotein encoded during late stages of the HIV-1 life cycle, is a notable example. Vif is conserved among all lentiviruses except equine infectious anemia virus, suggesting a prominent role in the life cycle of these retroviruses. Remarkably, its precise function remained mysterious for many years.
2) Early Vif observations
Soon after the discovery of HIV-1, it became apparent that Vif (viral infectivity factor) is required for HIV replication in some but not all cell types (Fisher et al., 1987; Gallo et al., 1988; Sodroski et al., 1986; Strebel et al., 1987). Specifically, HIV-1 virions lacking Vif (ΔVif HIV-1) can only spread in so-called “permissive” adherent cell cultures (e.g., HeLa and 293T) and various leukemic T-cell lines (e.g., CEM-SS and SupT1); they fail to spread in “nonpermissive” cells that include physiologically relevant primary CD4 T lymphocytes and macrophages, as well as various other T cell lines like CEM, HUT78 and H9. Of note, ΔVif virions are produced in normal numbers by these nonpermissive cells but the ability of these virions to productively infect the next target cell is greatly compromised (Courcoul et al., 1995; Fan and Peden, 1992; Fouchier et al., 1996; Gaddis et al., 2003; Goncalves et al., 1996; Hoglund et al., 1994; Simm et al., 1995; Simon and Malim, 1996; Simon et al., 1998b; Sova and Volsky, 1993; von Schwedler et al., 1993). Thus, Vif alters the quality but not quantity of HIV-1 virions produced by virus-infected CD4 T cells or macrophages (Gabuzda et al., 1992; Goncalves et al., 1996; Simon et al., 1998a; Simon and Malim, 1996; von Schwedler et al., 1993).
Why does replication of ΔVif HIV-1 in nonpermissive cell lines produces virions that cannot productively infect other cells? Two hypotheses have been offered: (1) the producer cells express an antiviral factor that Vif can restrict or (2) permissive cells express a proviral factor that can replace Vif and facilitate viral production. To determine which hypothesis was correct, heterokaryons were formed between permissive and nonpermissive cells and infected with ΔVif HIV-1. These heterokaryons yielded non-infectious virions, indicating that nonpermissive cells produce an HIV inhibitory factor whose action is circumvented by Vif (Madani and Kabat, 1998; Simon and Malim, 1996).
3) Discovery of A3G
Using a subtractive hybridization approach between the nonpermissive CEM cell line and the closely related but permissive cell line CEM-SS, Sheehy and colleagues identified this inhibitor as CEM-15 (Sheehy et al., 2002), now known as APOBEC3G (apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3G, or A3G), its expression in permissive cells is sufficient to render these cells nonpermissive. This finding showed that A3G can block HIV-1 replication in the absence of Vif (Fig. 1).
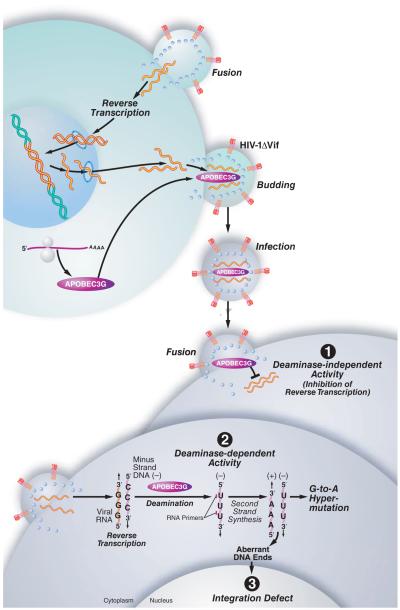
Figure 1. The impact of A3G on the lifecycle of ΔVif HIV-1.
In cells infected with ΔVif HIV-1, A3G is not degraded and is effectively incorporated into the budding virus and transferred to the next target cell, where it exerts antiviral effects at multiple levels. (1) A3G can inhibit the elongation of the reverse transcriptase in a deaminase-independent manner. Presumably, A3G binds directly to the viral RNA and thereby impairs the movement of the reverse transcriptase along the RNA template. (2) Most importantly, A3G can trigger massive deamination of dC to dU during synthesis of the viral minus strand. During synthesis of the DNA plus strand, adenosines are incorporated instead of the original guanines, resulting in G-to-A mutation. (3) Finally, A3G also inhibits viral DNA integration and provirus formation, probably by inducing defects in tRNA cleavage during plus-strand DNA transfer, leading to the formation of aberrant viral DNA ends. A3G also interacts with the integrase enzyme of HIV-1, which might interfere with the integrity of the integration complex, resulting in diminished integration rates. Figure was adapted from Chiu et al. 2008 with permission.
A3G belongs to a family of cytidine deaminases that includes seven family members (A3A–H), all located in a gene cluster on chromosome 22 (Conticello et al., 2005; Jarmuz et al., 2002). Of note, mice contain only a single A3 gene located at a synthetic region on chromosome 15. Forty-five percent of the human genome consists of endogenous mobile genetic elements or retroelements, such as short interspersed nuclear elements or long interspersed nuclear elements, which pose a potential threat to genome integrity. It is thought that the human A3 family arose by gene duplication by unequal crossover due to strong evolutionary pressure to control these elements (Conticello et al., 2003; Sawyer et al., 2004). A3G and A3F are best known for their antiviral activity against HIV-1 (Chiu and Greene, 2008), although A3A, A3B, A3C, A3DE, and A3H can also defend against HIV-1 to some extent (discussed below).
The common characteristic of all these enzymes is the presence of one (A3A, A3C, A3H) or two cytidine deaminase domains (CDAs) (A3B, A3DE, A3F, A3G) (Conticello et al., 2003; Jarmuz et al., 2002). Although the two CDAs are highly similar, they have different functions (Jarmuz et al., 2002). In primates, the N-terminal domain mediates RNA binding, whereas the C-terminal domain mediates sequence specific cytidine deamination of single-stranded DNA (Friew et al., 2009; Gooch and Cullen, 2008; Hache et al., 2005; Iwatani et al., 2006; Li et al., 2004; Navarro et al., 2005).
CDAs are characterized by a conserved zinc-binding motif (C/H)-X-E-X23~28-P-C-X2~4-C (Conticello et al., 2005; Jarmuz et al., 2002). This enzymatically active site catalyzes the hydrolytic deamination at the C4 position of 2′-deoxycytidine, resulting in a 2′-deoxyuridine. During the deamination reaction, the cysteines coordinate a single zinc ion, while the key glutamate is involved in proton shuttling (Betts et al., 1994).
Structural studies of the C-terminal CDA of A3G (Chen et al., 2008; Furukawa et al., 2009; Harjes et al., 2009; Holden et al., 2008; Zhang et al., 2007) and the single domain APOBEC2 protein (Prochnow et al., 2007) revealed that, the CDA consists of five β strands flanked by an α-helix on each side plus appropriate connecting loops.
4) Intravirion Packaging of A3G/F
In nonpermissive cells, inhibition of ΔVif HIV-1 by A3G only occurs during the next round of viral infection, indicating that A3G exerts its antiviral activity in the target cell. A3G is effectively incorporated into budding ΔVif virions, providing a potential mechanism for how A3G influences the infectivity of progeny virions in the next target cell (Gaddis et al., 2003; Sheehy et al., 2002; Suspene et al., 2004) (Fig.1). The amount of A3G molecules incorporated into the budding virus is proportional to its expression level in the virus-producing cell. Virus budded from activated peripheral blood mononuclear cells (PBMCs) contain approximately 7 (± 4) molecules of A3G (Xu et al., 2007); therefore, only a few A3G molecules are necessary to inhibit viral replication. For successful incorporation into ΔVif virions, A3G must interact with the N-terminal region of the nucleocapsid (NC) of Gag (Luo et al., 2004; Schafer et al., 2004; Svarovskaia et al., 2004; Zennou et al., 2004). HIV-1 virus-like particles that lack the nucleocapsid domain fail to package A3G (Schafer et al., 2004; Zennou et al., 2004).
A3G and NC have a strong propensity to bind RNA, which may serve an adaptor for binding, as co-immunoprecipitation of NC and A3G is RNase sensitive (Svarovskaia et al., 2004). Indeed, RNA binding is likely indispensable for the packaging of A3G into virions (Luo et al., 2004; Navarro et al., 2005; Schafer et al., 2004; Svarovskaia et al., 2004; Zennou et al., 2004). Mutations in the N-terminus (amino acids 124–127) of A3G impair its RNA binding properties, thus greatly compromising its encapsidation into budding virions. Such mutants of A3G function as very poor inhibitors of HIV-1 infection (Burnett and Spearman, 2007; Huthoff et al., 2009; Huthoff and Malim, 2007; Navarro et al., 2005). Therefore, A3G takes advantage of the fundamental property of retroviral RNA packaging to be incorporated into the virus (Zennou et al., 2004).
The nature of the RNA bound by A3G for encapsidation is unclear. Genomic HIV-1 RNA (Khan et al., 2007; Khan et al., 2005; Soros et al., 2007) was suggested, as well as 7SL RNA (a component of signal-recognition particles) (Wang et al., 2007). Recent results using an in vitro system showed that any single-stranded RNA (with G residues) can facilitate the formation of an A3G-RNA-NC complex (Bogerd and Cullen, 2008). Cellular A3G is mainly confined to high-molecular-mass (HMM) ribonucleoprotein complexes (Kreisberg et al., 2006; Stopak et al., 2003), which raises the question of how it is packaged into budding virions. Subsequent studies suggested that virion A3G is mainly recruited from the cellular pool of newly synthesized enzyme that is not yet assembled into HMM complexes (Soros et al., 2007). A second study suggested that “old” A3G was as efficiently packaged as “newly” synthesized A3G (Goila-Gaur et al., 2009); however, this study did not distinguish whether the old A3G emanated from a low or high molecular mass pool of A3G. A recent study showed that the N-terminal region of A3G including amino acids Y124 and W127 that are important for RNA binding, also facilitates lipid raft association. This finding reveals an interesting correlation between the ability of A3G to associate with lipid rafts and viral genomic RNA with its ability to be effectively encapsidated into budding virions (Khan et al., 2009).
5) Cytidine deaminase activity of A3G and A3F and inhibition of viral growth
Once incorporated into the virion, A3G is introduced into the next target cell as a result of virion fusion. Within the cell, the enzyme triggers massive deamination converting specific dC residues to dU during synthesis of the minus strand viral DNA (Suspene et al., 2004; Yu et al., 2004a) (Fig.1 point 2). During synthesis of the DNA plus strand, adenosines are incorporated instead of the original guanines, resulting in G-to-A mutation (Harris et al., 2003; Mangeat et al., 2003; Mariani et al., 2003; Zhang et al., 2003). During overexpression of A3G the level of these G-to-A mutations can exceed 10%. Accordingly, this phenomenon is often termed hypermutation. In vitro studies have shown that A3G displays a preference for deamination of 5′-CC (on the minus strand) (Bishop et al., 2004a; Harris et al., 2003; Suspene et al., 2004; Yu et al., 2004a), and up to 90% of sites containing the 5′-CCCA consensus sequence can be mutated (Yu et al., 2004a).
G-to-A mutations occur throughout the viral genome but in an unequal distribution (Kijak et al., 2008; Koulinska et al., 2003; Suspene et al., 2006; Suspene et al., 2004; Yu et al., 2004a). Most hypermutations are present in the envelope (Env) and Nef regions and decrease toward the 5′ UTR. This gradient of hypermutation is explained in part by the nature of the reverse transcription reaction. For HIV, this reaction begins with the binding of the tRNALys-3 to the primer-binding site of the viral RNA. The reverse transcriptase starts to synthesize minus-strand DNA, using the tRNALys-3 as a primer. The minus-strand strong-stop DNA is then transferred to 3′ of the viral RNA. RNaseH degrades the plus-strand RNA and restores the enzymatic activity of A3G, enabling it to act on the single-strand minus DNA and deaminate dC to dU. Plus-strand synthesis is initiated from two RNaseH-resistant polypurine tracts, cPPT and 3′PPT, which remain associated with the minus-strand cDNA to serve as initiation sites.
