Abstract
We have identified and characterized a zebrafish connexin, Cx30.3. Sequence similarity analyses suggested that Cx30.3 was orthologous to both mammalian Cx26 and Cx30, known to play important roles in the skin and inner ear of mammals. Analysis of mRNA expression showed that Cx30.3 was present in early embryos, and was highly abundant in skin, but also detected in other tissues including fins, inner ear, heart, and the retina. Injection of Cx30.3 cRNA into Xenopus oocytes elicited robust intercellular coupling with voltage gating sensitivity similar to mammalian Cx26 and Cx30. The similarities in functional properties and expression patterns suggest that Cx30.3, like mammalian Cx26 and Cx30, may play a significant role in skin development, hearing and balance in zebrafish. Thus, zebrafish could potentially serve as an excellent model to study disorders of the skin and deafness that result from human connexin mutations.
Keywords: connexin, gap junction, zebrafish, embryo, skin, inner ear
INTRODUCTION
Gap junction channels are highly specific cell membrane structures that allow direct cell-to-cell exchange of ions, small metabolites, and signaling molecules between adjacent cells, thus coordinating cellular activities during embryonic development and in adult tissues. Loss, or misregulation, of gap junctional communication can consequently lead to a variety of human diseases (White and Paul, 1999, Gerido and White, 2004, Dobrowolski and Willecke, 2009). The connexins (Cx) are a group of proteins originally identified as structural subunits of gap junctions (Goodenough, 1974). Each connexin forms channels with unique properties of size selectivity, ion permeability and gating (Bruzzone et al., 1996). The connexin genes belong to a large family with ≥20 members identified in the mouse and human genomes (Sohl and Willecke, 2004). Connexin genes show distinct, but overlapping, spatial-temporal expression patterns, as almost all cell types express connexins and one cell often expresses more than one type of connexin (Bruzzone et al., 1996, White, 2002, White, 2003, Oyamada et al., 2005).
We have cloned and characterized several zebrafish connexin genes in an attempt to unravel the molecular mechanisms regulating the expression and function of connexin genes during embryonic development (Cason et al., 2001, McLachlan et al., 2003, Cheng et al., 2004, Christie et al., 2004). As a part of this effort, we previously reported the isolation and characterization of zebrafish cx48.5 (Cheng et al., 2004). In the course of analyzing the regulatory elements involved in the transcription of cx48.5, we sequenced an additional ~12 kb upstream fragment that resulted in the discovery of an additional zebrafish connexin gene, cx30.3, which is the subject of the present report.
Abnormal function of specific connexin genes has been linked to several diseases, and mutations in human Cx26 and Cx30 cause genetic deafness and skin disease (Richard, 2000, Gerido and White, 2004). Phylogenetic comparison suggested that Cx30.3 was highly related to both mammalian Cx26 and Cx30. Analysis of mRNA expression showed that Cx30.3 was abundant in skin, but also present in the inner ear. Injection of Cx30.3 cRNA into Xenopus oocytes produced gap junctional coupling with functional properties similar to Cx26 and Cx30. The sequence homology, tissue expression and function properties of Cx30.3 show that it shares many features with Cx26 and Cx30, and suggests that it may have similar functions in the inner ear and skin. Thus, Cx30.3 could be a useful tool to model the role of human Cx26 and Cx30 mutations in the development and pathophysiology of the cochlea and epidermis.
RESULTS
Identification of the genomic and amino acid sequences of zebrafish Cx30.3
DNA sequencing of a 12 kb DNA fragment upstream of the cx48.5 gene revealed the presence of an additional coding region. A BLAST search indicated that this new gene matched a previously identified connexin gene, originally referred to as cx33.8 (GenBank accession numbers AY135446, BC095350, BC165699, NM212825), that had not been fully described. Further analysis of this novel clone confirmed that this connexin gene contains an open reading frame of 804 nucleotides, encoding a protein of 267 amino acids with a predicted molecular weight of 30,323 Da. (GenBank accession number GQ469999). Hereafter, we will refer to the gene and its protein as cx30.3 and Cx30.3, respectively, according to the conventional Cx molecular weight nomenclature (Beyer et al., 1990).
