Abstract
Regulatory T cells (Tregs) are required for the maintenance of immune homeostasis as first clearly described by Herman Waldmann’s laboratory. Dysfunction of Treg cells also leads to fatal autoimmunity in humans and mice. Conversely, the activation of different classes of Tregs operative systemically and within the cancer microenvironment can suppress host anti-tumor immune responses and promote tumor progression. Therefore, the development of new therapeutic approaches to regulate the activity of Treg cells may have considerable clinical potential.
FOXP3 is the key transcriptional regulator of Treg development and function. The activity of FOXP3 is regulated by acetylation, a process catalyzed by distinct types of histone/protein acetyltransferases (HATs) that regulate the functions of many transcription factors, independently of FOXP3, as well as non-histone proteins, in addition to their effects on chromatin accessibility. Interactions between FOXP3 and these enzymes determine the suppressive function of FOXP3. Clearly, small molecules targeting these enzymes are candidates for the regulation of Treg function in vaccines and tumor therapies.
Introduction
Regulatory T cells (Treg) constitute a small subset of CD4+ T cells that play an important role in maintaining the balance between immune tolerance and inflammation [1,2]. A transcription factor, FOXP3, is critical for Treg cell differentiation and maturation [3,4]. Through interactions with other transcription factors, FOXP3 suppresses the expression of a variety of inflammatory cytokines and promotes the expression of Treg-associated genes [5].
Genetic mutations in the FOXP3 locus lead to autoimmune diseases. Scurfy mice develop a fatal lymphoproliferative disorder due to a frameshift mutation of Foxp3 [6]. In human, dysfunction of FOXP3 is associated with immunodysregulation polyendocrinopathy enteropathy X-linked syndrome (IPEX) [7,8]. Evidence from clinical and experimental studies shows that Tregs contribute to the immunosuppression seen during cancer development, a process that prevents effective host clearance of cancer cells [9–12]. Regulation of FOXP3 function is a dominant research area and insights into FOXP3 functions represent opportunities for the development of new treatments of both human autoimmune diseases and cancer.
Our laboratory has focused on a detailed understanding of the biochemical modifications and atomic structures of the FOXP3 mediated regulatory complexes. We were able to show that FOXP3 undergoes post-translational changes as a consequence of extrinsic cellular signals. Various post-translational modifications of FOXP3, including acetylation, regulate Treg activity. Acetylation of FOXP3 affects its stability and promoter binding activity [13,14]. The acetylation of FOXP3 is regulated by components of a FOXP3-associated supermolecular complex containing multiple HATs, histone/protein deacetylases (HDACs) and other transcriptional coregulators [15,16]. We will focus our discussion on the interactions between FOXP3 and other transcription factors, especially HATs, and how these interactions affect post-translational modifications and activity of FOXP3. The possibility of regulating Treg function through regulating the activity of these enzymes is also discussed.
FOXP3
The role of Foxp3 in immune homeostasis was first identified in scurfy mice that display a phenotype similar to individuals afflicted with a clinical autoimmune lymphoproliferative disease [17]. Mutation of the Foxp3 gene was shown to be responsible for the scurfy phenotype and expression of the normal Foxp3 gene was capable of rescuing the mice from scurfy disease. A variety of studies demonstrated that FOXP3 was predominantly expressed in CD4+CD25+ Treg cells, a subset of T cells that had been shown able to suppress the proliferation of activated conventional CD4+ T cells [4,18]. Since CD25 is also expressed in activated CD4+ T cells, identification of FOXP3 provided a more reliable marker for Treg cells and has proven critical for further functional analysis of these cells. However it is now clear that the presence of FOXP3 alone is not sufficient to identify Treg cells as “functional” as post-translational changes in discrete residues of the protein lead to acquisition of function.
Domain structure of FOXP3
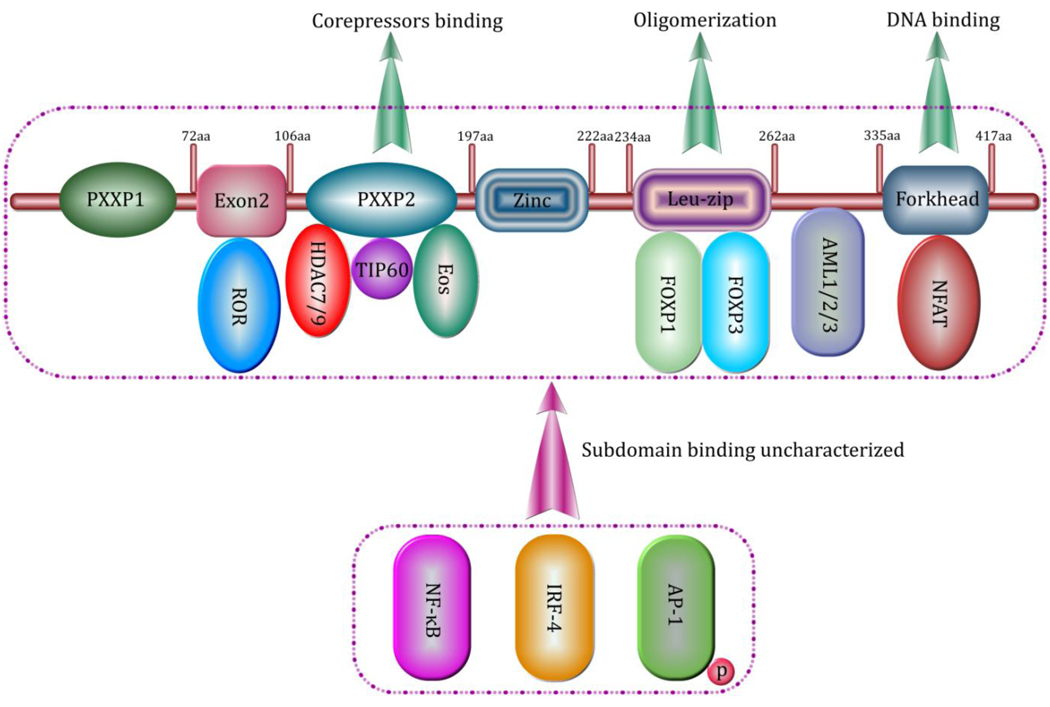
FOXP3 is a member of the forkhead/winged-helix box-containing protein P subfamily of transcription factors that include four subfamily members, FOXP1, FOXP2, FOXP3 and FOXP4. All four proteins contain highly conserved C terminal region containing zinc finger and leucine zipper oligomerization domain, and a forkhead DNA binding domain [19,20] (Figure 1). However, FOXP3 differs from other family members. FOXP3’s N terminal domain, which is proline-rich shares little similarity with the glutamine-rich N terminal domain of FOXP1, FOXP2 or FOXP4 [15]. This difference may contribute to the unique role of FOXP3 in regulating the development of Treg cells. It is clear that this structural region is required for recruitment of other coregulators essential for FOXP3 mediated repression of gene expression. Only one isoform of Foxp3 is found in mouse, while two FOXP3 isoforms are found in human, including FOXP3a and FOXP3b [21,22]. FOXP3b lacks the exon2 coding region that is critical for its association with transcription factor RORα and RORγ [19,21] (Figure 1).