The chance that a cytidine on the minus-strand will be deaminated correlates strongly with the time during which these DNA regions remain single stranded (Chelico et al., 2006; Suspene et al., 2006; Yu et al., 2004a). Therefore, the specificity of A3G for single-stranded DNA accounts for the highly polarized (5′→3′) mutational twin gradients within the viral genome, each containing the greatest number of mutations just 5′ to the cPPT and 3′PPT (Suspene et al., 2006). This deamination gradient could be further reinforced by a proposed model in which A3G, slides predominantly in the 3′→5′ direction on its minus strand DNA template (Chelico et al., 2006). This could also contribute to A3G’s preference for deaminating the 3′ C in the consensus sequence (5′-CCCA-3′) (Chelico et al., 2006). However, a more recent study suggests that A3G does not translocate on the DNA in a positionally correlated fashion by sliding or microscopic jumping but instead translocates in an uncorrelated fashion by macroscopic jumping or intersegmental transfer (Nowarski et al., 2008).
In rare cases, C→T mutations are detected in the U3 region of the 5′UTR, which reflects the deamination of a cytidine on the single plus strand. Of note, the primer binding site becomes single stranded through the action of RNaseH when it degrades the tRNALys-3 bound at this site (Yu et al., 2004a).
A3G incorporated into virions resides in the viral core, bound in a ribonucleoprotein complex containing the viral genomic RNA as well as the viral NC, IN and Vpr proteins. A3G assembly with HIV RNA results in an inhibition of its intrinsic deaminase activity (Soros et al., 2007). How, then, is the virion-incorporated A3G ultimately activated so that it mediate mutation of the newly synthesized minus-strand DNA? It appears that the action of the viral RNase H during reverse transcription frees the enzyme from its RNA inhibitor, allowing full expression of its deaminase activity (Soros et al., 2007). These findings highlight a rather unexpected host-pathogen interaction in which the anti-HIV activity of A3G depends on its activation by an HIV enzyme.
Although A3G mediates hypermutation of the viral cDNA, it was also clear that wildtype A3G diminishes the accumulation of early and the late reverse transcription products in newly infected cells in a dose-dependent manner, with the greatest effect observed for late products (Anderson and Hope, 2008; Bishop et al., 2006; Guo et al., 2006; Holmes et al., 2007; Iwatani et al., 2007; Kaiser and Emerman, 2006; Luo et al., 2007; Mangeat et al., 2003; Mbisa et al., 2007). One possible explanation for this finding is that cellular host proteins degrade the hypermutated viral cDNA products. The resulting massive dU mutations could trigger the removal of the uracil in the nascent viral DNA by uracil-DNA-glycosylases (UNG2 and SMUG1), leading to abasic sites, and degradation by apurinic/apyrimidinic endonucleases (Schrofelbauer et al., 2005; Yang et al., 2007). However, more recent studies have shown that cells deficient in UNG2 (Kaiser and Emerman, 2006) or both UNG2 and SMUG1 (Langlois and Neuberger, 2008) do not rescue the accumulation of defective viral DNA.
Thus, it seems that A3G decreases HIV-1 infectivity by inducing hypermutation of the viral genome, where dU-containing nascent viral DNA serves as a template for plus-strand DNA synthesis, resulting in G-to-A mutations and viral error catastrophe, where viral proteins are truncated or rendered nonfunctional. How A3G induces a reduction in viral cDNA products remained an unanswered question.
6) A3G and A3F exert antiviral effects independent of their deaminase activity
Although the antiviral activity of A3 proteins is clearly linked to their cytidine deaminase activity (Mangeat et al., 2003; Shindo et al., 2003; Zhang et al., 2003) (Fig. 2, point 2), increasing evidence suggests that the A3G and A3F proteins also exert antiviral activity independently of cytidine deamination (Fig. 1, point 1). Specifically, mutagenesis of key amino acids at the center of the enzymatically active CDA2 prevents deaminase activity (Navarro et al., 2005; Newman et al., 2005), yet these mutants continue to moderately impair infectivity of HIV-1 and reduce the amount of HIV-1 cDNA produced in new target cells (Newman et al., 2005). Of note, hypermutation levels do not directly correlate with the extent of HIV restriction or the amount of viral cDNA products (Bishop et al., 2006; Langlois and Neuberger, 2008).
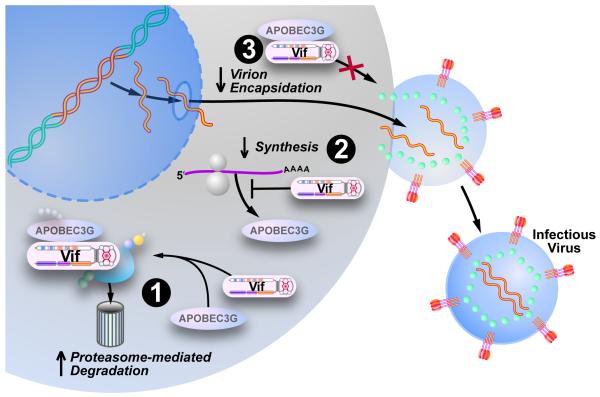
Figure 2. Vif diminishes the intracellular pool of A3G and A3F impairing virion encapsidation.
(1) Vif facilitates the degradation of the antiviral proteins A3G and A3F by hijacking the cellular ubiquitin-proteasome pathway. Vif connects the cellular E3 ubiquitin ligase with A3G/A3F, which initiates the polyubiquitination of the A3 proteins, followed by their degradation by the 26S proteasome. (2) Vif also impairs the translation of A3 mRNA and therefore reduces the cellular protein pool. (3) Finally, Vif may also interfere with the encapsidation of A3 proteins into the budding virus in the absence of A3G/F degradation. Adapted, with permission, from Chiu et al. 2008.
Many laboratories have investigated how this deaminase-independent antiviral activity of A3 proteins reduces the amount of viral cDNA. Various, sometimes contradictory, reports have emerged, suggesting that A3G can interfere with primer tRNA annealing, minus- and plus-strand DNA transfer, primer tRNA progression and removal, and DNA elongation (Anderson and Hope, 2008; Bishop et al., 2006; Guo et al., 2006; Guo et al., 2007; Guo et al., 2009; Holmes et al., 2007; Iwatani et al., 2007; Li et al., 2007; Luo et al., 2007; Mangeat et al., 2003; Mariani et al., 2003; Mbisa et al., 2007).
Recently, studies employing a cell-free reverse transcriptase assay, lacking any nucleases, showed that the reduced HIV-1 cDNA levels are due to A3G-mediated inhibition of RT elongation. This finding suggests that A3G can physically impair reverse transcription, further excluding the possibility that increased cDNA degradation accounts for the lower levels of HIV-1 cDNA (Bishop et al., 2008) (Fig. 1, point 1).
Expression of A3G also inhibits viral DNA integration and provirus formation, probably by inducing defects in tRNA cleavage during plus-strand DNA transfer, leading to the formation of aberrant viral DNA ends (Fig. 1, point 3). These aberrant structures presumably interfere with the chromosomal integration of the double-stranded viral DNA required for provirus formation. Of note, cytidine deaminase activity was necessary to inhibit provirus formation, although the mechanism remains unclear (Mbisa et al., 2007). Interestingly A3G, A3F, and noncatalytic A3G mutants interact with components of the HIV-1 preintegration complex, namely integrase. This interaction might interfere with the structural integrity of this complex, resulting in diminished nuclear import and a lower integration rate (Luo et al., 2007).
In general, most of the cytidine deaminase–independent actions of A3 proteins are dose-dependent, with the greatest effects occurring at higher levels of expression, which may exceed physiological concentrations of enzyme (Holmes et al., 2007; Miyagi et al., 2007; Schumacher et al., 2008). However, in the aforementioned in vitro RT study where a hypermutation-independent decrease in viral cDNA synthesis was observed, wildtype A3G was used at levels analogous to that found endogenously (Bishop et al., 2008).
Comparison of noncatalytic mutants of A3G and A3F revealed that both proteins can function as antiviral proteins in the absence of cytidine deaminase activity, although an important portion of their antiviral activity depends on this enzymatic activity Overall, the loss of deaminase activity appears to have a more pronounced effect on the antiviral activity of A3G than A3F (Holmes et al., 2007).
In addition to these described deaminase-independent antiviral activities of A3G, a post-entry restricting activity of low molecular mass A3G was reported in resting CD4 T cells (Chiu et al., 2005). These cells are normally refractory to HIV-1 infection due to a postentry block exerted at or shortly after reverse transcription (Stevenson et al., 1990; Zack et al., 1990). A role for A3G in this post-entry block was suggested by the finding that siRNA-mediated knockdown of A3G in resting CD4 T cells rendered these cells permissive to HIV infection. While a similar postentry restricting function for A3G has also been described in dendritc cells (Pion et al., 2006) and CCR6 expressing memory T cells (Lafferty et al., 2010), the initial results in resting T cells have proven difficult to reproduce (Kamata et al., 2009; Santoni de Sio and Trono, 2009). As such, the mechanisms underlying the nonpermissiveness of resting CD4 T cells to HIV infection remains unclear.
7) Vif-mediated degradation of A3G
A3G can effectively inhibit the spread of ΔVif HIV-1 but not wildtype HIV-1. A principal function of the Vif protein is to circumvent the action of A3G (Fig. 2). After the discovery of A3G, it was observed that Vif reduces A3G incorporation into the budding virion by ~99% (Kao et al., 2003; Mariani et al., 2003; Marin et al., 2003; Mehle et al., 2004b; Sheehy et al., 2003; Stopak et al., 2003), suggesting that Vif inhibits the packaging of A3G into virions. To achieve this, Vif could directly inhibit the encapsidation of A3G or deplete the cellular pool of A3G. While there is evidence for both mechanisms, the depletion of cellular A3G levels has been shown to be paramount. Vif accomplishes this by hijacking a cellular E3 ubiquitin ligase complex. Vif binds to A3G and simultaneously to an E3 ubiqutin ligase complex consisting of cullin5 (Cul5), elonginB, elonginC, and a Ring finger protein (Rbx). Rbx binds to an unknown, E2 ubiquitin-conjugating enzyme (E2). Vif therefore connects the ligase complex and its substrate A3G (Yu et al., 2003). Recruitment of the E3 ubiquitin ligase complex induces the polyubiquitination of A3G and directs it to the 26S proteasome for degradation (Conticello et al., 2003; Kobayashi et al., 2005; Marin et al., 2003; Mehle et al., 2004b; Sheehy et al., 2003; Stopak et al., 2003; Yu et al., 2003) (Fig. 2 point 1 and Fig. 3B).
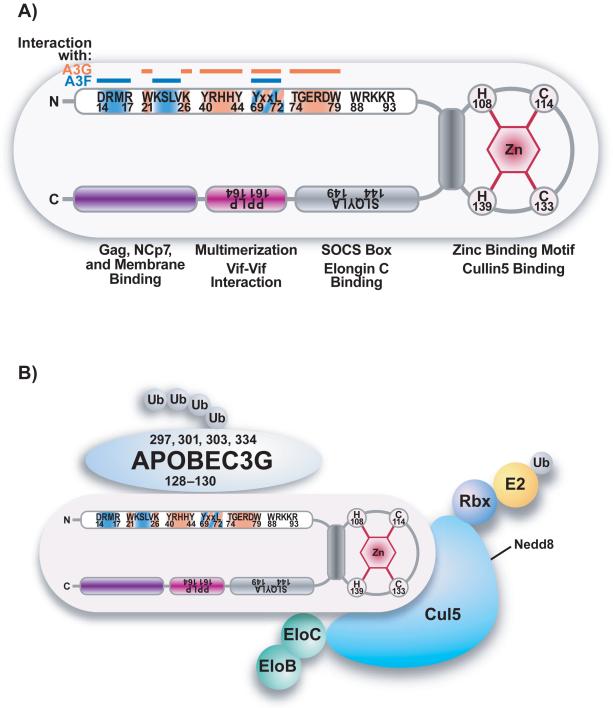
Figure 3. Summary of HIV-1 Vif domain structure and a model depicting the interaction of HIV-1 Vif, cullin5, elongin BC, and A3G.