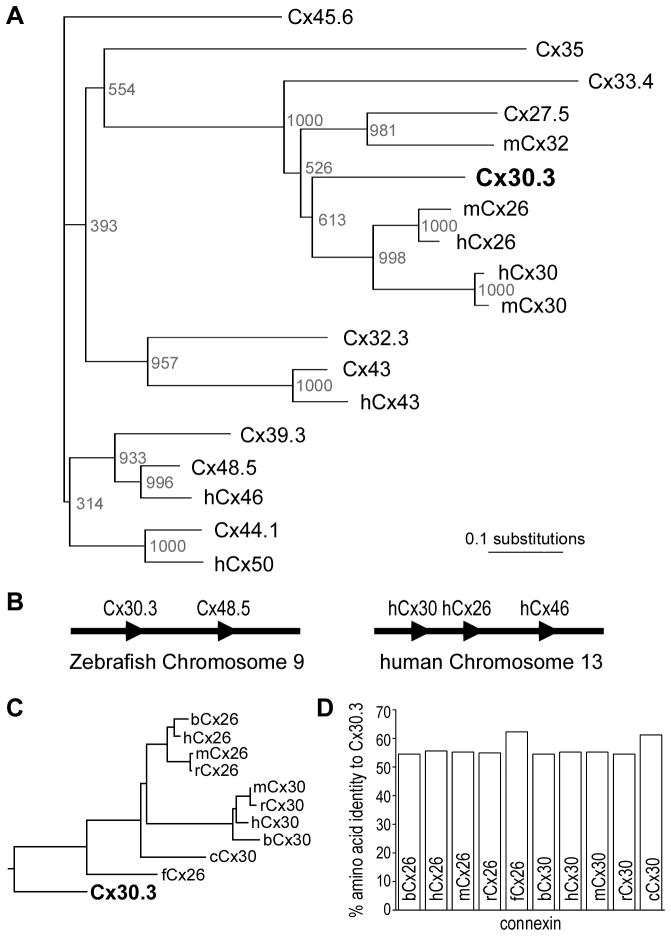
Using transmembrane domain prediction software (DAS, Cserzo et al., 1997), the amino acid sequence of Cx30.3 was found to have a similar topology to that of other connexins, with four transmembrane domains, two extracellular loops, one intracellular loop, and cytoplasmic N-and C- termini (John and Revel, 1991, Rahman and Evans, 1991, Dahl et al., 1992, Unger et al., 1999). Each of the two extracellular loops contains three conserved cysteines (Fig. 1). Multiple protein sequence comparisons showed that Cx30.3 was most closely related to mammalian Cx26 and Cx30 (Fig. 2A), as had also been suggested by other researchers (Cruciani and Mikalsen, 2006, Eastman et al., 2006, Cruciani and Mikalsen, 2007).
Fig. 1. Amino acid sequence alignment of Cx30.3.
Zebrafish Cx30.3, and two closely related sequences, human Cx26 (hCx26) and Cx30 (hCx30) were aligned by the CLUSTAL W method. Solid lines under the sequences indicate the four predicted transmembrane domains (M1, M2, M3, and M4). The asterisks indicate the six conserved cysteine residues within the two predicted extracellular loops of Cx30.3. Yellow shading highlights residues identical to Cx30.3.
Fig. 2. Phylogenetic analysis and genomic organization of zebrafish and mammalian connexins.
(A). A neighbour-joining tree shows that the zebrafish Cx30.3 amino acid sequence is most closely related to human Cx26 and Cx30. Bootstrap values were based on 1000 neighboring replicates and indicated as node labelling. (B). Similar to a cluster of three connexins (Cx30, Cx26, Cx46) on human chromosome 13, cx48.5 (the zebrafish ortholog of Cx46) is closely linked to cx30.3 on chromosome 9. Connexin genes are represented by arrowheads; gene distance is not drawn in scale. (C). When sequence comparison was limited to Cx26 and Cx30 orthologs, Cx30.3 was most closely related to chicken Cx30 and frog Cx26 sequences, and equally related to mammalian orthologs from bovine, human, mouse and rat. (D). Pair wise sequence comparison of Cx30.3 to both Cx26 and Cx30 orthologs was equally high, and ranged from 55 to 62% identity.