Figure 1. The FOXP3 ensemble of transcription factors and corepressors in T cell regulation.
The exon2 encoding region of FOXP3 interacts with RORα and RORγ. The leucine zipper motif interacts with FOXP1 and/or FOXP3. The linker region between leucine zipper and forkhead DNA binding domain interacts with AML1/2/3. The second proline-rich domain between exon2 encoding region and zinc finger motif interacts with enzymatic transcriptional corepressors including TIP60 and HDAC7/9 as well as transcription factor Eos. Other FOXP3-binding transcription factors include NFκB, IRF-4 and phosphorylated AP-1.
Interactions of FOXP3 with other transcription factors
FOXP3 acts as a transcriptional regulator during both Treg development and as a functional regulatory element in the peripheral suppressive T cell pool. An important effect of FOXP3 is to regulate the production of a variety of cytokines through interacting with a range of partner proteins, including transcription factors, the nuclear factor of activated T cells (NFAT), NF-κB and Runx1/AML1 [15,23]. Both NFAT and Runx1/AML1 appear to be required for IL-2 expression after T cell receptor (TCR) stimulation [24,25]. Association of FOXP3 and NFAT and Runx1/AML1 transcription factors represses the activation of IL-2, IL-4 and IFN-γ promoters. Interactions between FOXP3 and NFAT are also required for the upregulation of Treg markers, such as CD25 and CTLA-4.
FOXP3 interacts with other members of the FOXP family to associate with its target promoters by forming a complex that is capable of actively repressing the expression of target genes. Discrete residues of FOXP3 are determining. For instance, deletion of a single amino acid E251 within the leucine zipper region of FOXP3 causes the X-linked autoimmunity-allergic dysregulation (XLAAD) or IPEX syndrome [16,26,27]. We have found that this type of mutations disrupt both the homo-oligomerization of FOXP3 and its hetero association with FOXP1. As a consequence, the mutant FOXP3 fails to associate with the IL-2 promoter and cannot repress IL-2 transcription upon T cell activation [28]. Impaired DNA binding property of FOXP3 mutants are considered responsible for the phenotype of patients with XLAAD/IPEX autoimmune syndromes.
Acetylation of FOXP3 regulates both its activity and stability
Our laboratory found that FOXP3 exists in Tregs as part of a large HAT/HDAC complex which regulates the acetylation status of FOXP3 protein. Discovery of acetylation dates from the 1960s, when core histones were first discovered to be acetylated at the ε-amino nitrogen specific lysine residues located in the amino terminal tails [29,30]. Protein acetylation is an important post-translational modification for regulating protein function. Acetylation of core histones is important to chromatin remodeling and gene activation [31,32]. However only recently have several non-histone proteins found to be acetylated by HATs. Acetylated proteins include the DNA binding transcription factors p53, NF-κB, TFIIE, signaling regulator Smad3 and cytoskeleton protein α-tubulin [33–36]. In contrast to acetylation that occurs in co-translational manner, lysine acetylation is a reversible post-translational process in which the dynamic equilibrium is governed by the competing functions of HATs and HDACs [37,38].
We identified FOXP3 acetylation as an important and required post-translational modification that is regulated by components of the HAT/HDAC complex present in Tregs [15,16]. Acetylation has multiple effects on FOXP3. First, acetylated FOXP3 appears to be more stable as HDAC inhibitors (HDACi) or HAT enzymes inhibit or limit FOXP3 degradation [14]. The activities of the proteasome inhibitor, MG132, prevent FOXP3 degradation, consistent with the concept that FOXP3 turnover is affected by post-translational modifications. These modifications include acetylation and ubiquitination of FOXP3 key lysine residues. When all the lysines in FOXP3 are mutated to arginines, the stability of the FOXP3 mutant is greatly increased, in fact to a level comparable to acetylated FOXP3 despite the protein’s overall deficiency in acetylation [14]. Second, the amount of FOXP3 binding to chromatin is significantly increased when it is acetylated, indicating that FOXP3 acetylation promotes DNA binding [13].
When FOXP3 is in complex with TIP60, acetylated FOXP3 is readily detected and overall transcriptional repression ability on the IL-2 promoter is greatly enhanced. These interactions indicate a correlation between TIP60 associated FOXP3 acetylation and Treg activity [16].
p300 also promotes the repressive activity of FOXP3. It is likely that TIP60 and p300 acetylate discrete sets of lysines of FOXP3. Site specific lysine to arginine mutants and anti-actylated lysine antibodies will prove useful in further investigations of the effects of acetylation on FOXP3 function.
Characterization of HATs involved in FOXP3 acetylation
Transcriptional co-activators, including PCAF, p300 (E1A-associated 300 kD protein) and CBP (CREB-binding protein), have intrinsic acetyltransferase activity [39–41]. Many other proteins have been found to possess acetyltransferase activities, making the HAT community a very diverse protein family. Based on sequence similarity, known HATs can be divided into three different groups [42], namely the Gcn5/PCAF family, which includes Gcn5, PCAF and Gcn5L; the p300/CBP family, which includes CBP and p300; and the MYST family, which includes Esa1, MOF and TIP60.