(A) Schematic structure of the Vif domains. The N-terminal A3G binding sites are indicated in orange, A3F binding sites in blue. The structure of the H108x5Cx17-18Cx3-5H139 zinc finger motif that connects Vif to cullin5 is shown in gray as well as the viral BC-box (S144LQYLA149) that facilitates the binding to elongin C (EloC). Vif multimerization is facilitated by the hydrophobic amino acids P161PLP164 (purple). At the C-terminus the Gag, NC, and membrane binding domain is located (indicated in violet). B) Vif initiates the degradation of A3G by binding to the cytidine deaminase and simultaneously to an E3 ubiquitin ligase complex. This complex consists of cullin5 (Cul5), elonginB (EloB), elonginC (EloC), and a ring finger protein (Rbx). Rbx binds to an unknown E2 ubiquitin-conjugating enzyme (E2). Therefore Vif connects the ligase complex and its A3 protein substrates to initiate polyubiquitination and proteasome-mediated degradation of A3 proteins.
A recent report identified four critical lysine residues (K297, K301, K303, and K334) in the C-terminal region of A3G that are required for Vif-mediated degradation (Iwatani et al., 2009). Mutation of these residues restored the antiviral function of A3G in the presence of Vif (Iwatani et al., 2009). However, another study showed evidence for the polyubiquitination and degradation of a lysine-free A3G in the presence of HIV-1 Vif (Shao et al, 2010). Other data suggest that the polyubiquitination of Vif is required for A3G degradation, rather than A3G polyubiquitination (Dang et al., 2008b). Therefore, it remains unresolved which residues in A3G, if any, are important for A3G polyubiquitination and proteasomal degradation.
HIV-1 Vif binds the E3 ubiquitin ligase complex through at least two interaction sites: it binds to the elongin C protein through its SOCS box, a S144LQYLA149 motif, (Mehle et al., 2004a; Yu et al., 2004b), and to cullin5 through a zinc-binding H108x5Cx17-18Cx3-5H139 motif (Luo et al., 2005; Mehle et al., 2006) (Fig. 3B). Interestingly, the SOCS box of Vif is the most conserved region in different Vif proteins (Oberste and Gonda, 1992), indicating that this motif is crucial for the function of Vif. In addition, post-translational modifications may influence Vif’s function. Vif’s binding to elonginC can be negatively regulated by the phosphorylation of S144 in the SOCS box motif (Mehle et al., 2004a). An S144A mutation that prevents phosphorylation leads to the production of virions with poor infectivity, although this mutant effectively depletes A3G.
The integrity of the zinc-binding motif is also indispensable for Vif function. Any mutations that alter the arrangement of the zinc-binding domain or the spacing within the HCCH motif markedly compromise the ability of Vif to recruit A3G to the E3 ubiquitin ligase (Paul et al., 2006; Xiao et al., 2006). The binding of zinc to the HCCH leads to a conformational change in Vif, rendering it capable of forming high-order protein assemblies, probably by altering the protein conformation to expose protein-protein interaction sites (Giri and Maynard, 2009; Paul et al., 2006). Therefore, chelation of zinc by a cell-permeable zinc chelator inhibits Vif’s function in infectivity assays, allowing the virus to become sensitive to the antiviral activity of A3G (Xiao et al., 2007).
8) Vif facilitates HIV-1 in addition to promoting the degradation of APOBEC3
Several mechanisms have been proposed to explain how Vif increases the viral infectivity of HIV-1 besides degrading A3 proteins. For instance, Vif can deplete intracellular A3G by impairing the translation of its mRNA (Kao et al., 2003; Mariani et al., 2003; Stopak et al., 2003). Vif achieves this by binding to the 3′UTR and the 5′UTR of the A3G mRNA (Mercenne et al., 2010) (Fig. 2, point 2).
Vif also directly prevents the encapsidation of A3G, as shown by Vif’s ability to inhibit the packaging of the degradation-resistant A3G mutant C97A (Opi et al., 2007) and by the greater reduction of A3G levels in the virion than in the cell (Kao et al., 2007; Mariani et al., 2003; Schrofelbauer et al., 2004) (Fig. 2, point 3). These observations could indicate that Vif competes for the A3G binding site on viral genomic RNA or components of Gag, both of which are needed for A3G encapsidation (Kao et al., 2004; Mariani et al., 2003; Opi et al., 2007).
Furthermore, both Vif and Vpr, another HIV-1 accessory protein, can induce G2 cell-cycle delay by recruiting the same E3 ubiqutin ligase complex that targets A3 proteins for degradation (DeHart et al., 2008; Sakai et al., 2006). To add even more complexity to its mode of action, Vif also induces the degradation of Vpr (Wang et al., 2008) and therefore alters Vpr-induced G2 arrest in HIV-1-infected cells. These findings suggest that Vif alters the activity of A3G by means beyond its degradation in 26S proteasome, although this remains the dominant mechanism by which Vif counters the action of A3G.
9) Interaction between Vif, A3G, and A3F
Because it is difficult to express soluble full-length A3G or Vif at high levels in prokaryotic cells or insect cells, data on the three-dimensional structure of the A3G–Vif interaction are not yet available (Auclair et al., 2007; Iwatani et al., 2006; Reingewertz et al., 2009; Stanley et al., 2008). However, considerable information has emerged from the analysis of mutants in Vif-A3G interaction assays (Fig. 3).
The interaction between A3G and Vif is critically dependent on amino acids 128–130 in A3G (Huthoff and Malim, 2007; Russell et al., 2009b) (Fig. 3B). Their importance is shown by the fact that the species specificity of HIV/SIV Vif is determined by D128 in A3G. More precisely, the African green monkey (agm) A3G protein contains a lysine (K) in lieu of aspartic acid (D) at position 128 and is not degraded by HIV-1 Vif. Substitution of D for K in agm A3G renders the enzyme sensitive to HIV-1 Vif, while mutation of D to K in human A3G makes it resistant to HIV-1 Vif but sensitive to SIV-1 Vif (Bogerd et al., 2004; Mangeat et al., 2004; Schrofelbauer et al., 2004; Xu et al., 2004). The corresponding sequence in Vif that likely interacts with the D128 region of A3G is D14RMR17. Substitution of Vif D14RMR17 with S14ERQ17 or S14EMQ17, the sequence found in agm Vif, allows the mutant HIV-1 Vif to degrade agm A3G but not human A3G. This species specificity was attributed to two positively charged amino acids (15 and 17) in Vif and their interaction with negatively charged D128 in human A3G (Schrofelbauer et al., 2006). Another region in Vif that is essential for the degradation of A3G is facilitated by amino acids 40–44 (Y40RHHY44) (Russell and Pathak, 2007; Yamashita et al., 2008). Of note, co-immunopreciptation experiments demonstrated that the Y40RHHY44 domain is actually the region necessary for the binding of Vif to A3G, while the D14RMR17 domain is more involved in a secondary step necessary for A3G degradation (Russell and Pathak, 2007). Indeed, Vif can bind the D128K A3G mutant but cannot facilitate its degradation (Russell and Pathak, 2007; Xu et al., 2004). Another region important for A3G binding to Vif is the amino acid stretch between 82 and 99 (Santa-Marta et al., 2005).
Other A3 family members besides A3G are also active against HIV-1, as will be discussed below. The most important is A3F, which is expressed in the natural target cells of HIV-1, but is produced at lower levels than A3G and is a less potent inhibitor of HIV-1 (Koning et al., 2009; Liddament et al., 2004; Simon et al., 2005; Xu et al., 2004; Zennou and Bieniasz, 2006). Vif can also degrade A3F, but with lower efficiency (Liddament et al., 2004; Zennou and Bieniasz, 2006).
Vif binds to A3F via the D14RMR17 domain, but not the Y40RHHY44 domain, which is essential for A3G degradation. Therefore Vif has two different binding sites—the Y40RHHY44 site, which is critical for A3G binding, and the D14RMR17 site, which is critical for A3F binding (Russell and Pathak, 2007). Interestingly, mutations in the A3G binding domain of Vif increase its sensitivity to A3F. Hence, it seems that Vif evolved to target the more potent A3G, even though Vif can reduce A3F with lower efficiency (Russell et al., 2009b) (Fig. 3A).
Other amino acids involved in the binding of Vif to A3F, but not to A3G, are W11, Q12, 74-76, and W79 (He et al., 2008; Russell and Pathak, 2007; Simon et al., 2005; Tian et al., 2006). In contrast, I9, K22, E45, Y40, and N48 are critical for Vif’s ability to degrade A3G, but not A3F (Russell and Pathak, 2007; Simon et al., 2005; Wichroski et al., 2005). The amino acids Y69xxl72 (He et al., 2008; Pery et al., 2009; Yamashita et al., 2008) and the residues W5, W21, W38, N48, and W89 are needed to degrade A3G as well as A3F (Tian et al., 2006). Recently two groups identified W21KSLVK26 as a novel motif in Vif critical for neutralizing A3G and A3F (Chen et al., 2009; Dang et al., 2009) with K22 and K26 playing especially important roles in this neutralization. However, these groups disagreed about the importance of S23LV25 residues for neutralization of A3F and A3G. One group identified SLV as important for both A3G and A3F neutralization (Dang et al., 2009), while the other found only L24 to be important for both A3G and A3F neutralization, and V25 important for the degradation of only A3F (Chen et al., 2009) (Fig. 3A). Recently two additional domains in Vif, L81GxGxSIEW89 and E171DRW174, were shown to be important for Vif neutralization of A3G and A3F. In the L81GxGxSIEW89 domain, residues S86IEW89 are involved in Vif binding to A3G, A3F and Cul5; residue G84 is important for Vif binding to both A3G and A3F; and residues L81, G82 are involved in Vif binding to A3F. The E171DRW174 domain is critical for Vif neutralization of A3F (Dang et al., 2010).
A3G and A3F also bind to different regions of Vif. A3G binds to the amino acid stretch 128–130 (Huthoff and Malim, 2007; Russell et al., 2009b), whereas A3F binds to amino acids 283–300 (Russell et al., 2009b).
Two additional domains are important for the steady-state levels of Vif and for interactions with tyrosine kinases—the E88WRKKR93 site (Fujita et al., 2004; Fujita et al., 2003) and the proline-rich P161PLP164 rich region (Donahue et al., 2008) (Fig. 3A). Mutating these sites results in reduced Vif expression (Fujita et al., 2004; Fujita et al., 2003), diminished Vif-Vif multimerization (Yang et al., 2003; Yang et al., 2001), loss of viral infectivity (Yang et al., 2001), and reduced binding to A3G (Donahue et al., 2008) and HIV-1 RT (Kataropoulou et al., 2009). Interestingly, Vif containing mutations or deletions in the PPLP motif (and in the HCCH and SOCS-box motifs) functions as a dominant negative inhibiting wild-type Vif from excluding A3G from virion encapsidation. Such dominant negative versions of Vif decrease virion infectivity (Walker et al. 2010).
10) Other A3 proteins
As noted above, the A3 family is comprised of seven members, A3A–H (Conticello et al., 2005). A3G and A3F have been the most intensively studied members due to their ability to restrict HIV spread. These proteins are over 50% identical, and both use the same conserved amino acids to bind RNA and to facilitate cytidine deamination. Like A3G, A3F acts on the minus-strand DNA, resulting in G-to-A mutation on the plus strand. However, unlike A3G, whose target sequence is CC, the target sequence of A3F is TC. A3F is expressed in the same cells as A3G, but at lower levels, and it is less potent than A3G (Bishop et al., 2004a; Liddament et al., 2004; Wiegand et al., 2004; Zennou and Bieniasz, 2006; Zheng et al., 2004). Nevertheless, G-to-A mutations induced by A3F can be easily identified in HIV-1 isolates from infected individuals (Simon et al., 2005). HIV-1 Vif degrades A3F and A3G by similar mechanisms, although the binding sites for these two proteins differ, and A3F is less sensitive to Vif-mediated proteasomal degradation than A3G (Liddament et al., 2004).
A3B has a moderate activity against HIV-1. A3B is highly similar to A3G and A3F, and like A3G and A3F, it contains two CDAs. It can also bind to the nucleocapsid of HIV-1 Gag and therefore is packaged into the budding virus. Unlike A3G and A3F, A3B is resistant to Vif-induced degradation. However, A3B is expressed at extremely low levels in the natural target cells of HIV-1 and thus is unlikely to exert antiviral effects against HIV-1 under normal conditions (Bishop et al., 2004b; Bogerd et al., 2007; Doehle et al., 2005).
A3DE corresponds to another A3 protein with two CDA domains. It displays moderate activity against HIV-1, but Vif can facilitate its degradation through the 26S proteasome. In the absence of Vif, A3DE is encapsidated into virions and can deaminate CG on the minus-strand DNA, resulting in GC-to-AC mutations on the viral plus strand (Dang et al., 2006).