Examination of the genomic organization of zebrafish and human connexin genes revealed that similar clusters of connexins had been maintained during evolution. In humans, Cx30, Cx26, and Cx46 are all found physically linked in the same gene orientation, on chromosome 13. In zebrafish, cx30.3 and cx48.5 (the ortholog of human Cx46) are physically linked on chromosome 9, in the same manner (Fig. 2B). These similarities (orthologous connexin genes in genomic clusters) suggest that the Cx30.3, Cx26 and Cx30 most likely evolved from a common ancestor, as had also been observed by others (Cruciani and Mikalsen, 2006, Eastman et al., 2006).
Further alignment of Cx30.3’s amino acid sequence with ten different Cx26 and Cx30 orthologs from the bovine, human, mouse, rat, chicken and frog genomes revealed an equally high degree of identity with both proteins (Fig. 2C). Pair wise analysis showed ~55% identity with any of the mammalian Cx26 or Cx30 orthologs, which increased to ≥60% identity when the Cx30.3 sequence was compared to the chicken and frog proteins. Taken together, these data suggested that zebrafish Cx30.3 is equally orthologous to Cx26 as it is to Cx30.
Identification of the transcriptional start sites and UTRs of zebrafish cx30.3
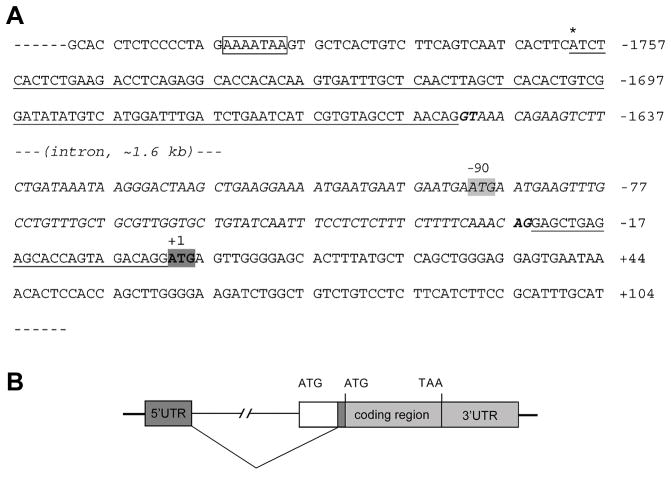
Using the rapid amplification of cDNA ends method (RACE), we identified the transcription start site and 5′ and 3′ untranslated regions (UTRs) of zebrafish cx30.3. The 5′ RACE nested PCR generated a single band of ~250 bp in size; and for 3′ RACE, the nested PCR produced a single band of ~1.4 kb in size (data not shown). Three independent positive clones were sequenced for each of the 5′ and 3′ UTRs. RNA sequencing results were then aligned against the genomic DNA sequence, resulting in the identification of the transcriptional start site and the intro-exon genomic structure of zebrafish cx30.3 (Fig. 3). The 5′ RACE results identified a single transcriptional start site for cx30.3, which is located at a position −1760 nucleotides upstream of the ATG translation start codon (the adenine in the ATG codon was assigned as position +1). The 5′ UTR of cx30.3 is interrupted by an intron from position −1561 to −24. Pre-mRNA splicing uses the conserved “GU-AG” rule. The 3′ UTR is 1308 nucleotides in size, from position +805 to position +2113 downstream of the ATG codon, and contains the conserved hexamer “AAUAAA” poly (A) signal (Nag et al., 2006). The 5′ and 3′ RACE data suggest that cx30.3 gene contains two exons, with an intron interrupting its 5′ UTR. The complete coding region and 3′ UTR is located in exon 2.
Fig. 3.
Intron/Exon Structure of Zebrafish cx30.3 (A). Sequence of the 5′UTR (underlined) and part of the coding region. The translational start site (ATG) identified in the present study is highlighted in a dark box (GenBank accession number: GQ469999). The position of the adenine nucleotide is designated as +1, with its upstream nucleotide as −1 and downstream as +2. The transcriptional start site is indicated by asterisk (*) above the sequence at −1760, and a “TATA” like element was found upstream in the core promoter (indicated in a box). The 5′UTR of cx30.3 is interrupted by a single intron of about 1.6 kb in size (italic letters). The previously reported translational start site (ATG) is highlighted in a gray box (GenBank accession number: AY135446), with its adenine nucleotide located at −90, which is within the intron region identified in the present study. (B). Sketch of intron/exon genomic structure. Filled boxes represent both exons of zebrafish cx30.3 transcript, while solid lines represent genomic sequences, or the intron (not drawn on scale). The blank box represents the sequence between the previously reported translational start site and the second exon identified in the present study.