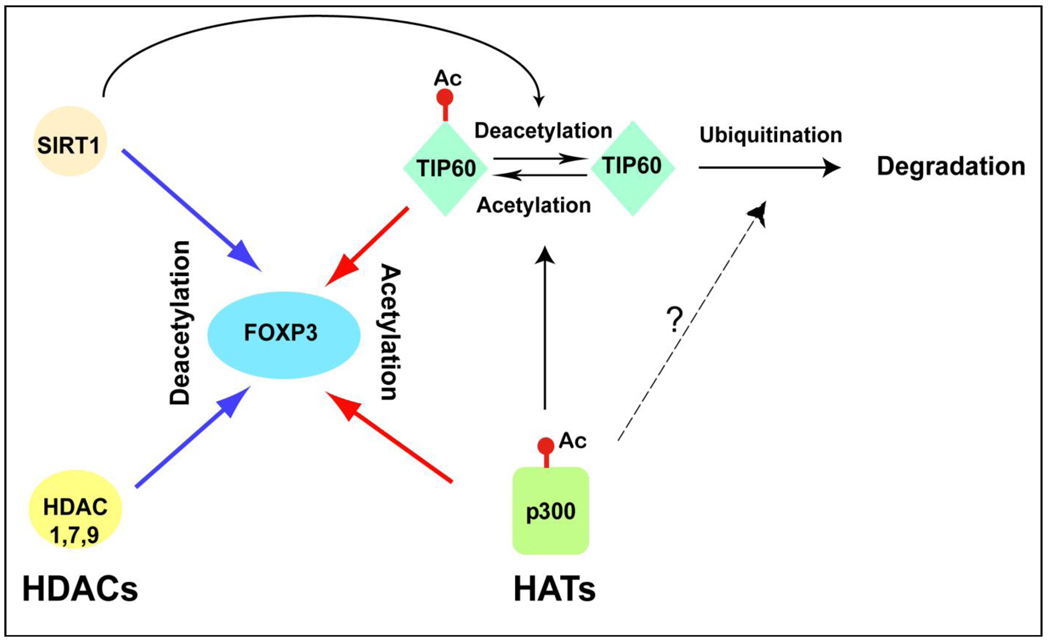
We have studied the p300 and the MYST family of transferases as well as certain deacetylases. Proteins that modify the acetylation level of FOXP3 include TIP60, p300, HDAC7, HDAC9 and SIRT1 [14,16] (Figure 2). We have also identified HDAC1 as a component of the FOXP3 mediated larger complex. These proteins, upon activation by discrete extrinsic signals, act to regulate the acetylation levels of FOXP3.
Figure 2. Regulation of FOXP3 acetylation by HATs and HDACs.
HATs and HDACs have contrasting roles in the regulation of FOXP3 acetylation. Acetylation of FOXP3 relies on not only its interaction with HATs (TIP60 and p300) or HDACs (HDAC1, HDAC7, HDAC9 and Sirt1) but also the presence of active forms of enzymes. Both TIP60 and p300 have higher acetyltransferase activity when they are acetylated. Acetylation also appears to avert TIP60 from ubiquitination and subsequent proteasomal degradation. p300 is known to induce acetylation of TIP60, but might also be involved in the degradation of TIP60 under certain circumstance (e.g. HIV infection). As a deacetylase, Sirt1 directly regulates acetylation and activity of TIP60. HDAC7 can also interact with TIP60. The dynamic interplays among HDACs and HATs decide the acetylation status of FOXP3 and affect the function of Treg cells.
Among these proteins, TIP60 of the MYST family and p300 of the p300/CBP family positively regulate the acetylation of FOXP3, while SIRT1 negatively regulates this process. The roles of HDAC7 and HDAC9 in FOXP3 acetylation are relevant but less defined, since class IIa HDACs are thought to lack functional intrinsic catalytic activity and likely act by recruiting class I HDACs such as HDAC3 to the complex. The effects of HDAC on FOXP3 expression are complex and individual HDACs may differentially affect the function of FOXP3 [43].
Histone acetyltransferases
TIP60 is first known as HIV-1 Tat-interactive protein 60 kD and possesses cellular acetyltransferase activity [44]. There are three isoforms of TIP60 which arise as a consequence of alternative splicing: TIP60 isoform1, TIP60 isoform2 (TIP60α) and TIP60 isoform3 (TIP60β) [45,46]. Isoform2 of TIP60 is the most completely characterized biochemically.
TIP60 possesses several functional domains. A MYST domain, composed of a conserved HAT domain and a C2HC zinc finger domain, is conserved in all MYST family members and is responsible for the catalytic activity and substrate binding [47]. Two mutations in the HAT domain, Q377E /G380E, abolish TIP60 binding to acetyl-CoA, resulting in profound deficiency of HAT activity [48]. The C2HC zinc finger located in MYST domain is essential for TIP60 to interact with other proteins [49,50]. Interaction of TIP60 and p300 is dependent on residues located in this region and this interaction is essential for TIP60 acetylation that is directly promoted by p300 [51]. The N-terminal region of TIP60 has a chromodomain that mediates interactions of associated proteins with methylated lysines of histone or RNA molecules in other regulatory proteins [52], but the actual function of the chromodomain of TIP60 remains to be established. A nuclear receptor interaction box (NR box) has been identified at the C terminal of TIP60 [53].
TIP60 was first found to acetylate histones and modify nucleosomes assembly in vitro [48,54]. Other transcriptional regulators have been identified as the substrate of TIP60, including transcription factors such as androgen receptor (AR), c-Myc, p53 and ATM[55–57]. However, studies of the lysine site acetylated by TIP60 have not revealed an apparent consensus motif. Cellular TIP60 forms a large stable protein complex with many other proteins, including the scaffold protein transformation/transcription domain-associated protein (TRRAP) which is also found in other acetyltransferase complexes, and p400/Domono which has chromatin remodeling activity [48]. In most cases, TIP60 functions as a transcription activator, but it also represses the transcription of certain genes, indicating a selective co-regulatory role of TIP60 in specific gene transcription. TIP60 can enhance the activity of transcription factors directly or serve as a co-activator. It also mediates transcriptional repression by recruitment of other protein complexes. For example, upon IL-9 stimulation, TIP60 recruits HDAC7 to the TIP60/STAT-3 complex in the nucleus, therefore modulating STAT-3 activity and repressing STAT-3 regulated gene transcription [50].
Post-translational modifications are important in regulating the activity of TIP60. HIV-1 transactivator Tat uses p300/CBP to stimulate polyubiquitination of TIP60. Ubiquitination promotes the proteasomal degradation of TIP60, resulting in reduced apoptosis in response to genotoxic processes in cells infected by HIV [51]. In addition to ubiquitination, activity of TIP60 is affected by acetylation, including auto-acetylation and acetylation mediated by other acetyltransferase like p300/CBP. TIP60 is more active in its acetylated form, which can be stimulated by UV irritation [58]. Ubiquitination and acetylation target different domains of TIP60, and the acetylation deficient form of TIP60 can still be ubiquinated, indicating that these two post-translational modifications are independent [51].