A3 protein family members containing only a single CDA domain (A3A, A3C, and A3H) may also impair HIV-1. A3C is expressed in CD4 T cells and can introduce limited G-to-A mutations (Bourara et al., 2007). A3A is active against HIV-1 and is expressed at higher levels in monocytes than in macrophages (Peng et al., 2007). It was suspected that A3A is partly responsible for the relative resistance of monocytes to HIV-1 infection, as silencing of A3A increases their susceptibility to infection (Peng et al., 2007). A3H was thought to be unstable due to two different mutations (Dang et al., 2008a; OhAinle et al., 2008; OhAinle et al., 2006). However, the re-mutated stable form of A3H can restrict HIV-1 up to 150-fold. Whether A3H is associated with G-to-A hypermutation is still controversial (Dang et al., 2008a; Harari et al., 2009). Of note, a haplotype of A3H, hapII-RDD, is stably expressed in cells and is active against HIV-1. Although this haplotype is common in people of African descent (Harari et al., 2009), stable endogenous expression could not be shown in PBMCs, the natural target cells of HIV-1 (Li et al., 2009).
11) Intravirion encapsidation of Vif
Vif is an RNA-binding protein, but it binds RNA in a relatively nonspecific manner. This is illustrated by its effective binding to homopolymeric RNA (Zhang et al., 2000). Importantly, Vif interacts with HIV-1 genomic RNA in the cytoplasm of infected cells (Dettenhofer and Yu, 1999; Khan et al., 2001; Zhang et al., 2000). In this case, Vif binds in a cooperative manner to the 5′-untranslated region of HIV-1 RNA and Gag (Bernacchi et al., 2007). More precisely, it binds with strong affinity to the apical loop of TAR and to a short region in Gag and with lower affinity to the neighboring primer-binding site and the whole leader region (Bernacchi et al., 2007; Henriet et al., 2005). The fact that Vif binds with different affinities to two sites suggests that when Vif is expressed at high levels, it will bind to the whole 5′ UTR region, leading to effective encapsidation into budding virions. Conversely, when Vif is expressed at low levels in the virus-producing cell, only the high-affinity binding sites will be engaged and much less Vif will be present in the virion. Thus, intravirion Vif levels correlate with intracellular expression levels (Simon et al., 1998b).
Vif also interacts with Pr55gag (Simon et al., 1999). Vif was found in a complex together with Gag and the Gag-Pol precursor (Zimmerman et al., 2002). Therefore, Vif’s encapsidation into the virus might be mediated by an interaction with genomic RNA and partly by interaction with Gag and Gag-Pol (Bardy et al., 2001; Khan et al., 2001). In addition, a recent report suggests that virion encapsidation of Vif is dependent on Vif binding to A3G and A3F (Yamashita et al. 2010).
It is not entirely clear whether Vif is encapsidated into virions under normal conditions and if so in what amounts. An initial report suggested that 60–100 copies of Vif are incorporated per virion (Liu et al., 1995). Later studies lowered the amount to 20 copies or even none (Camaur and Trono, 1996; Dettenhofer and Yu, 1999; Simon et al., 1998b). These discrepancies were attributed to contamination during the virus preparation with Vif-containing microvesicles or secreted proteins and to nonspecific encapsidation due to overexpression of Vif (which overestimates copy number) or degradation of virion-incorporated Vif (which underestimates copy number), especially as virus-associated Vif is unstable (Henriet et al., 2009; Kao et al., 2003).
12) Naturally occurring variations of Vif, Cullin5, and A3G/F affect the fitness of HIV-1
The finding that ΔVif HIV-1 cannot effectively spread in cultures of CD4 T cells—a natural target of HIV-1 in vivo—has been principally attributed to the antiviral action of A3G and A3F proteins. Overexpression of A3G can modestly suppress wildtype HIV-1 (Mangeat et al., 2003; Sheehy et al., 2002; Zhang et al., 2003), probably by overwhelming the intracellular Vif block. These facts make the Vif-APOBEC3 interaction a worthwhile pharmacological target. Although current antiretroviral drugs effectively suppress viral replication and entry, this may not be true of viral escape mutants. Therefore, explorations of new drug targets are constantly needed.
G-to-A hypermutations were first recognized in the env gene region during propagation of HIV-1 in vitro (Vartanian et al., 1991). Periodic G-to-A hypermutations are easily detectable in clinical samples from HIV-1 infected subjects and can account for >9% of the integrated viral DNA in cells (Gandhi et al., 2008; Kieffer et al., 2005; Land et al., 2008; Simon et al., 2005). Hypermutated virus could not be found in patient plasma, indicating that A3G/F function in vivo gives rise to hypermutated viral genomes that can be integrated but do not produce progeny viruses (Kieffer et al., 2005). Infection studies with ΔVif HIV-1 in CEM cells (which express A3G) and CEM-SS cells (which do not express A3G) show high levels of G-to-A mutations in viral DNA, as expected. However, significantly fewer mutations were detected in intracellular viral RNA, and even fewer in virion RNA (Russell et al., 2009a).
Genetic polymorphisms in A3G, A3F, or cullin5 and natural variations in their levels of expression could also affect HIV-1 pathogenesis. Indeed, in HIV-1-infected patients, A3G mRNA levels correlate inversely with HIV-1 viral loads and positively with CD4 cell counts (Jin et al., 2005), A3G mRNA levels are higher in long-term nonprogressors than in uninfected controls, and the lowest A3G mRNA levels are found in HIV progressors (Jin et al., 2005). Another study has described a significant increase in A3G expression in PBMCs from HIV-1-exposed seronegative individuals compared to HIV-1-seropositive patients or healthy controls (Biasin et al., 2007).
Specific single nucleotide polymorphisms (SNPs) in A3G and cullin5 can affect the rapidity of HIV-1 disease progression. An SNP in A3G (H186R), which is highly abundant in African Americans (37%) but rare in Europeans or European Americans (5%, <3%), is strongly associated with a more rapid decline in the number of CD4 T cells and accelerated progression to AIDS (An et al., 2004; Reddy, 2010). This can also be seen with a codon-changing variant in the fourth exon of A3G and a 3′ extragenetic mutation (Reddy et al., 2010). The cullin5 gene exhibits several different SNPs, which can be classified into two clusters or haplotypes. Interestingly, the two haplotypes produce opposing effects on CD4 T-cell counts during HIV-1 infection; the loss of these cells was delayed by cluster I and accelerated by cluster II. Cullin5 polymorphisms linked to accelerated CD4 T cell decline are associated with increased HIV-1 viral load (An et al., 2007). These findings raise the possibility that the Vif–cullin5 interface might represent a tractable target for small molecule development
Other studies indicate that A3G/F hypermutation of HIV-1 is associated with increased CD4 T-cells count (Land et al., 2008) and a 0.7 log10 reduction in viremia (Pace et al., 2006). However, some studies have not found a correlation between A3G/F mRNA levels or polymorphisms and viral load and/or CD4 T-cell counts (Cho et al., 2006; Do et al., 2005; Gandhi et al., 2008; Piantadosi et al., 2009; Reddy, 2010).
Interestingly, defective Vif alleles that cannot effectively neutralize A3G/F are readily detected in infected patients (Simon et al., 2005). Unopposed but nonlethal A3G/F action could promote viral sequence diversification in these patients. This raises the question of whether these G-to-A mutations might actually be beneficial for the virus, for example by promoting HIV-1 drug escape mutants or using A3G-induced mutations as a means to achieve rapid viral evolution.
Low-level mutation and single nucleotide variations of HIV-1 caused by A3G/F have been identified (Keele et al., 2008), and increased drug resistance to 3TC induced by suboptimal A3G concentrations has been reported (Mulder et al., 2008). Although A3G/F hypermutations can contribute to HIV-1 evolution, mutations introduced by HIV-1 reverse transcriptase are more likely to be important in the evolution of drug-resistance variants (Berkhout and de Ronde, 2004).
13) The Vif – A3G interaction as a pharmacological target
Recently, increased efforts were made to block the A3G interaction with Vif. For example, inhibition of the Vif–Vif interaction with peptides that interfere with the PPLP motif of Vif restored A3G encapsidation in nonpermissive cells and reduced HIV-1 infectivity (Miller et al., 2007; Yang et al., 2003). While peptides will not emerge as viable Vif antagonists, these studies supporting the notion that true Vif antagonists could be an exciting new class of antiviral drugs. The fact that the membrane-permeable zinc chelator (TPEN) prevents Vif function in nonpermissive cells by inhibiting cullin5 recruitment and A3G degradation (Xiao et al., 2007) provides yet an additional protein-protein interface for pharmaceutical targeting.
Without question, a potent, orally bioavailable small-molecule inhibitor of Vif would be an exciting medicinal lead. We and others have used different approaches to find an inhibitor of the interactions of Vif with A3G, cullin5, and elonginC (Cao et al., 2005; Nowotny et al., 2010)(W. Yonomoto, and W. C. Greene, unpublished data). Using YFP-tagged A3G and HIV-1 vectors with and without Vif, Rana and colleagues screened a library of 30,000 small molecules and identified 66 compounds, which were also scored in a secondary screen. The most promising was RN-18, which enhances Vif degradation in the presence of A3G, increases A3G virion encapsidation, and enhances hypermutation (Nathans et al., 2008). Recently, Cen et al. reported two compounds, IMB-26 and IMB-35, that protect A3G from Vif induced proteasomal degradation and recover A3G virion incorporation in the presence of Vif. These two compounds bind directly to A3G and thereby suppress the Vif/A3G interaction without inhibiting A3G’s catalytic activity (Cen et al., 2010).
Acknowledgments
We thank John Carroll for assistance with the graphics, Stephen Ordway for editorial assistance, and Robin Givens and Sue Cammack for administrative support. The research in our laboratory is supported by funding from the National Institutes of Health (1P01 AI083050-01 and R01 AI065329 to W.C.G.) and California Institute of Regenerative Medicine (TRI-01227 to W.C.G. and TG2/01160 to S.W.).
Biography
Vitae
Warner C. Greene, MD, PhD is Director of the Gladstone Institute of Virology and Immunology and the Nick and Sue Hellmann Distinguished Professor of Translational Medicine. In addition, he is Professor of Medicine, Microbiology and Immunology at the University of California, San Francisco. Dr. Greene also serves as President of the Accordia Global Health Foundation and Co-Director of the UCSF-GIVI Center for AIDS Research. The author of more than 330 publications, Dr. Greene’s research focuses on human retroviral pathogenesis.
Silke Wissing, PhD., was born in Emsdetten, Germany. She attended the University of Tuebingen, Germany, where she obtained a Diploma degree in Biochemistry in 2001. She then joined the group of Frank Madeo at the University of Tuebingen, where she worked on the molecular mechanisms of apoptosis in Saccharomyces cerevisiae and earned her PhD in 2005. She then joined the laboratory of Warner C. Greene at the Gladstone Institute of Virology and Immunology in San Francisco, USA, working as a postdoctoral researcher. Her current research focuses on the protective role of APOBEC3 proteins against exogenous and endogenous retroviruses.