mRNA expression of zebrafish cx30.3
Using RT-PCR, we studied the mRNA expression of cx30.3 in zebrafish embryos in stages spanning from 0.5 to 5 days post fertilization (dpf). RNA samples were isolated from two independently collected batches of embryos, and reverse transcribed to obtain independent cDNA samples. Results from these parallel RT-PCR experiments were consistent with each other. The zebrafish elongation factor 1α (ef1α) gene was used as a positive control, and the primers were designed to anneal to different exons, so that amplification from cDNA templates will result in shorter PCR fragments than amplification from genomic DNA templates (569 versus 733 bp). Therefore, the parallel amplification of ef1α also serves as an indicator for the integrity and non-genomic-contamination of the cDNA samples (Cheng et al., 2004, Christie et al., 2004). As shown in Fig. 4A, cx30.3 transcripts were not detected in 0.5 dpf embryos, but were readily detected at 1 dpf and later stages.
Fig. 4. Temporal and spatial analyses of cx30.3 mRNA expression.
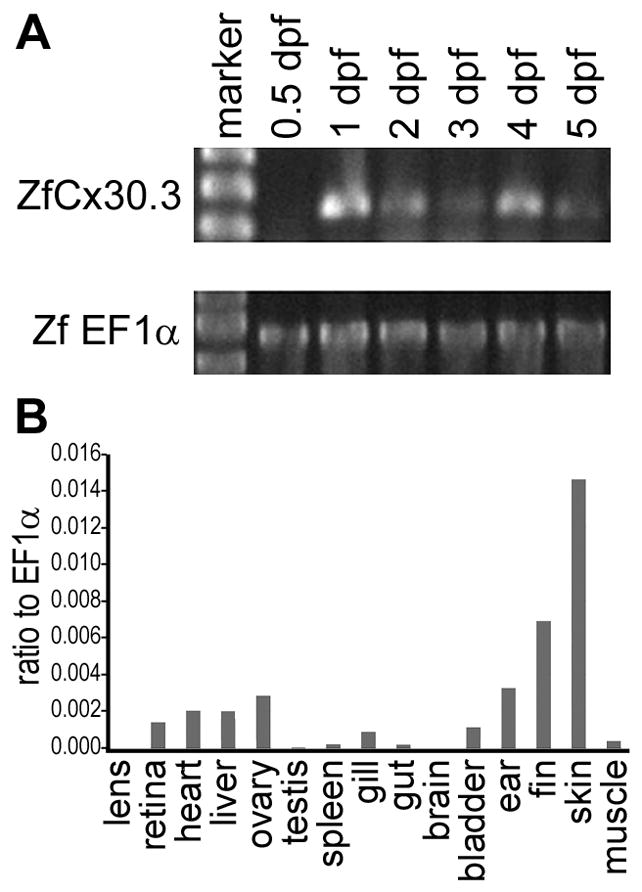
(A) Expression of cx30.3 was detected at the 1 day post fertilization (dpf) stage, and thereafter maintained throughout embryonic development. The zebrafish ef1α gene was used as positive control to demonstrate the integrity and genomic-DNA-contamination-free status of the cDNA samples. (B) cx30.3 mRNA is predominantly expressed in the adult skin, fin, inner ear, ovary, liver, heart, retina, while a relatively low level of cx30.3 expression was also detected in the gill, swim bladder, skeletal muscle, spleen and gut. The relative level of mRNA expression was calculated by comparing ΔCt between mean Cts of zebrafish cx30.3 and ef1α.
Using real-time RT-PCR, we further analyzed the tissue distribution pattern of cx30.3 mRNA expression among different tissues in adult zebrafish. We again used ef1α as a reference gene, as ef1α has been shown to be a suitable reference gene for zebrafish real-time PCR analyses, both in time-course and tissue based experiments (Tang et al., 2007). cx30.3 expression levels were highest in the skin, fins, and inner ear of adult zebrafish (Fig. 4B). Low levels of expression were also detected in the heart, retina, liver, ovary, swim bladder, and the gill.