SIRT1, a member of HDAC family, inhibits auto-acetylation of TIP60 and decreases the expression level of TIP60 [58,59]. Our unpublished data indicate that p300 can greatly increase the expression level of TIP60. Taken together, these data suggest that the acetylation level of TIP60 may be dynamically regulated by p300 and Sirt1, two independently activated proteins that collectively regulate the protein level of TIP60 (Figure 2). Regulation reflects the competing effects of acetylation/deacetylation versus ubiquitination and proteasomal degradation, as is the case for other transcription factors.
TIP60 actively takes part in various cellular processes such as p53 pathway, DNA damage repair, and cell apoptosis [48,51,60]. Binding of TIP60 to the androgen receptor (AR) and the acetylation of AR by TIP60 indicate that TIP60 may also play important role in cancer development [55,61]. Our studies showed that TIP60 promotes FOXP3 acetylation and is involved in FOXP3 mediated transcription repression, defining a role for TIP60 in Treg cell development and regulation of immune responses [16].
We and others have found that p300 is able to promote FOXP3 acetylation [14]. Both p300 and CBP can acetylate histones and non-histone proteins. p300 and CBP participate in many physiological processes such as cell proliferation, differentiation and apoptosis [62]. p300 and CBP were identified through their ability to interact with the adenovirus E1A oncoprotein or cAMP-regulated transcripction factor CREB respectively [63,64]. p300 and CBP have not only overlapping but also different functions. Most transcription factors acetylated by p300 can also be acetylated by CBP. Mice with mutations in p300 or CBP show similar phenotypes, indicating an overlapping role of p300 and CBP during development [65]. CBP but not p300 is required for normal haematopoietic differentiation [66]. Likewise, p300 mRNA ribozymes but not CBP mRNA ribozyme are able to inhibit retinoic-acid-induced differentiation of embryonal carcinoma cells [67]. The basis of these biological differences in activity is unclear.
The genes encoding CBP/p300 are conserved from worms to humans and p300 and CBP share extensive homology. The conserved regions constitute the most functional domains of the two proteins. They contain one bromodomain frequently found in mammalian HAT, three cysteine-histine (CH) rich domain, a KIX domain and glutamine-rich C terminus domain that all serve as docking modules for many transcriptional factors, and an acetyltransferase domain located in the middle of CBP/p300 [68]. A proteolytically sensitive loop in HAT domain (~ residues 1520–1560) has a role in regulating the activity of p300, by serving as a pseudosubstrate and auto-inhibit p300 activity. Acetylation of this loop by p300 itself or other HAT dislodges this loop from the enzyme active centre, permitting increased p300 activity [69]. The catalytic activity of this domain is not required for the activation of all the CBP/p300 binding transcription factors, suggesting that in addition to directly acetylating transcription factors, CBP/p300 causes transcriptional activation through interactions with other transcription co-activators. CBP/p300 may serve as a bridge to connect transcription factors to components of basal transcription machinery [70,71]. CBP/p300 may act as a scaffold for the assembly of transcription factors, thereby facilitating protein-protein or protein-DNA interactions to confer transcription activation [72,73].
Effect of TIP60 and p300 on FOXP3 acetylation
TIP60 is expressed in both CD25+ and CD25− T cells and co-localized with FOXP3 in the nucleus of human Treg cells, indicating the likely role of TIP60 in regulating FOXP3 acetylation under physiological conditions. In 293T cells co-transfected with TIP60 and FOXP3, TIP60 co-immunoprecipitated with FOXP3 and promoted FOXP3 acetylation in a dose-dependent manner. The N terminal of FOXP3 (106–190 aa), which defines FOXP3 uniquely in the FOXP subfamily, is required for FOXP3 binding to TIP60. Deletion of this region or the use of the HAT-deficient form of TIP60 abolishes FOXP3 mediated transcription repression, indicating that the presence of catalytic active TIP60 is essential for the normal function of this repression complex.
HDAC7, which is always in complex with TIP60, also interacts with FOXP3 and as a complex participates in FOXP3 mediated transcription repression. Combinations of TIP60 and HDAC7 are needed for maximum FOXP3 mediated transcription repression of the IL-2 promoter. In vivo over expression of TIP60 or HDAC7 alone actually reduces the total repression efficiency, and knockdown of endogenous TIP60 limits FOXP3-mediated transcription repression [16]. HDAC7, even though needed for optimal suppression, may play a more complex role in FOXP3 function. It is possible that HDAC7 targets other component of the complex FOXP3 ensemble and affects FOXP3 activity indirectly.
After the discovery of FOXP3 acetylation by TIP60, several lines of evidence indicated that another HAT, p300, also contributed to regulation of FOXP3 acetylation [14]. First, p300 co-localizes with FOXP3 in the nucleus. Second, FOXP3 acetylation is increased in the presence of p300. Third, in vitro acetylation assays indicate that FOXP3 acetylation can be mediated by p300.
Acetylation of FOXP3 by p300 is not affected by FOXP3 dimer formation. It is not known whether FOXP3 interacts with TIP60 and p300 in the same way, and the exact acetylation sites of FOXP3 targeted by TIP60 versus p300 have not been identified. Acetylation site of Histone H4 by TIP60 are the GK/AK sites. Interestingly, human FOXP3 also has two GK sites, and one of them is located at the K8 site, which was recently identified by MS sequencing as an acetylated site in FOXP3-transfected Jurkat T cells (Yayi Gao, Mark I. Greene and Bin Li, unpublished data). In 293T cells cotransfected with FOXP3 and p300, FOXP3 acetylation level was not affect by the K8R mutation, but was significantly decreased by K250R or K252R mutations (Yan Xiao and Mark I. Greene, unpublished data).
Acetylation at different sites by different HATs may result in different biological functions. Acetylation of p53 at K164 and six C terminal lysines by p300 increases activity and stability of p53 [74], while acetylation of K120 located in DNA binding domain by TIP60 promotes p53 mediated cell apoptosis [60,75]. A similar mechanism may exist in regulating FOXP3 activity through acetylation. Different acetyltransferase may acetylate different sites of FOXP3 and lead to different consequences of regulation of FOXP3 activity. Stability of FOXP3 is remarkably increased in the presence of p300, while TIP60 does not discernibly affect its stability [14,16].
Structural analysis predicts that K250 and K252 play a role in regulating FOXP3 dimer formation and acetylation of these two sites may alter the stability of the FOXP3 dimer. Mutation in K250 and K252 decreases FOXP3 dimer formation (Zhaocai Zhou and Mark I. Greene, unpublished data). Acetylation of these two sites by p300 may serve as a molecular switch that regulates the complex FOXP3 forms either with itself or with other transcription factors.