Nicole L. K. Galloway, B.A., was born in Portland, Oregon. She attended Bowdoin College in Maine, where she earned her B.A. diploma with Honors in Biochemistry in 2005. Following graduation, she worked in the laboratory of Tom J. Hope at Northwestern University in Chicago, where she researched the early events of HIV-1 transmission. She currently is in graduate school at the University of California, San Francisco and joined Warner C. Greene’s Lab at the Gladstone Institute of Virology and Immunology. Here she focuses on the two HIV-1 non-structural proteins, Vif and Vpr.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- An P, Bleiber G, Duggal P, Nelson G, May M, Mangeat B, Alobwede I, Trono D, Vlahov D, Donfield S, et al. APOBEC3G genetic variants and their influence on the progression to AIDS. J Virol. 2004;78:11070–11076. doi: 10.1128/JVI.78.20.11070-11076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An P, Duggal P, Wang LH, O’Brien SJ, Donfield S, Goedert JJ, Phair J, Buchbinder S, Kirk GD, Winkler CA. Polymorphisms of CUL5 are associated with CD4+ T cell loss in HIV-1 infected individuals. PLoS Genet. 2007;3:e19. doi: 10.1371/journal.pgen.0030019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JL, Hope TJ. APOBEC3G restricts early HIV-1 replication in the cytoplasm of target cells. Virology. 2008;375:1–12. doi: 10.1016/j.virol.2008.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auclair JR, Green KM, Shandilya S, Evans JE, Somasundaran M, Schiffer CA. Mass spectrometry analysis of HIV-1 Vif reveals an increase in ordered structure upon oligomerization in regions necessary for viral infectivity. Proteins. 2007;69:270–284. doi: 10.1002/prot.21471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardy M, Gay B, Pebernard S, Chazal N, Courcoul M, Vigne R, Decroly E, Boulanger P. Interaction of human immunodeficiency virus type 1 Vif with Gag and Gag-Pol precursors: co-encapsidation and interference with viral protease-mediated Gag processing. J Gen Virol. 2001;82:2719–2733. doi: 10.1099/0022-1317-82-11-2719. [DOI] [PubMed] [Google Scholar]
- Berkhout B, de Ronde A. APOBEC3G versus reverse transcriptase in the generation of HIV-1 drug-resistance mutations. AIDS. 2004;18:1861–1863. doi: 10.1097/00002030-200409030-00022. [DOI] [PubMed] [Google Scholar]
- Bernacchi S, Henriet S, Dumas P, Paillart JC, Marquet R. RNA and DNA binding properties of HIV-1 Vif protein: a fluorescence study. J Biol Chem. 2007;282:26361–26368. doi: 10.1074/jbc.M703122200. [DOI] [PubMed] [Google Scholar]
- Betts L, Xiang S, Short SA, Wolfenden R, Carter CW., Jr. Cytidine deaminase. The 2.3 A crystal structure of an enzyme: transition-state analog complex. J Mol Biol. 1994;235:635–656. doi: 10.1006/jmbi.1994.1018. [DOI] [PubMed] [Google Scholar]
- Biasin M, Piacentini L, Lo Caputo S, Kanari Y, Magri G, Trabattoni D, Naddeo V, Lopalco L, Clivio A, Cesana E, et al. Apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3G: a possible role in the resistance to HIV of HIV-exposed seronegative individuals. J Infect Dis. 2007;195:960–964. doi: 10.1086/511988. [DOI] [PubMed] [Google Scholar]
- Bieniasz PD. Intrinsic immunity: a front-line defense against viral attack. Nat Immunol. 2004;5:1109–1115. doi: 10.1038/ni1125. [DOI] [PubMed] [Google Scholar]
- Bishop KN, Holmes RK, Malim MH. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J Virol. 2006;80:8450–8458. doi: 10.1128/JVI.00839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol. 2004a;14:1392–1396. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- Bishop KN, Holmes RK, Sheehy AM, Malim MH. APOBEC-mediated editing of viral RNA. Science. 2004b;305:645. doi: 10.1126/science.1100658. [DOI] [PubMed] [Google Scholar]
- Bishop KN, Verma M, Kim EY, Wolinsky SM, Malim MH. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 2008;4:e1000231. doi: 10.1371/journal.ppat.1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Cullen BR. Single-stranded RNA facilitates nucleocapsid: APOBEC3G complex formation. RNA. 2008;14:1228–1236. doi: 10.1261/rna.964708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Doehle BP, Wiegand HL, Cullen BR. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc Natl Acad Sci U S A. 2004;101:3770–3774. doi: 10.1073/pnas.0307713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Wiegand HL, Doehle BP, Cullen BR. The intrinsic antiretroviral factor APOBEC3B contains two enzymatically active cytidine deaminase domains. Virology. 2007;364:486–493. doi: 10.1016/j.virol.2007.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourara K, Liegler TJ, Grant RM. Target cell APOBEC3C can induce limited G-to-A mutation in HIV-1. PLoS Pathog. 2007;3:1477–1485. doi: 10.1371/journal.ppat.0030153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett A, Spearman P. APOBEC3G multimers are recruited to the plasma membrane for packaging into human immunodeficiency virus type 1 virus-like particles in an RNA-dependent process requiring the NC basic linker. J Virol. 2007;81:5000–5013. doi: 10.1128/JVI.02237-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camaur D, Trono D. Characterization of human immunodeficiency virus type 1 Vif particle incorporation. J Virol. 1996;70:6106–6111. doi: 10.1128/jvi.70.9.6106-6111.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Isaacson J, Patick AK, Blair WS. High-throughput human immunodeficiency virus type 1 (HIV-1) full replication assay that includes HIV-1 Vif as an antiviral target. Antimicrob Agents Chemother. 2005;49:3833–3841. doi: 10.1128/AAC.49.9.3833-3841.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cen S, Peng Z, Li X, Li Z, Ma J, Wang Y, Bo Fan X-FY, Wang Yu-Ping, Liu Fei, Shao Rong-Guang, Zhao Li-Xun, Yu Liyan, Jiang Jian-Dong. Small molecular inhibitors for HIV-1 replication through specifically stabilizing APOBEC3G. J Biol Chem. 2010 April 2; doi: 10.1074/jbc.M109.085308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelico L, Pham P, Calabrese P, Goodman MF. APOBEC3G DNA deaminase acts processively 3′ --> 5′ on single-stranded DNA. Nat Struct Mol Biol. 2006;13:392–399. doi: 10.1038/nsmb1086. [DOI] [PubMed] [Google Scholar]
- Chen G, He Z, Wang T, Xu R, Yu XF. A patch of positively charged amino acids surrounding the human immunodeficiency virus type 1 Vif SLVx4Yx9Y motif influences its interaction with APOBEC3G. J Virol. 2009;83:8674–8682. doi: 10.1128/JVI.00653-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KM, Harjes E, Gross PJ, Fahmy A, Lu Y, Shindo K, Harris RS, Matsuo H. Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature. 2008;452:116–119. doi: 10.1038/nature06638. [DOI] [PubMed] [Google Scholar]
- Chiu YL, Greene WC. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu Rev Immunol. 2008;26:317–353. doi: 10.1146/annurev.immunol.26.021607.090350. [DOI] [PubMed] [Google Scholar]
- Chiu YL, Soros VB, Kreisberg JF, Stopak K, Yonemoto W, Greene WC. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature. 2005;435:108–114. doi: 10.1038/nature03493. [DOI] [PubMed] [Google Scholar]
- Cho SJ, Drechsler H, Burke RC, Arens MQ, Powderly W, Davidson NO. APOBEC3F and APOBEC3G mRNA levels do not correlate with human immunodeficiency virus type 1 plasma viremia or CD4+ T-cell count. J Virol. 2006;80:2069–2072. doi: 10.1128/JVI.80.4.2069-2072.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conticello SG, Harris RS, Neuberger MS. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr Biol. 2003;13:2009–2013. doi: 10.1016/j.cub.2003.10.034. [DOI] [PubMed] [Google Scholar]
- Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol. 2005;22:367–377. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- Courcoul M, Patience C, Rey F, Blanc D, Harmache A, Sire J, Vigne R, Spire B. Peripheral blood mononuclear cells produce normal amounts of defective Vif- human immunodeficiency virus type 1 particles which are restricted for the preretrotranscription steps. J Virol. 1995;69:2068–2074. doi: 10.1128/jvi.69.4.2068-2074.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Davis RW, York IA, Zheng YH. Identification of 81LGxGxxIxW89 and 171EDRW174 domains from human immunodeficiency virus type 1 virus Vif that regulate APOBEC3G and APOBEC3F neutralizing activity. J Virol. 2010;84:5741–5750. doi: 10.1128/JVI.00079-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Siew LM, Wang X, Han Y, Lampen R, Zheng YH. Human cytidine deaminase APOBEC3H restricts HIV-1 replication. J Biol Chem. 2008a;283:11606–11614. doi: 10.1074/jbc.M707586200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Siew LM, Zheng YH. APOBEC3G is degraded by the proteasomal pathway in a Vif-dependent manner without being polyubiquitylated. J Biol Chem. 2008b;283:13124–13131. doi: 10.1074/jbc.M708728200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Wang X, Esselman WJ, Zheng YH. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J Virol. 2006;80:10522–10533. doi: 10.1128/JVI.01123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Wang X, Zhou T, York IA, Zheng YH. Identification of a novel WxSLVK motif in the N terminus of human immunodeficiency virus and simian immunodeficiency virus Vif that is critical for APOBEC3G and APOBEC3F neutralization. J Virol. 2009;83:8544–8552. doi: 10.1128/JVI.00651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeHart JL, Bosque A, Harris RS, Planelles V. Human immunodeficiency virus type 1 Vif induces cell cycle delay via recruitment of the same E3 ubiquitin ligase complex that targets APOBEC3 proteins for degradation. J Virol. 2008;82:9265–9272. doi: 10.1128/JVI.00377-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettenhofer M, Yu XF. Highly purified human immunodeficiency virus type 1 reveals a virtual absence of Vif in virions. J Virol. 1999;73:1460–1467. doi: 10.1128/jvi.73.2.1460-1467.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do H, Vasilescu A, Diop G, Hirtzig T, Heath SC, Coulonges C, Rappaport J, Therwath A, Lathrop M, Matsuda F, et al. Exhaustive genotyping of the CEM15 (APOBEC3G) gene and absence of association with AIDS progression in a French cohort. J Infect Dis. 2005;191:159–163. doi: 10.1086/426826. [DOI] [PubMed] [Google Scholar]
- Doehle BP, Schafer A, Cullen BR. Human APOBEC3B is a potent inhibitor of HIV-1 infectivity and is resistant to HIV-1 Vif. Virology. 2005;339:281–288. doi: 10.1016/j.virol.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Donahue JP, Vetter ML, Mukhtar NA, D’Aquila RT. The HIV-1 motif is necessary for human APOBEC3G binding and degradation. Virology. 2008;377:49–53. doi: 10.1016/j.virol.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Peden K. Cell-free transmission of Vif mutants of HIV-1. Virology. 1992;190:19–29. doi: 10.1016/0042-6822(92)91188-z. [DOI] [PubMed] [Google Scholar]
- Fisher AG, Ensoli B, Ivanoff L, Chamberlain M, Petteway S, Ratner L, Gallo RC, Wong-Staal F. The sor gene of HIV-1 is required for efficient virus transmission in vitro. Science. 1987;237:888–893. doi: 10.1126/science.3497453. [DOI] [PubMed] [Google Scholar]
- Fouchier RA, Simon JH, Jaffe AB, Malim MH. Human immunodeficiency virus type 1 Vif does not influence expression or virion incorporation of gag-, pol-, and envencoded proteins. J Virol. 1996;70:8263–8269. doi: 10.1128/jvi.70.12.8263-8269.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friew YN, Boyko V, Hu WS, Pathak VK. Intracellular interactions between APOBEC3G, RNA, and HIV-1 Gag: APOBEC3G multimerization is dependent on its association with RNA. Retrovirology. 2009;6:56. doi: 10.1186/1742-4690-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Akari H, Sakurai A, Yoshida A, Chiba T, Tanaka K, Strebel K, Adachi A. Expression of HIV-1 accessory protein Vif is controlled uniquely to be low and optimal by proteasome degradation. Microbes and Infection. 2004;6:791–798. doi: 10.1016/j.micinf.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Fujita M, Sakurai A, Yoshida A, Miyaura M, Koyama AH, Sakai K, Adachi A. Amino acid residues 88 and 89 in the central hydrophilic region of human immunodeficiency virus type 1 Vif are critical for viral infectivity by enhancing the steady-state expression of Vif. J Virol. 2003;77:1626–1632. doi: 10.1128/JVI.77.2.1626-1632.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa A, Nagata T, Matsugami A, Habu Y, Sugiyama R, Hayashi F, Kobayashi N, Yokoyama S, Takaku H, Katahira M. Structure and real-time monitoring of the enzymatic reaction of APOBEC3G which is involved in anti-HIV activity. Nucleic Acids Symp Ser. 2009:87–88. doi: 10.1093/nass/nrp044. [DOI] [PubMed] [Google Scholar]