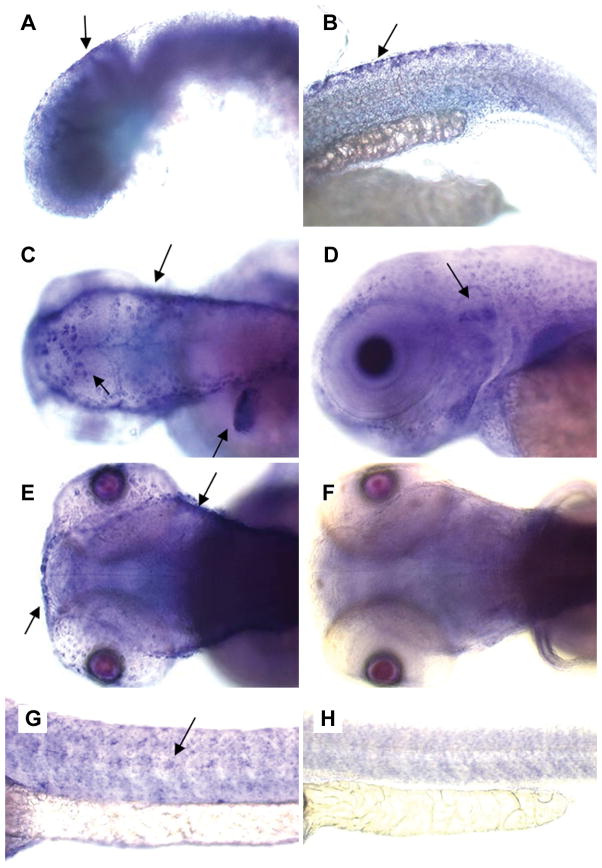
Whole mount in situ hybridization was performed with embryos at different embryonic stages from 1–5 dpf. The anti-sense and sense probes were ~1.4 kb in size, and were designed to target the 3′UTR plus a short fragment from the 3′ end of the coding region. These connexin sequences are usually less conserved than other regions and provide higher specificity for mRNA hybridization. A cx30.3-specific in situ hybridization signal was detected in 1 dpf stage embryos, in the otic vesicle (Fig. 5, A) and dorsal fins (Fig. 5, B). At 2 and 3 dpf, cx30.3-specific mRNA expression was readily detected in the skin (Fig. 5, C), inner ear (Fig. 5, D), and the lateral line (Fig. 5, E & G). This pattern remained unchanged at 4 and 5 dpf (data not shown). In order to obtain a more detailed picture regarding the internal expression pattern, we embedded embryos in plastic and cut 5 μm sections with a microtome. Images taken from 2 dpf stage embryos are presented in Fig. 6, and the staining patterns observed in later stage embryos were similar. As seen in these images, cx30.3 mRNA signals were strongly detected in the epithelial cells of the skin, in addition to cells in the inner ear. Signals were not detected in the heart or other embryonic tissues, although the more sensitive technique of real-time PCR, suggested that cx30.3 mRNA expression occurs in a number of these adult tissues (Fig. 4B). Either cx30.3 is not expressed during 1–5 dpf of development in these organs, or the transcript levels are simply too low to be detected by our in situ hybridization protocol.
Fig. 5. In situ hybridization of Cx30.3 mRNA in embryos.
Embryos were stained by whole-mount in situ hybridization with a cx30.3 anti-sense riboprobe. Signal was detected (and highlighted with arrows) in 1 dpf stage embryos, in the otic vesicle (A) and dorsal fins (B). At 2 dpf (C, G, H) or 3 dpf (D, E, F), cx30.3 mRNA signal was readily detected in the skin (C), the inner ear (D), the lateral line (E, G), and the fins (C). Sense-probe controls are shown in (F) and (H).
Fig. 6. Sections of cx30.3 in situ hybridization stained embryos.