Our recent data showed that combination of TIP60 and p300 promoted FOXP3 acetylation while either TIP60 or p300 alone only result in weak FOXP3 acetylation (Yan Xiao and Mark Greene, unpublished data).
Since acetylation is also required for the HAT activity of TIP60 and p300 (Figure 2), a process which is achieved either by auto-acetylation or acetylation by other HATs [59,76], one consideration is that TIP60 and p300 may cooperate with each other to facilitate FOXP3 acetylation. p300 promotes TIP60 acetylation, which in turn enhances the HAT activity of TIP60, and this process appears responsible for the increased acetylation level of FOXP3. On the other hand, it is also possible that TIP60 promotes the acetylation of p300, therefore increasing the activity of p300 and facilitating FOXP3 acetylation.
This complicated interplay is currently under investigation in our laboratory. In addition to HAT, extracellular stimuli such as TGF-β can increase the levels of acetylated FOXP3 [13]. However IL-6 diminishes FOXP3 levels and acetylation and the ability of the FOXP3 to interact with DNA. One scenario is that the complex FOXP3 ensembles of HATs/HDACs is regulated in response to these diverse sets of signals. Consequently the altered interaction of FOXP3 and these enzymes regulate FOXP3 at a post-translational level. The function of Treg is thereby regulated in part by extracellular stimuli which alter the discrete pattern of modifications by constituents of the regulatory complex.
Significance of regulating FOXP3 acetylation and activity in disease control
Since HAT/HDAC play defining roles in the regulation of FOXP3 activity, it is possible to regulate Treg function by modulating the activity of HAT/HDAC complex [43]. In fact, several HDAC inhibitors have proved effective as therapeutic agents in murine models of collagen-induced arthritis, allograft rejection and colitis [77–79]. HDAC inhibitor treatment in these mouse models not only increased Foxp3 expression in Treg cells but also enhanced the number and suppressive function of Treg cells, leading to the prevention and improved treatment of autoimmune diseases.
Our laboratory originally described the negative role mediated by Treg during cancer development [11,12,80]. Increasing evidence indicated that FOXP3 may inhibit host immune responses directed at cancer cells [81,82]. Treg cells can be either recruited or converted from naïve T cells and become an obstacle to targeted monoclonal antibody therapy for cancer treatment, a therapy our laboratory introduced for the treatment of HER2/neu mediated human cancers [83–86].
Our early efforts were to down regulate T suppressive activities using passively administered antibodies [87,88]. Downregulating the activity of Treg cells may be an effective anti-cancer treatment which can be achieved by targeting specific enzymes. We can now focus on small molecules that disable specific histone acetyltransfereases. Since acetylation of FOXP3 is important to suppressive functions, small molecules inhibiting the activity of HATs would be attractive therapeutics to downregulate Treg activity and enhance the efficacy of targeted cancer therapies.
Conclusion
Our studies have been focused on defining the atomic feature of the FOXP3 protein and how FOXP3 acquires functionality as a consequence of post-translational modifications. Diverse extrinsic signals modify these processes by acting on enzymes within the Treg cell. Acetylation level of FOXP3 determines its function and this process is regulated through its interaction with HAT/HDAC complexes.
It will be important to define the range of signals that can differentially activate single or multiples HATs that are responsible for direct acetylation of FOXP3 in vivo. It is also important to define how this normally controlled set of modifications becomes overwhelmed to adversely affect the function of FOXP3 in the development of autoimmune diseases and cancer. Answers to these biological and biochemical problems will not only broaden our understanding about how FOXP3 regulates Treg development and function, but will also lead to new treatment of autoimmune diseases and cancers.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
* of special interest
** of outstanding interest
- 1.Maloy KJ, Powrie F. Regulatory T cells in the control of immune pathology. Nat Immunol. 2001;2:816–822. doi: 10.1038/ni0901-816. [DOI] [PubMed] [Google Scholar]
- 2.Sakaguchi S, Miyara M, Costantino CM, Hafler DA. FOXP3(+) regulatory T cells in the human immune system. Nat Rev Immunol. 10:490–500. doi: 10.1038/nri2785. [DOI] [PubMed] [Google Scholar]
- 3.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 4. Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. In this study, Foxp3 was identified as the key transcription regulator specifically expressed in naturally arising Treg cells. Expression of Foxp3 was responsible for converting naïve T cells towards a Treg cell phenotype, revealing a mechanism for development of Treg cells.
- 5.Schubert LA, Jeffery E, Zhang Y, Ramsdell F, Ziegler SF. Scurfin (FOXP3) acts as a repressor of transcription and regulates T cell activation. J Biol Chem. 2001;276:37672–37679. doi: 10.1074/jbc.M104521200. [DOI] [PubMed] [Google Scholar]
- 6.Clark LB, Appleby MW, Brunkow ME, Wilkinson JE, Ziegler SF, Ramsdell F. Cellular and molecular characterization of the scurfy mouse mutant. J Immunol. 1999;162:2546–2554. [PubMed] [Google Scholar]
- 7.Bennett CL, Christie J, Ramsdell F, Brunkow ME, Ferguson PJ, Whitesell L, Kelly TE, Saulsbury FT, Chance PF, Ochs HD. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet. 2001;27:20–21. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- 8.Wildin RS, Ramsdell F, Peake J, Faravelli F, Casanova JL, Buist N, Levy-Lahad E, Mazzella M, Goulet O, Perroni L, et al. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat Genet. 2001;27:18–20. doi: 10.1038/83707. [DOI] [PubMed] [Google Scholar]
- 9.Ormandy LA, Hillemann T, Wedemeyer H, Manns MP, Greten TF, Korangy F. Increased populations of regulatory T cells in peripheral blood of patients with hepatocellular carcinoma. Cancer Res. 2005;65:2457–2464. doi: 10.1158/0008-5472.CAN-04-3232. [DOI] [PubMed] [Google Scholar]
- 10.Wang HY, Wang RF. Regulatory T cells and cancer. Curr Opin Immunol. 2007;19:217–223. doi: 10.1016/j.coi.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 11.Fujimoto S, Greene M, Sehon AH. Immunosuppressor T cells in tumor bearing host. Immunol Commun. 1975;4:201–217. doi: 10.3109/08820137409055774. [DOI] [PubMed] [Google Scholar]
- 12.Perry LL, Greene MI. T cell subset interactions in the regulation of syngeneic tumor immunity. Fed Proc. 1981;40:39–44. [PubMed] [Google Scholar]
- 13. Samanta A, Li B, Song X, Bembas K, Zhang G, Katsumata M, Saouaf SJ, Wang Q, Hancock WW, Shen Y, et al. TGF-beta and IL-6 signals modulate chromatin binding and promoter occupancy by acetylated FOXP3. Proc Natl Acad Sci U S A. 2008;105:14023–14027. doi: 10.1073/pnas.0806726105. This article provides compelling evidence that Foxp3 function is regulated in response to extracellular stimuli. TGF-β increased the level of acetylated FOXP3 and enhanced chromatin bound FOXP3. Proinflammatory cytokines Il-6 together with TGF-β limit or reverse FOXP3 function.