- Gabuzda DH, Lawrence K, Langhoff E, Terwilliger E, Dorfman T, Haseltine WA, Sodroski J. Role of vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J Virol. 1992;66:6489–6495. doi: 10.1128/jvi.66.11.6489-6495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaddis NC, Chertova E, Sheehy AM, Henderson LE, Malim MH. Comprehensive investigation of the molecular defect in vif-deficient human immunodeficiency virus type 1 virions. J Virol. 2003;77:5810–5820. doi: 10.1128/JVI.77.10.5810-5820.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo R, Wong-Staal F, Montagnier L, Haseltine WA, Yoshida M. HIV/HTLV gene nomenclature. Nature. 1988;333:504. doi: 10.1038/333504a0. [DOI] [PubMed] [Google Scholar]
- Gandhi SK, Siliciano JD, Bailey JR, Siliciano RF, Blankson JN. Role of APOBEC3G/F-mediated hypermutation in the control of human immunodeficiency virus type 1 in elite suppressors. J Virol. 2008;82:3125–3130. doi: 10.1128/JVI.01533-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri K, Maynard EL. Conformational analysis of a peptide approximating the HCCH motif in HIV-1 Vif. Biopolymers. 2009;92:417–425. doi: 10.1002/bip.21209. [DOI] [PubMed] [Google Scholar]
- Goila-Gaur R, Khan MA, Miyagi E, Strebel K. Differential sensitivity of “old” versus “new” APOBEC3G to human immunodeficiency virus type 1 vif. J Virol. 2009;83:1156–1160. doi: 10.1128/JVI.01734-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves J, Korin Y, Zack J, Gabuzda D. Role of Vif in human immunodeficiency virus type 1 reverse transcription. J Virol. 1996;70:8701–8709. doi: 10.1128/jvi.70.12.8701-8709.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooch BD, Cullen BR. Functional domain organization of human APOBEC3G. Virology. 2008;379:118–124. doi: 10.1016/j.virol.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Cen S, Niu M, Saadatmand J, Kleiman L. Inhibition of formula-primed reverse transcription by human APOBEC3G during human immunodeficiency virus type 1 replication. J Virol. 2006;80:11710–11722. doi: 10.1128/JVI.01038-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Cen S, Niu M, Yang Y, Gorelick RJ, Kleiman L. The interaction of APOBEC3G with human immunodeficiency virus type 1 nucleocapsid inhibits tRNA3Lys annealing to viral RNA. J Virol. 2007;81:11322–11331. doi: 10.1128/JVI.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Saadatmand J, Niu M, Kleiman L. Roles of Gag and NCp7 in facilitating tRNA(Lys)(3) Annealing to viral RNA in human immunodeficiency virus type 1. J Virol. 2009;83:8099–8107. doi: 10.1128/JVI.00488-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hache G, Liddament MT, Harris RS. The retroviral hypermutation specificity of APOBEC3F and APOBEC3G is governed by the C-terminal DNA cytosine deaminase domain. J Biol Chem. 2005;280:10920–10924. doi: 10.1074/jbc.M500382200. [DOI] [PubMed] [Google Scholar]
- Harari A, Ooms M, Mulder LC, Simon V. Polymorphisms and splice variants influence the antiretroviral activity of human APOBEC3H. J Virol. 2009;83:295–303. doi: 10.1128/JVI.01665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harjes E, Gross PJ, Chen KM, Lu Y, Shindo K, Nowarski R, Gross JD, Kotler M, Harris RS, Matsuo H. An extended structure of the APOBEC3G catalytic domain suggests a unique holoenzyme model. J Mol Biol. 2009;389:819–832. doi: 10.1016/j.jmb.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- He Z, Zhang W, Chen G, Xu R, Yu XF. Characterization of conserved motifs in HIV-1 Vif required for APOBEC3G and APOBEC3F interaction. J Mol Biol. 2008;381:1000–1011. doi: 10.1016/j.jmb.2008.06.061. [DOI] [PubMed] [Google Scholar]
- Henriet S, Mercenne G, Bernacchi S, Paillart JC, Marquet R. Tumultuous relationship between the human immunodeficiency virus type 1 viral infectivity factor (Vif) and the human APOBEC-3G and APOBEC-3F restriction factors. Microbiol Mol Biol Rev. 2009;73:211–232. doi: 10.1128/MMBR.00040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriet S, Richer D, Bernacchi S, Decroly E, Vigne R, Ehresmann B, Ehresmann C, Paillart JC, Marquet R. Cooperative and specific binding of Vif to the 5′ region of HIV-1 genomic RNA. J Mol Biol. 2005;354:55–72. doi: 10.1016/j.jmb.2005.09.025. [DOI] [PubMed] [Google Scholar]
- Hoglund S, Ohagen A, Lawrence K, Gabuzda D. Role of vif during packing of the core of HIV-1. Virology. 1994;201:349–355. doi: 10.1006/viro.1994.1300. [DOI] [PubMed] [Google Scholar]
- Holden LG, Prochnow C, Chang YP, Bransteitter R, Chelico L, Sen U, Stevens RC, Goodman MF, Chen XS. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature. 2008;456:121–124. doi: 10.1038/nature07357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes RK, Koning FA, Bishop KN, Malim MH. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J Biol Chem. 2007;282:2587–2595. doi: 10.1074/jbc.M607298200. [DOI] [PubMed] [Google Scholar]
- Huthoff H, Autore F, Gallois-Montbrun S, Fraternali F, Malim MH. RNA-dependent oligomerization of APOBEC3G is required for restriction of HIV-1. PLoS Pathog. 2009;5:e1000330. doi: 10.1371/journal.ppat.1000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huthoff H, Malim MH. Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and Virion encapsidation. J Virol. 2007;81:3807–3815. doi: 10.1128/JVI.02795-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatani Y, Chan DS, Liu L, Yoshii H, Shibata J, Yamamoto N, Levin JG, Gronenborn AM, Sugiura W. HIV-1 Vif-mediated ubiquitination/degradation of APOBEC3G involves four critical lysine residues in its C-terminal domain. Proc Natl Acad Sci U S A. 2009;106:19539–19544. doi: 10.1073/pnas.0906652106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatani Y, Chan DS, Wang F, Maynard KS, Sugiura W, Gronenborn AM, Rouzina I, Williams MC, Musier-Forsyth K, Levin JG. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 2007;35:7096–7108. doi: 10.1093/nar/gkm750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatani Y, Takeuchi H, Strebel K, Levin JG. Biochemical activities of highly purified, catalytically active human APOBEC3G: correlation with antiviral effect. J Virol. 2006;80:5992–6002. doi: 10.1128/JVI.02680-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79:285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- Jin X, Brooks A, Chen H, Bennett R, Reichman R, Smith H. APOBEC3G/CEM15 (hA3G) mRNA levels associate inversely with human immunodeficiency virus viremia. J Virol. 2005;79:11513–11516. doi: 10.1128/JVI.79.17.11513-11516.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser SM, Emerman M. Uracil DNA glycosylase is dispensable for human immunodeficiency virus type 1 replication and does not contribute to the antiviral effects of the cytidine deaminase Apobec3G. J Virol. 2006;80:875–882. doi: 10.1128/JVI.80.2.875-882.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamata M, Nagaoka Y, Chen IS. Reassessing the role of APOBEC3G in human immunodeficiency virus type 1 infection of quiescent CD4+ T-cells. PLoS Pathog. 2009;5:e1000342. doi: 10.1371/journal.ppat.1000342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao S, Goila-Gaur R, Miyagi E, Khan MA, Opi S, Takeuchi H, Strebel K. Production of infectious virus and degradation of APOBEC3G are separable functional properties of human immunodeficiency virus type 1 Vif. Virology. 2007;369:329–339. doi: 10.1016/j.virol.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao S, Khan MA, Miyagi E, Plishka R, Buckler-White A, Strebel K. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J Virol. 2003;77:11398–11407. doi: 10.1128/JVI.77.21.11398-11407.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao S, Miyagi E, Khan MA, Takeuchi H, Opi S, Goila-Gaur R, Strebel K. Production of infectious human immunodeficiency virus type 1 does not require depletion of APOBEC3G from virus-producing cells. Retrovirology. 2004;1:27. doi: 10.1186/1742-4690-1-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataropoulou A, Bovolenta C, Belfiore A, Trabatti S, Garbelli A, Porcellini S, Lupo R, Maga G. Mutational analysis of the HIV-1 auxiliary protein Vif identifies independent domains important for the physical and functional interaction with HIV-1 reverse transcriptase. Nucleic Acids Res. 2009;37:3660–3669. doi: 10.1093/nar/gkp226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MA, Aberham C, Kao S, Akari H, Gorelick R, Bour S, Strebel K. Human immunodeficiency virus type 1 Vif protein is packaged into the nucleoprotein complex through an interaction with viral genomic RNA. J Virol. 2001;75:7252–7265. doi: 10.1128/JVI.75.16.7252-7265.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MA, Goila-Gaur R, Kao S, Miyagi E, Walker RC, Jr., Strebel K. Encapsidation of APOBEC3G into HIV-1 virions involves lipid raft association and does not correlate with APOBEC3G oligomerization. Retrovirology. 2009;6:99. doi: 10.1186/1742-4690-6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MA, Goila-Gaur R, Opi S, Miyagi E, Takeuchi H, Kao S, Strebel K. Analysis of the contribution of cellular and viral RNA to the packaging of APOBEC3G into HIV-1 virions. Retrovirology. 2007;4:48. doi: 10.1186/1742-4690-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MA, Kao S, Miyagi E, Takeuchi H, Goila-Gaur R, Opi S, Gipson CL, Parslow TG, Ly H, Strebel K. Viral RNA is required for the association of APOBEC3G with human immunodeficiency virus type 1 nucleoprotein complexes. J Virol. 2005;79:5870–5874. doi: 10.1128/JVI.79.9.5870-5874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer TL, Kwon P, Nettles RE, Han Y, Ray SC, Siliciano RF. G-->A hypermutation in protease and reverse transcriptase regions of human immunodeficiency virus type 1 residing in resting CD4+ T cells in vivo. J Virol. 2005;79:1975–1980. doi: 10.1128/JVI.79.3.1975-1980.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijak GH, Janini LM, Tovanabutra S, Sanders-Buell E, Arroyo MA, Robb ML, Michael NL, Birx DL, McCutchan FE. Variable contexts and levels of hypermutation in HIV-1 proviral genomes recovered from primary peripheral blood mononuclear cells. Virology. 2008;376:101–111. doi: 10.1016/j.virol.2008.03.017. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Takaori-Kondo A, Miyauchi Y, Iwai K, Uchiyama T. Ubiquitination of APOBEC3G by an HIV-1 Vif-Cullin5-Elongin B-Elongin C complex is essential for Vif function. J Biol Chem. 2005;280:18573–18578. doi: 10.1074/jbc.C500082200. [DOI] [PubMed] [Google Scholar]
- Koning FA, Newman EN, Kim EY, Kunstman KJ, Wolinsky SM, Malim MH. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J Virol. 2009;83:9474–9485. doi: 10.1128/JVI.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koulinska IN, Chaplin B, Mwakagile D, Essex M, Renjifo B. Hypermutation of HIV type 1 genomes isolated from infants soon after vertical infection. AIDS Res Hum Retroviruses. 2003;19:1115–1123. doi: 10.1089/088922203771881211. [DOI] [PubMed] [Google Scholar]
- Kreisberg JF, Yonemoto W, Greene WC. Endogenous factors enhance HIV infection of tissue naive CD4 T cells by stimulating high molecular mass APOBEC3G complex formation. J Exp Med. 2006;203:865–870. doi: 10.1084/jem.20051856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafferty MK, Sun L, DeMasi L, Lu W, Garzino-Demo A. CCR6 ligands inhibit HIV by inducing APOBEC3G. Blood. 2010;115:1564–1571. doi: 10.1182/blood-2009-06-226423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land AM, Ball TB, Luo M, Pilon R, Sandstrom P, Embree JE, Wachihi C, Kimani J, Plummer FA. Human immunodeficiency virus (HIV) type 1 proviral hypermutation correlates with CD4 count in HIV-infected women from Kenya. J Virol. 2008;82:8172–8182. doi: 10.1128/JVI.01115-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois MA, Neuberger MS. Human APOBEC3G can restrict retroviral infection in avian cells and acts independently of both UNG and SMUG1. J Virol. 2008;82:4660–4664. doi: 10.1128/JVI.02469-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Potash MJ, Volsky DJ. Functional domains of APOBEC3G required for antiviral activity. J Cell Biochem. 2004;92:560–572. doi: 10.1002/jcb.20082. [DOI] [PubMed] [Google Scholar]