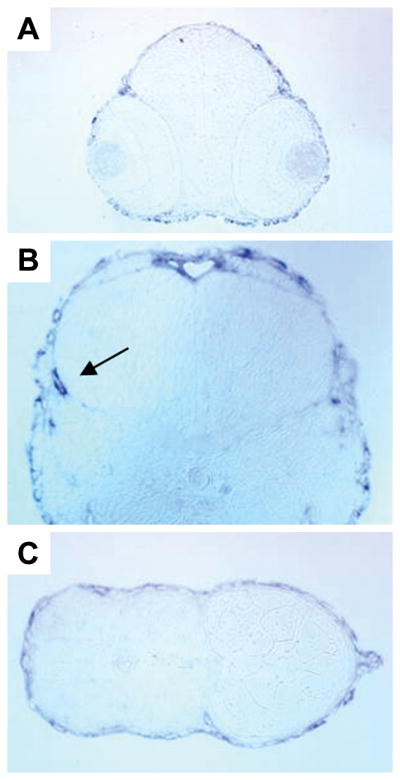
Embryos were stained by whole-mount in situ hybridization with cx30.3 anti-senses riboprobe, embedded in plastic and 5 μm sections were cut with a microtome. Only sections from embryos at 2 dpf are shown, the staining pattern in later stage embryos is similar. (A) Transverse cross-section of embryonic eyes, showing a lack of staining of the probe. (B) The developing inner ear with probe labeling the otic vesicle (arrow). (C) A transverse section of the embryonic fish tail and fin, with probe accumulating around the peripheral skin and on the fin (far right).
Functional analysis of zebrafish Cx30.3 channels
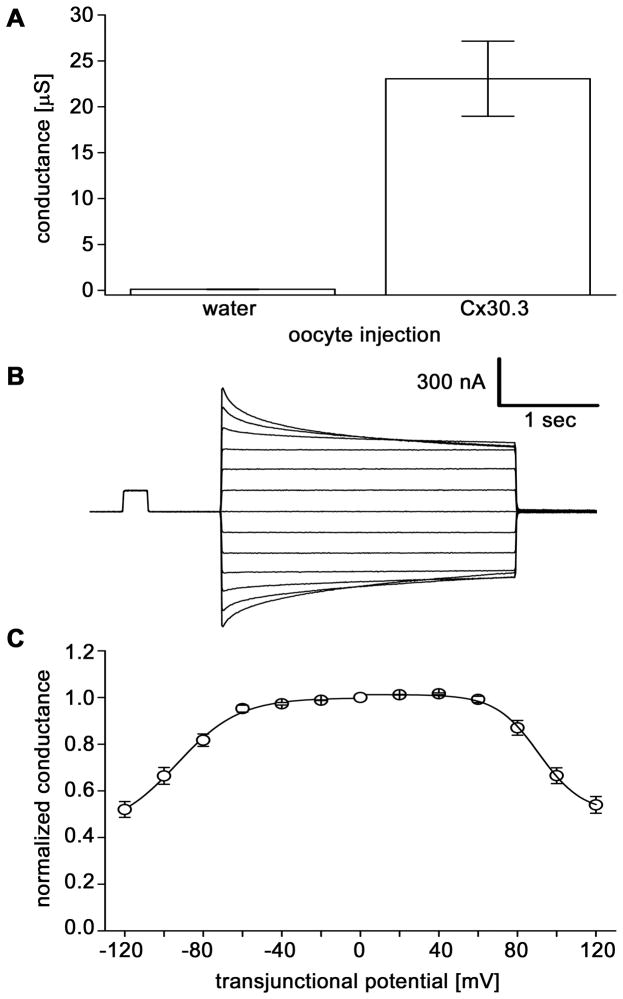
To determine whether Cx30.3 could form intercellular channels, we used the paired Xenopus oocyte expression system (Dahl, et al., 1987). Oocytes were clamped at −40 mV for measurements of junctional conductance (Gj). As shown in Fig. 7A, Cx30.3 induced high levels of electrical coupling 24–48 hours after RNA injection. The mean conductance of paired Cx30.3-injected cells was 23.1 ± 4.1 μS (mean ± SE, n=13). In contrast, the mean background conductance value, measured between oligo-injected control oocytes, was nearly 200 fold lower at 0.12 ± 0.03 μS (n=19). Thus, the expression of Cx30.3 resulted in the formation of functional gap junction channels.
Fig. 7. Cx30.3 forms functional channels that are gated by transjunctional voltage.