- 14. van Loosdregt J, Vercoulen Y, Guichelaar T, Gent YY, Beekman JM, van Beekum O, Brenkman AB, Hijnen DJ, Mutis T, Kalkhoven E, et al. Regulation of Treg functionality by acetylation-mediated Foxp3 protein stabilization. Blood. 2010;115:965–974. doi: 10.1182/blood-2009-02-207118. This article shows p300 and SIRT1 are involved in regulating the dynamic process of Foxp3 acetylation, underlying a mechanism of how HAT or HDAC inhibitors may upregulate the function of Foxp3 and Treg cells.
- 15.Li B, Greene MI. FOXP3 actively represses transcription by recruiting the HAT/HDAC complex. Cell Cycle. 2007;6:1432–1436. [PubMed] [Google Scholar]
- 16. Li B, Samanta A, Song X, Iacono KT, Bembas K, Tao R, Basu S, Riley JL, Hancock WW, Shen Y, et al. FOXP3 interactions with histone acetyltransferase and class II histone deacetylases are required for repression. Proc Natl Acad Sci U S A. 2007;104:4571–4576. doi: 10.1073/pnas.0700298104. This is the first article identifying that activity of FOXP3 is regulated by TIP60 mediated acetylation, revealing the molecular mechanism by which FOXP3 mediated transcription repression may occur.
- 17.Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, Yasayko SA, Wilkinson JE, Galas D, Ziegler SF, Ramsdell F. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27:68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- 18.Walker MR, Kasprowicz DJ, Gersuk VH, Benard A, Van Landeghen M, Buckner JH, Ziegler SF. Induction of FoxP3 and acquisition of T regulatory activity by stimulated human CD4+CD25−T cells. J Clin Invest. 2003;112:1437–1443. doi: 10.1172/JCI19441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Z, Song X, Li B, Greene MI. FOXP3 and its partners: structural and biochemical insights into the regulation of FOXP3 activity. Immunol Res. 2008;42:19–28. doi: 10.1007/s12026-008-8029-x. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Z, Song X, Berezov A, Li B, Greene MI. Structural aspects of the FOXP3 regulatory complex as an immunopharmacological target. Int Immunopharmacol. 2009;9:518–520. doi: 10.1016/j.intimp.2009.01.018. [DOI] [PubMed] [Google Scholar]
- 21.Ziegler SF. FOXP3: of mice and men. Annu Rev Immunol. 2006;24:209–226. doi: 10.1146/annurev.immunol.24.021605.090547. [DOI] [PubMed] [Google Scholar]
- 22.Allan SE, Passerini L, Bacchetta R, Crellin N, Dai M, Orban PC, Ziegler SF, Roncarolo MG, Levings MK. The role of 2 FOXP3 isoforms in the generation of human CD4+ Tregs. J Clin Invest. 2005;115:3276–3284. doi: 10.1172/JCI24685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ono M, Yaguchi H, Ohkura N, Kitabayashi I, Nagamura Y, Nomura T, Miyachi Y, Tsukada T, Sakaguchi S. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature. 2007;446:685–689. doi: 10.1038/nature05673. [DOI] [PubMed] [Google Scholar]
- 24.Jain J, Loh C, Rao A. Transcriptional regulation of the IL-2 gene. Curr Opin Immunol. 1995;7:333–342. doi: 10.1016/0952-7915(95)80107-3. [DOI] [PubMed] [Google Scholar]
- 25.Rooney JW, Sun YL, Glimcher LH, Hoey T. Novel NFAT sites that mediate activation of the interleukin-2 promoter in response to T-cell receptor stimulation. Mol Cell Biol. 1995;15:6299–6310. doi: 10.1128/mcb.15.11.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chatila TA, Blaeser F, Ho N, Lederman HM, Voulgaropoulos C, Helms C, Bowcock AM. JM2, encoding a fork head-related protein, is mutated in X-linked autoimmunity-allergic disregulation syndrome. J Clin Invest. 2000;106:R75–R81. doi: 10.1172/JCI11679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wildin RS, Smyk-Pearson S, Filipovich AH. Clinical and molecular features of the immunodysregulation, polyendocrinopathy, enteropathy, X linked (IPEX) syndrome. J Med Genet. 2002;39:537–545. doi: 10.1136/jmg.39.8.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li B, Samanta A, Song X, Iacono KT, Brennan P, Chatila TA, Roncador G, Banham AH, Riley JL, Wang Q, et al. FOXP3 is a homo-oligomer and a component of a supramolecular regulatory complex disabled in the human XLAAD/IPEX autoimmune disease. Int Immunol. 2007;19:825–835. doi: 10.1093/intimm/dxm043. This study illustrates the molecular mechanism of how FOXP3 mutation leads to IPEX syndrome in human patients. Failure in repressive complex formation due to mutation within the FOXP3 leucine zipper region results in deficient promoter binding and repression of IL-2 production.