- Li MM, Wu LI, Emerman M. The Range of Human APOBEC3H Sensitivity to Lentiviral Vif Proteins. J Virol. 2009;84:88–95. doi: 10.1128/JVI.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, Guo F, Zhang L, Kleiman L, Cen S. APOBEC3G inhibits DNA strand transfer during HIV-1 reverse transcription. J Biol Chem. 2007;282:32065–32074. doi: 10.1074/jbc.M703423200. [DOI] [PubMed] [Google Scholar]
- Liddament MT, Brown WL, Schumacher AJ, Harris RS. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr Biol. 2004;14:1385–1391. doi: 10.1016/j.cub.2004.06.050. [DOI] [PubMed] [Google Scholar]
- Liu H, Wu X, Newman M, Shaw GM, Hahn BH, Kappes JC. The Vif protein of human and simian immunodeficiency viruses is packaged into virions and associates with viral core structures. J Virol. 1995;69:7630–7638. doi: 10.1128/jvi.69.12.7630-7638.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K, Liu B, Xiao Z, Yu Y, Yu X, Gorelick R, Yu XF. Amino-terminal region of the human immunodeficiency virus type 1 nucleocapsid is required for human APOBEC3G packaging. J Virol. 2004;78:11841–11852. doi: 10.1128/JVI.78.21.11841-11852.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K, Wang T, Liu B, Tian C, Xiao Z, Kappes J, Yu XF. Cytidine deaminases APOBEC3G and APOBEC3F interact with human immunodeficiency virus type 1 integrase and inhibit proviral DNA formation. J Virol. 2007;81:7238–7248. doi: 10.1128/JVI.02584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K, Xiao Z, Ehrlich E, Yu Y, Liu B, Zheng S, Yu XF. Primate lentiviral virion infectivity factors are substrate receptors that assemble with cullin 5-E3 ligase through a HCCH motif to suppress APOBEC3G. Proc Natl Acad Sci U S A. 2005;102:11444–11449. doi: 10.1073/pnas.0502440102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madani N, Kabat D. An endogenous inhibitor of human immunodeficiency virus in human lymphocytes is overcome by the viral Vif protein. J Virol. 1998;72:10251–10255. doi: 10.1128/jvi.72.12.10251-10255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim MH, Emerman M. HIV-1 accessory proteins--ensuring viral survival in a hostile environment. Cell Host Microbe. 2008;3:388–398. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- Mangeat B, Turelli P, Liao S, Trono D. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J Biol Chem. 2004;279:14481–14483. doi: 10.1074/jbc.C400060200. [DOI] [PubMed] [Google Scholar]
- Mariani R, Chen D, Schrofelbauer B, Navarro F, Konig R, Bollman B, Munk C, Nymark-McMahon H, Landau NR. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell. 2003;114:21–31. doi: 10.1016/s0092-8674(03)00515-4. [DOI] [PubMed] [Google Scholar]
- Marin M, Rose KM, Kozak SL, Kabat D. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat Med. 2003;9:1398–1403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- Mbisa JL, Barr R, Thomas JA, Vandegraaff N, Dorweiler IJ, Svarovskaia ES, Brown WL, Mansky LM, Gorelick RJ, Harris RS, et al. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J Virol. 2007;81:7099–7110. doi: 10.1128/JVI.00272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehle A, Goncalves J, Santa-Marta M, McPike M, Gabuzda D. Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-Cul5 complex that promotes APOBEC3G degradation. Genes Dev. 2004a;18:2861–2866. doi: 10.1101/gad.1249904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehle A, Strack B, Ancuta P, Zhang C, McPike M, Gabuzda D. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J Biol Chem. 2004b;279:7792–7798. doi: 10.1074/jbc.M313093200. [DOI] [PubMed] [Google Scholar]
- Mehle A, Thomas ER, Rajendran KS, Gabuzda D. A zinc-binding region in Vif binds Cul5 and determines cullin selection. J Biol Chem. 2006;281:17259–17265. doi: 10.1074/jbc.M602413200. [DOI] [PubMed] [Google Scholar]
- Mercenne G, Bernacchi S, Richer D, Bec G, Henriet S, Paillart JC, Marquet R. HIV-1 Vif binds to APOBEC3G mRNA and inhibits its translation. Nucleic Acids Res. 2010;38:633–646. doi: 10.1093/nar/gkp1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH, Presnyak V, Smith HC. The dimerization domain of HIV-1 viral infectivity factor Vif is required to block virion incorporation of APOBEC3G. Retrovirology. 2007;4:81. doi: 10.1186/1742-4690-4-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagi E, Opi S, Takeuchi H, Khan M, Goila-Gaur R, Kao S, Strebel K. Enzymatically active APOBEC3G is required for efficient inhibition of human immunodeficiency virus type 1. J Virol. 2007;81:13346–13353. doi: 10.1128/JVI.01361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder LC, Harari A, Simon V. Cytidine deamination induced HIV-1 drug resistance. Proc Natl Acad Sci U S A. 2008;105:5501–5506. doi: 10.1073/pnas.0710190105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans R, Cao H, Sharova N, Ali A, Sharkey M, Stranska R, Stevenson M, Rana TM. Small-molecule inhibition of HIV-1 Vif. Nat Biotechnol. 2008;26:1187–1192. doi: 10.1038/nbt.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro F, Bollman B, Chen H, Konig R, Yu Q, Chiles K, Landau NR. Complementary function of the two catalytic domains of APOBEC3G. Virology. 2005;333:374–386. doi: 10.1016/j.virol.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Newman EN, Holmes RK, Craig HM, Klein KC, Lingappa JR, Malim MH, Sheehy AM. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr Biol. 2005;15:166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- Nowarski R, Britan-Rosich E, Shiloach T, Kotler M. Hypermutation by intersegmental transfer of APOBEC3G cytidine deaminase. Nat Struct Mol Biol. 2008;15:1059–1066. doi: 10.1038/nsmb.1495. [DOI] [PubMed] [Google Scholar]
- Nowotny B, Schneider T, Pradel G, Schirmeister T, Rethwilm A, Kirschner M. Inducible APOBEC3G-Vif double stable cell line as a high throughput screening platform to identify antiviral compounds. Antimicrob Agents Chemother. 2010;54:78–87. doi: 10.1128/AAC.00775-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberste MS, Gonda MA. Conservation of amino-acid sequence motifs in lentivirus Vif proteins. Virus Genes. 1992;6:95–102. doi: 10.1007/BF01703760. [DOI] [PubMed] [Google Scholar]
- OhAinle M, Kerns JA, Li MM, Malik HS, Emerman M. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe. 2008;4:249–259. doi: 10.1016/j.chom.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OhAinle M, Kerns JA, Malik HS, Emerman M. Adaptive evolution and antiviral activity of the conserved mammalian cytidine deaminase APOBEC3H. J Virol. 2006;80:3853–3862. doi: 10.1128/JVI.80.8.3853-3862.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opi S, Kao S, Goila-Gaur R, Khan MA, Miyagi E, Takeuchi H, Strebel K. Human immunodeficiency virus type 1 Vif inhibits packaging and antiviral activity of a degradation-resistant APOBEC3G variant. J Virol. 2007;81:8236–8246. doi: 10.1128/JVI.02694-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace C, Keller J, Nolan D, James I, Gaudieri S, Moore C, Mallal S. Population level analysis of human immunodeficiency virus type 1 hypermutation and its relationship with APOBEC3G and vif genetic variation. J Virol. 2006;80:9259–9269. doi: 10.1128/JVI.00888-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul I, Cui J, Maynard EL. Zinc binding to the HCCH motif of HIV-1 virion infectivity factor induces a conformational change that mediates protein-protein interactions. Proc Natl Acad Sci U S A. 2006;103:18475–18480. doi: 10.1073/pnas.0604150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G, Greenwell-Wild T, Nares S, Jin W, Lei KJ, Rangel ZG, Munson PJ, Wahl SM. Myeloid differentiation and susceptibility to HIV-1 are linked to APOBEC3 expression. Blood. 2007;110:393–400. doi: 10.1182/blood-2006-10-051763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pery E, Rajendran KS, Brazier AJ, Gabuzda D. Regulation of APOBEC3 proteins by a novel YXXL motif in human immunodeficiency virus type 1 Vif and simian immunodeficiency virus SIVagm Vif. J Virol. 2009;83:2374–2381. doi: 10.1128/JVI.01898-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piantadosi A, Humes D, Chohan B, McClelland RS, Overbaugh J. Analysis of the percentage of human immunodeficiency virus type 1 sequences that are hypermutated and markers of disease progression in a longitudinal cohort, including one individual with a partially defective Vif. J Virol. 2009;83:7805–7814. doi: 10.1128/JVI.00280-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pion M, Granelli-Piperno A, Mangeat B, Stalder R, Correa R, Steinman RM, Piguet V. APOBEC3G/3F mediates intrinsic resistance of monocyte-derived dendritic cells to HIV-1 infection. J Exp Med. 2006;203:2887–2893. doi: 10.1084/jem.20061519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochnow C, Bransteitter R, Klein MG, Goodman MF, Chen XS. The APOBEC-2 crystal structure and functional implications for the deaminase AID. Nature. 2007;445:447–451. doi: 10.1038/nature05492. [DOI] [PubMed] [Google Scholar]
- Reddy KW, Cheryl A, Werner Lise, Mlisana Koleka, Karim Abdool, Salim S, Ndung’u Thumbi, the CAPRISA Acute Infection Study Team APOBEC3G expression is dysregulated in primary HIV-1 infection and polymorphic variants influence CD4+ T-cell counts and plasma viral load. AIDS. 2010;24:195–204. doi: 10.1097/QAD.0b013e3283353bba. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reingewertz TH, Benyamini H, Lebendiker M, Shalev DE, Friedler A. The C-terminal domain of the HIV-1 Vif protein is natively unfolded in its unbound state. Protein Eng Des Sel. 2009;22:281–287. doi: 10.1093/protein/gzp004. [DOI] [PubMed] [Google Scholar]
- Russell RA, Moore MD, Hu WS, Pathak VK. APOBEC3G induces a hypermutation gradient: purifying selection at multiple steps during HIV-1 replication results in levels of G-to-A mutations that are high in DNA, intermediate in cellular viral RNA, and low in virion RNA. Retrovirology. 2009a;6:16. doi: 10.1186/1742-4690-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RA, Pathak VK. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J Virol. 2007;81:8201–8210. doi: 10.1128/JVI.00395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RA, Smith J, Barr R, Bhattacharyya D, Pathak VK. Distinct domains within APOBEC3G and APOBEC3F interact with separate regions of human immunodeficiency virus type 1 Vif. J Virol. 2009b;83:1992–2003. doi: 10.1128/JVI.01621-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai K, Dimas J, Lenardo MJ. The Vif and Vpr accessory proteins independently cause HIV-1-induced T cell cytopathicity and cell cycle arrest. Proc Natl Acad Sci U S A. 2006;103:3369–3374. doi: 10.1073/pnas.0509417103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santa-Marta M, da Silva FA, Fonseca AM, Goncalves J. HIV-1 Vif can directly inhibit apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G-mediated cytidine deamination by using a single amino acid interaction and without protein degradation. J Biol Chem. 2005;280:8765–8775. doi: 10.1074/jbc.M409309200. [DOI] [PubMed] [Google Scholar]