- 29.Vidali G, Gershey EL, Allfrey VG. Chemical studies of histone acetylation. The distribution of epsilon-N-acetyllysine in calf thymus histones. J Biol Chem. 1968;243:6361–6366. [PubMed] [Google Scholar]
- 30.Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci U S A. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turner BM. Cellular memory and the histone code. Cell. 2002;111:285–291. doi: 10.1016/s0092-8674(02)01080-2. [DOI] [PubMed] [Google Scholar]
- 32.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 33.Gronroos E, Hellman U, Heldin CH, Ericsson J. Control of Smad7 stability by competition between acetylation and ubiquitination. Mol Cell. 2002;10:483–493. doi: 10.1016/s1097-2765(02)00639-1. [DOI] [PubMed] [Google Scholar]
- 34.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 35.Imhof A, Yang XJ, Ogryzko VV, Nakatani Y, Wolffe AP, Ge H. Acetylation of general transcription factors by histone acetyltransferases. Curr Biol. 1997;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 36.LeDizet M, Piperno G. Identification of an acetylation site of Chlamydomonas alpha-tubulin. Proc Natl Acad Sci U S A. 1987;84:5720–5724. doi: 10.1073/pnas.84.16.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carrozza MJ, Utley RT, Workman JL, Cote J. The diverse functions of histone acetyltransferase complexes. Trends Genet. 2003;19:321–329. doi: 10.1016/S0168-9525(03)00115-X. [DOI] [PubMed] [Google Scholar]
- 38.Polevoda B, Sherman F. N-alpha-terminal acetylation of eukaryotic proteins. J Biol Chem. 2000;275:36479–36482. doi: 10.1074/jbc.R000023200. [DOI] [PubMed] [Google Scholar]
- 39.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 40.Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 41.Yang XJ, Ogryzko VV, Nishikawa J, Howard BH, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 42. Yang XJ. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 2004;32:959–976. doi: 10.1093/nar/gkh252. This paper provides a comprehensive overview about the structure and function of histone acetyltrasferases and their involvement in oncogenesis.
- 43.Tao R, de Zoeten EF, Ozkaynak E, Chen C, Wang L, Porrett PM, Li B, Turka LA, Olson EN, Greene MI, et al. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat Med. 2007;13:1299–1307. doi: 10.1038/nm1652. [DOI] [PubMed] [Google Scholar]
- 44.Kamine J, Elangovan B, Subramanian T, Coleman D, Chinnadurai G. Identification of a cellular protein that specifically interacts with the essential cysteine region of the HIV-1 Tat transactivator. Virology. 1996;216:357–366. doi: 10.1006/viro.1996.0071. [DOI] [PubMed] [Google Scholar]
- 45.Legube G, Trouche D. Identification of a larger form of the histone acetyl transferase Tip60. Gene. 2003;310:161–168. doi: 10.1016/s0378-1119(03)00547-x. [DOI] [PubMed] [Google Scholar]
- 46.Ran Q, Pereira-Smith OM. Identification of an alternatively spliced form of the Tat interactive protein (Tip60), Tip60(beta) Gene. 2000;258:141–146. doi: 10.1016/s0378-1119(00)00410-8. [DOI] [PubMed] [Google Scholar]
- 47.Utley RT, Cote J. The MYST family of histone acetyltransferases. Curr Top Microbiol Immunol. 2003;274:203–236. doi: 10.1007/978-3-642-55747-7_8. [DOI] [PubMed] [Google Scholar]
- 48.Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 49.Nordentoft I, Jorgensen P. The acetyltransferase 60 kDa trans-acting regulatory protein of HIV type 1-interacting protein (Tip60) interacts with the translocation E26 transforming-specific leukaemia gene (TEL) and functions as a transcriptional co-repressor. Biochem J. 2003;374:165–173. doi: 10.1042/BJ20030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao H, Chung J, Kao HY, Yang YC. Tip60 is a co-repressor for STAT3. J Biol Chem. 2003;278:11197–11204. doi: 10.1074/jbc.M210816200. [DOI] [PubMed] [Google Scholar]
- 51. Col E, Caron C, Chable-Bessia C, Legube G, Gazzeri S, Komatsu Y, Yoshida M, Benkirane M, Trouche D, Khochbin S. HIV-1 Tat targets Tip60 to impair the apoptotic cell response to genotoxic stresses. EMBO J. 2005;24:2634–2645. doi: 10.1038/sj.emboj.7600734. In this study, Tip60 was identified as a substrate for CBP/p300 acetylase activity and CBP/p300 assiciated E4-type ubiquitin-ligase activity. Tat may use this mechanism to induce degradation of Tip60, impairing Tip60 mediated cell apoptosis.
- 52.Akhtar A, Zink D, Becker PB. Chromodomains are protein-RNA interaction modules. Nature. 2000;407:405–409. doi: 10.1038/35030169. [DOI] [PubMed] [Google Scholar]
- 53.Gaughan L, Brady ME, Cook D, Neal DE, Robson CN. Tip60 is a co-activator specific for class I nuclear hormone receptors. J Biol Chem. 2001;276:46841–46848. doi: 10.1074/jbc.M103710200. [DOI] [PubMed] [Google Scholar]
- 54.Kimura A, Horikoshi M. Tip60 acetylates six lysines of a specific class in core histones in vitro. Genes Cells. 1998;3:789–800. doi: 10.1046/j.1365-2443.1998.00229.x. [DOI] [PubMed] [Google Scholar]
- 55.Gaughan L, Logan IR, Cook S, Neal DE, Robson CN. Tip60 and histone deacetylase 1 regulate androgen receptor activity through changes to the acetylation status of the receptor. J Biol Chem. 2002;277:25904–25913. doi: 10.1074/jbc.M203423200. [DOI] [PubMed] [Google Scholar]
- 56.Patel JH, Du Y, Ard PG, Phillips C, Carella B, Chen CJ, Rakowski C, Chatterjee C, Lieberman PM, Lane WS, et al. The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol Cell Biol. 2004;24:10826–10834. doi: 10.1128/MCB.24.24.10826-10834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci U S A. 2005;102:13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang J, Chen J. SIRT1 regulates autoacetylation and histone acetyltransferase activity of TIP60. J Biol Chem. 2010;285:11458–11464. doi: 10.1074/jbc.M109.087585. This paper shows that auto-acetylation of Tip60 is promoted in response to UV damage and decreased in the presence of SIRT1. The mechanism of how acetylation status affects the function of Tip60 is revealed.
- 59.Yamagata K, Kitabayashi I. Sirt1 physically interacts with Tip60 and negatively regulates Tip60-mediated acetylation of H2AX. Biochem Biophys Res Commun. 2009;390:1355–1360. doi: 10.1016/j.bbrc.2009.10.156. [DOI] [PubMed] [Google Scholar]
- 60.Tang Y, Luo J, Zhang W, Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell. 2006;24:827–839. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 61.Brady ME, Ozanne DM, Gaughan L, Waite I, Cook S, Neal DE, Robson CN. Tip60 is a nuclear hormone receptor coactivator. J Biol Chem. 1999;274:17599–17604. doi: 10.1074/jbc.274.25.17599. [DOI] [PubMed] [Google Scholar]
- 62.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 63.Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 64.Eckner R, Ewen ME, Newsome D, Gerdes M, DeCaprio JA, Lawrence JB, Livingston DM. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 65.Yao TP, Oh SP, Fuchs M, Zhou ND, Ch'ng LE, Newsome D, Bronson RT, Li E, Livingston DM, Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 66.Kung AL, Rebel VI, Bronson RT, Ch'ng LE, Sieff CA, Livingston DM, Yao TP. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev. 2000;14:272–277. [PMC free article] [PubMed] [Google Scholar]
- 67.Kawasaki H, Eckner R, Yao TP, Taira K, Chiu R, Livingston DM, Yokoyama KK. Distinct roles of the co-activators p300 and CBP in retinoic-acid-induced F9-cell differentiation. Nature. 1998;393:284–289. doi: 10.1038/30538. [DOI] [PubMed] [Google Scholar]
- 68.Arany Z, Sellers WR, Livingston DM, Eckner R. E1A-associated p300 and CREB-associated CBP belong to a conserved family of coactivators. Cell. 1994;77:799–800. doi: 10.1016/0092-8674(94)90127-9. [DOI] [PubMed] [Google Scholar]
- 69. Thompson PR, Wang D, Wang L, Fulco M, Pediconi N, Zhang D, An W, Ge Q, Roeder RG, Wong J, et al. Regulation of the p300 HAT domain via a novel activation loop. Nat Struct Mol Biol. 2004;11:308–315. doi: 10.1038/nsmb740. In this study, a novel mechanism identifying the regulation of p300 activity via an autoregulatory loop is described. Acetylation of this loop relieves its repression of p300 activity.
- 70.Kwok RP, Lundblad JR, Chrivia JC, Richards JP, Bachinger HP, Brennan RG, Roberts SG, Green MR, Goodman RH. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 71.Blobel GA. CREB-binding protein and p300: molecular integrators of hematopoietic transcription. Blood. 2000;95:745–755. [PubMed] [Google Scholar]
- 72.Xu L, Glass CK, Rosenfeld MG. Coactivator and corepressor complexes in nuclear receptor function. Curr Opin Genet Dev. 1999;9:140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- 73.Korzus E, Torchia J, Rose DW, Xu L, Kurokawa R, McInerney EM, Mullen TM, Glass CK, Rosenfeld MG. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 74.Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133:612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, McMahon SB. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24:841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Black JC, Mosley A, Kitada T, Washburn M, Carey M. The SIRT2 deacetylase regulates autoacetylation of p300. Mol Cell. 2008;32:449–455. doi: 10.1016/j.molcel.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.de Zoeten EF, Wang L, Sai H, Dillmann WH, Hancock WW. Inhibition of HDAC9 increases T regulatory cell function and prevents colitis in mice. Gastroenterology. 138:583–594. doi: 10.1053/j.gastro.2009.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saouaf SJ, Li B, Zhang G, Shen Y, Furuuchi N, Hancock WW, Greene MI. Deacetylase inhibition increases regulatory T cell function and decreases incidence and severity of collagen-induced arthritis. Exp Mol Pathol. 2009;87:99–104. doi: 10.1016/j.yexmp.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wang L, de Zoeten EF, Greene MI, Hancock WW. Immunomodulatory effects of deacetylase inhibitors: therapeutic targeting of FOXP3+ regulatory T cells. Nat Rev Drug Discov. 2009;8:969–981. doi: 10.1038/nrd3031. This paper provides an excellent overview of the effect of HDAC inhibitors in promoting the production and repressive function of Treg cells, which may be of therapeutic benefit in cancer treatment, immuno-inflammatory disorders and organ/tissue transplantation.
- 80.Fujimoto S, Greene MI, Sehon AH. Regulation of the immune response to tumor antigens. II. The nature of immunosuppressor cells in tumor-bearing hosts. J Immunol. 1976;116:800–806. [PubMed] [Google Scholar]
- 81.Betts G, Twohig J, Van den Broek M, Sierro S, Godkin A, Gallimore A. The impact of regulatory T cells on carcinogen-induced sarcogenesis. Br J Cancer. 2007;96:1849–1854. doi: 10.1038/sj.bjc.6603824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Curiel TJ, Coukos G, Zou L, Alvarez X, Cheng P, Mottram P, Evdemon-Hogan M, Conejo-Garcia JR, Zhang L, Burow M, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med. 2004;10:942–949. doi: 10.1038/nm1093. [DOI] [PubMed] [Google Scholar]
- 83.Liu VC, Wong LY, Jang T, Shah AH, Park I, Yang X, Zhang Q, Lonning S, Teicher BA, Lee C. Tumor evasion of the immune system by converting CD4+CD25− T cells into CD4+CD25+ T regulatory cells: role of tumor-derived TGF-beta. J Immunol. 2007;178:2883–2892. doi: 10.4049/jimmunol.178.5.2883. [DOI] [PubMed] [Google Scholar]
- 84.Valzasina B, Piconese S, Guiducci C, Colombo MP. Tumor-induced expansion of regulatory T cells by conversion of CD4+CD25− lymphocytes is thymus and proliferation independent. Cancer Res. 2006;66:4488–4495. doi: 10.1158/0008-5472.CAN-05-4217. [DOI] [PubMed] [Google Scholar]
- 85.Ghiringhelli F, Puig PE, Roux S, Parcellier A, Schmitt E, Solary E, Kroemer G, Martin F, Chauffert B, Zitvogel L. Tumor cells convert immature myeloid dendritic cells into TGF-beta-secreting cells inducing CD4+CD25+ regulatory T cell proliferation. J Exp Med. 2005;202:919–929. doi: 10.1084/jem.20050463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Beyer M, Kochanek M, Giese T, Endl E, Weihrauch MR, Knolle PA, Classen S, Schultze JL. In vivo peripheral expansion of naive CD4+CD25high FoxP3+ regulatory T cells in patients with multiple myeloma. Blood. 2006;107:3940–3949. doi: 10.1182/blood-2005-09-3671. [DOI] [PubMed] [Google Scholar]
- 87.Perry LL, Kripke ML, Benacerraf B, Dorf ME, Greene MI. Regulation of the immune response to tumor antigen. VIII. The effects of host specific anti-I-J antibodies on the immune response to tumors of different origin. Cell Immunol. 1980;51:349–359. doi: 10.1016/0008-8749(80)90266-x. [DOI] [PubMed] [Google Scholar]
- 88.Drebin JA, Waltenbaugh C, Schatten S, Benacerraf B, Greene MI. Inhibition of tumor growth by monoclonal anti-I-J antibodies. J Immunol. 1983;130:506–509. [PubMed] [Google Scholar]