- Santoni de Sio FR, Trono D. APOBEC3G-depleted resting CD4+ T cells remain refractory to HIV1 infection. PLoS One. 2009;4:e6571. doi: 10.1371/journal.pone.0006571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Emerman M, Malik HS. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2004;2:E275. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer A, Bogerd HP, Cullen BR. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology. 2004;328:163–168. doi: 10.1016/j.virol.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Schrofelbauer B, Chen D, Landau NR. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif) Proc Natl Acad Sci U S A. 2004;101:3927–3932. doi: 10.1073/pnas.0307132101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrofelbauer B, Senger T, Manning G, Landau NR. Mutational alteration of human immunodeficiency virus type 1 Vif allows for functional interaction with nonhuman primate APOBEC3G. J Virol. 2006;80:5984–5991. doi: 10.1128/JVI.00388-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrofelbauer B, Yu Q, Zeitlin SG, Landau NR. Human immunodeficiency virus type 1 Vpr induces the degradation of the UNG and SMUG uracil-DNA glycosylases. J Virol. 2005;79:10978–10987. doi: 10.1128/JVI.79.17.10978-10987.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher AJ, Hache G, Macduff DA, Brown WL, Harris RS. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD, and human immunodeficiency virus type 1 restriction. J Virol. 2008;82:2652–2660. doi: 10.1128/JVI.02391-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Q, Wang Yudi, Hildreth James E. K., Liu Bindong. Polyubiquitination of APOBEC3G Is Essential for Its Degradation by HIV-1 Vif. J Virol. 2010;84:4840–4844. doi: 10.1128/JVI.01911-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Malim MH. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat Med. 2003;9:1404–1407. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- Shindo K, Takaori-Kondo A, Kobayashi M, Abudu A, Fukunaga K, Uchiyama T. The enzymatic activity of CEM15/Apobec-3G is essential for the regulation of the infectivity of HIV-1 virion but not a sole determinant of its antiviral activity. J Biol Chem. 2003;278:44412–44416. doi: 10.1074/jbc.C300376200. [DOI] [PubMed] [Google Scholar]
- Simm M, Shahabuddin M, Chao W, Allan JS, Volsky DJ. Aberrant Gag protein composition of a human immunodeficiency virus type 1 vif mutant produced in primary lymphocytes. J Virol. 1995;69:4582–4586. doi: 10.1128/jvi.69.7.4582-4586.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JH, Carpenter EA, Fouchier RA, Malim MH. Vif and the p55(Gag) polyprotein of human immunodeficiency virus type 1 are present in colocalizing membrane-free cytoplasmic complexes. J Virol. 1999;73:2667–2674. doi: 10.1128/jvi.73.4.2667-2674.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JH, Gaddis NC, Fouchier RA, Malim MH. Evidence for a newly discovered cellular anti-HIV-1 phenotype. Nat Med. 1998a;4:1397–1400. doi: 10.1038/3987. [DOI] [PubMed] [Google Scholar]
- Simon JH, Malim MH. The human immunodeficiency virus type 1 Vif protein modulates the postpenetration stability of viral nucleoprotein complexes. J Virol. 1996;70:5297–5305. doi: 10.1128/jvi.70.8.5297-5305.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JH, Miller DL, Fouchier RA, Malim MH. Virion incorporation of human immunodeficiency virus type-1 Vif is determined by intracellular expression level and may not be necessary for function. Virology. 1998b;248:182–187. doi: 10.1006/viro.1998.9296. [DOI] [PubMed] [Google Scholar]
- Simon V, Zennou V, Murray D, Huang Y, Ho DD, Bieniasz PD. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 2005;1:e6. doi: 10.1371/journal.ppat.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodroski J, Goh WC, Rosen C, Tartar A, Portetelle D, Burny A, Haseltine W. Replicative and cytopathic potential of HTLV-III/LAV with sor gene deletions. Science. 1986:1549–1553. doi: 10.1126/science.3006244. [DOI] [PubMed] [Google Scholar]
- Soros VB, Yonemoto W, Greene WC. Newly synthesized APOBEC3G is incorporated into HIV virions, inhibited by HIV RNA, and subsequently activated by RNase H. PLoS Pathog. 2007;3:e15. doi: 10.1371/journal.ppat.0030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sova P, Volsky DJ. Efficiency of viral DNA synthesis during infection of permissive and nonpermissive cells with vif-negative human immunodeficiency virus type 1. J Virol. 1993;67:6322–6326. doi: 10.1128/jvi.67.10.6322-6326.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley BJ, Ehrlich ES, Short L, Yu Y, Xiao Z, Yu XF, Xiong Y. Structural insight into the human immunodeficiency virus Vif SOCS box and its role in human E3 ubiquitin ligase assembly. J Virol. 2008;82:8656–8663. doi: 10.1128/JVI.00767-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson M, Stanwick TL, Dempsey MP, Lamonica CA. HIV-1 replication is controlled at the level of T cell activation and proviral integration. EMBO J. 1990;9:1551–1560. doi: 10.1002/j.1460-2075.1990.tb08274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopak K, de Noronha C, Yonemoto W, Greene WC. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol Cell. 2003;12:591–601. doi: 10.1016/s1097-2765(03)00353-8. [DOI] [PubMed] [Google Scholar]
- Strebel K, Daugherty D, Clouse K, Cohen D, Folks T, Martin MA. The HIV ‘A’ (sor) gene product is essential for virus infectivity. Nature. 1987;328:728–730. doi: 10.1038/328728a0. [DOI] [PubMed] [Google Scholar]
- Suspene R, Rusniok C, Vartanian JP, Wain-Hobson S. Twin gradients in APOBEC3 edited HIV-1 DNA reflect the dynamics of lentiviral replication. Nucleic Acids Res. 2006;34:4677–4684. doi: 10.1093/nar/gkl555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suspene R, Sommer P, Henry M, Ferris S, Guetard D, Pochet S, Chester A, Navaratnam N, Wain-Hobson S, Vartanian JP. APOBEC3G is a single-stranded DNA cytidine deaminase and functions independently of HIV reverse transcriptase. Nucleic Acids Res. 2004;32:2421–2429. doi: 10.1093/nar/gkh554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svarovskaia ES, Xu H, Mbisa JL, Barr R, Gorelick RJ, Ono A, Freed EO, Hu WS, Pathak VK. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J Biol Chem. 2004;279:35822–35828. doi: 10.1074/jbc.M405761200. [DOI] [PubMed] [Google Scholar]
- Tian C, Yu X, Zhang W, Wang T, Xu R, Yu XF. Differential requirement for conserved tryptophans in human immunodeficiency virus type 1 Vif for the selective suppression of APOBEC3G and APOBEC3F. J Virol. 2006;80:3112–3115. doi: 10.1128/JVI.80.6.3112-3115.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartanian JP, Meyerhans A, Asjo B, Wain-Hobson S. Selection, recombination, and G----A hypermutation of human immunodeficiency virus type 1 genomes. J Virol. 1991;65:1779–1788. doi: 10.1128/jvi.65.4.1779-1788.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schwedler U, Song J, Aiken C, Trono D. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J Virol. 1993;67:4945–4955. doi: 10.1128/jvi.67.8.4945-4955.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker RC, Jr., Khan MA, Kao S, Goila-Gaur R, Miyagi E, Strebel K. Identification of dominant negative human immunodeficiency virus type 1 Vif mutants that interfere with the functional inactivation of APOBEC3G by virus-encoded Vif. J Virol. 2010;84:5201–5211. doi: 10.1128/JVI.02318-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Shackelford JM, Selliah N, Shivers DK, O’Neill E, Garcia JV, Muthumani K, Weiner D, Yu XF, Gabuzda D, et al. The HIV-1 Vif protein mediates degradation of Vpr and reduces Vpr-induced cell cycle arrest. DNA Cell Biol. 2008;27:267–277. doi: 10.1089/dna.2007.0707. [DOI] [PubMed] [Google Scholar]
- Wang T, Tian C, Zhang W, Luo K, Sarkis PT, Yu L, Liu B, Yu Y, Yu XF. 7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G. J Virol. 2007;81:13112–13124. doi: 10.1128/JVI.00892-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichroski MJ, Ichiyama K, Rana TM. Analysis of HIV-1 viral infectivity factor-mediated proteasome-dependent depletion of APOBEC3G: correlating function and subcellular localization. J Biol Chem. 2005;280:8387–8396. doi: 10.1074/jbc.M408048200. [DOI] [PubMed] [Google Scholar]
- Wiegand HL, Doehle BP, Bogerd HP, Cullen BR. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 2004;23:2451–2458. doi: 10.1038/sj.emboj.7600246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Ehrlich E, Luo K, Xiong Y, Yu XF. Zinc chelation inhibits HIV Vif activity and liberates antiviral function of the cytidine deaminase APOBEC3G. FASEB J. 2007;21:217–222. doi: 10.1096/fj.06-6773com. [DOI] [PubMed] [Google Scholar]
- Xiao Z, Ehrlich E, Yu Y, Luo K, Wang T, Tian C, Yu XF. Assembly of HIV-1 Vif-Cul5 E3 ubiquitin ligase through a novel zinc-binding domain-stabilized hydrophobic interface in Vif. Virology. 2006;349:290–299. doi: 10.1016/j.virol.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Xu H, Chertova E, Chen J, Ott DE, Roser JD, Hu WS, Pathak VK. Stoichiometry of the antiviral protein APOBEC3G in HIV-1 virions. Virology. 2007;360:247–256. doi: 10.1016/j.virol.2006.10.036. [DOI] [PubMed] [Google Scholar]
- Xu H, Svarovskaia ES, Barr R, Zhang Y, Khan MA, Strebel K, Pathak VK. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc Natl Acad Sci U S A. 2004;101:5652–5657. doi: 10.1073/pnas.0400830101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T, Kamada K, Hatcho K, Adachi A, Nomaguchi M. Identification of amino acid residues in HIV-1 Vif critical for binding and exclusion of APOBEC3G/F. Microbes Infect. 2008;10:1142–1149. doi: 10.1016/j.micinf.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Yamashita T, Nomaguchi M, Miyake A, Uchiyama T, Adachi A. Status of APOBEC3G/F in cells and progeny virions modulated by Vif determines HIV-1 infectivity. Microbes Infect. 2010;12:166–171. doi: 10.1016/j.micinf.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Yang B, Chen K, Zhang C, Huang S, Zhang H. Virion-associated uracil DNA glycosylase-2 and apurinic/apyrimidinic endonuclease are involved in the degradation of APOBEC3G-edited nascent HIV-1 DNA. J Biol Chem. 2007;282:11667–11675. doi: 10.1074/jbc.M606864200. [DOI] [PubMed] [Google Scholar]
- Yang B, Gao L, Li L, Lu Z, Fan X, Patel CA, Pomerantz RJ, DuBois GC, Zhang H. Potent suppression of viral infectivity by the peptides that inhibit multimerization of human immunodeficiency virus type 1 (HIV-1) Vif proteins. J Biol Chem. 2003;278:6596–6602. doi: 10.1074/jbc.M210164200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Sun Y, Zhang H. The multimerization of human immunodeficiency virus type I Vif protein: a requirement for Vif function in the viral life cycle. J Biol Chem. 2001;276:4889–4893. doi: 10.1074/jbc.M004895200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Konig R, Pillai S, Chiles K, Kearney M, Palmer S, Richman D, Coffin JM, Landau NR. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat Struct Mol Biol. 2004a;11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- Yu Y, Xiao Z, Ehrlich ES, Yu X, Yu XF. Selective assembly of HIV-1 Vif-Cul5-ElonginB-ElonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 2004b;18:2867–2872. doi: 10.1101/gad.1250204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack JA, Arrigo SJ, Weitsman SR, Go AS, Haislip A, Chen IS. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- Zennou V, Bieniasz PD. Comparative analysis of the antiretroviral activity of APOBEC3G and APOBEC3F from primates. Virology. 2006;349:31–40. doi: 10.1016/j.virol.2005.12.035. [DOI] [PubMed] [Google Scholar]
- Zennou V, Perez-Caballero D, Gottlinger H, Bieniasz PD. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J Virol. 2004;78:12058–12061. doi: 10.1128/JVI.78.21.12058-12061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Pomerantz RJ, Dornadula G, Sun Y. Human immunodeficiency virus type 1 Vif protein is an integral component of an mRNP complex of viral RNA and could be involved in the viral RNA folding and packaging process. J Virol. 2000;74:8252–8261. doi: 10.1128/jvi.74.18.8252-8261.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang KL, Mangeat B, Ortiz M, Zoete V, Trono D, Telenti A, Michielin O. Model structure of human APOBEC3G. PLoS One. 2007;2:e378. doi: 10.1371/journal.pone.0000378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng YH, Irwin D, Kurosu T, Tokunaga K, Sata T, Peterlin BM. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J Virol. 2004;78:6073–6076. doi: 10.1128/JVI.78.11.6073-6076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman C, Klein KC, Kiser PK, Singh AR, Firestein BL, Riba SC, Lingappa JR. Identification of a host protein essential for assembly of immature HIV-1 capsids. Nature. 2002;415:88–92. doi: 10.1038/415088a. [DOI] [PubMed] [Google Scholar]