(A). Junctional conductance (Gj) developed between pairs of Xenopus oocytes as measured by dual voltage clamp. Bars show the mean ± SE of 13–19 pairs. (B). Voltage gating behavior of gap junction channels formed by Cx30.3. A time dependent decay of junctional currents (Ij) was induced by transjunctional voltage (Vj) steps. At Vj steps > ± 60 mV, Ij decayed symmetrically over the time course of the voltage step. (C). The relationship of Vj to steady-state junctional conductance (Gjss) normalized to the values obtained at ± 20 mV for Cx30.3. Solid lines represent the best fits to Boltzmann equations, whose parameters are: V0=91, Gmin=0.51, A=0.09 for +Vjs and V0=−93, Gmin=0.43, A=0.06 for −Vjs. The voltage gating of intercellular channels composed of zebrafish Cx30.3 displayed a high degree of conservation to human and rat Cx26.
To characterize the gating behavior of channels comprised of Cx30.3, we analyzed their voltage dependence. A representative family of junctional currents (Ij) evoked by voltage steps of opposite polarities and increasing amplitude (Fig. 7B) shows that Ij decreased in a time and voltage-dependent manner for transjunctional voltages > ± 60 mV. The steady state voltage dependence of Cx30.3 channels was analyzed by plotting junctional conductance as a function of transjunctional potential (Vj, Fig. 7C). Gj values for steady-state junctional conductance (Gjss) were normalized to the maximal conductance measured at the lowest Vj (=20 mV). No fast gating effects (< 5ms) of voltage on these channels were observed (data not shown). In contrast, Gjss was dependent on voltage and this plot was fitted to a Boltzmann relation (Spray et al., 1981) whose parameters were comparable to data generated with rat and human Cx26 in paired oocytes (Barrio et al., 1991;Mese et al., 2004). In contrast, mammalian Cx30 showed greater voltage dependence than Cx30.3 (Dahl et al., 1996). This analysis clearly showed that zebrafish Cx30.3 channels display weak voltage sensitivity like their mammalian Cx26 orthologs.
DISCUSSION
Cx30.3 is the zebrafish ortholog of mammalian Cx26 and Cx30
We have identified a new connexin gene, cx30.3, by genomic sequencing. A BLAST search revealed that a similar sequence had initially been deposited in GenBank, and named cx33.8 (accession number AY135446). Using 5′ and 3′ RACE, we identified the open reading frame (ORF) of this novel connexin gene and following careful comparison of the ORF with that of the previously deposited cx33.8 in GenBank, we found that an upstream ATG sequence located in the first intron was mistakenly considered as the translation start codon of cx33.8, thus giving an inaccurate prediction of its molecular weight and an erroneous name. The incorrect translation start codon was also noted by others, who then used the corrected translation for comparative analysis (Cruciani and Mikalsen, 2006, Eastman et al., 2006, Cruciani and Mikalsen, 2007). The predicted amino acid sequence of Cx30.3 is consistent with all the features of a typical connexin gene, having four transmembrane domains and two extracellular loops with three highly conserved cysteine residues in each. Phylogenetic analyses suggest that zebrafish Cx30.3 is equally orthologous to both mammalian Cx26 and Cx30 (Fig. 2C). Interestingly, the chicken and frog sequences did not cluster within the mammalian Cx26 and Cx30 groups, but were closer to zebrafish Cx30.3.
Using RNA sequencing data generated from RACE analysis, we have verified both the correct translation start codon, the 5′ and 3′ UTRs, and the intron-exon structure of the zebrafish cx30.3 gene. The 5′RACE strategy takes advantage of the 5′ cap to selectively capture full-length 5′ ends of mRNAs, thus making it feasible to map the transcriptional start sites of a gene. This method has been demonstrated to be comparable and, in many cases, more accurate than S1 nuclease protection, RNase protection, and primer extension approaches (Liu and Gorovsky, 1993, Schaefer, 1995). 5′ RACE identified a single transcriptional start site for cx30.3, and a TATA box located 28 bp upstream appears to be responsible for the transcriptional initiation. 3′ RACE identified the complete 3′ UTR sequence of cx30.3. These RACE experiments suggest that cx30.3 gene contains two exons, with its 5′ UTR interrupted by an intron. A two-exon genomic structure module has been described as the common gene structure for the vast majority of connexin genes (Sohl and Willecke, 2004), and zebrafish cx30.3 follows this rule. We have deposited the full-length cDNAsequence of cx30.3 in GenBank (accession number GQ469999).
The zebrafish Cx30.3 expression pattern is similar to mammalian Cx26 and Cx30
Using RT-PCR and in situ hybridization, we investigated the transcription of zebrafish cx30.3 during embryonic development and in adult tissues. These data clearly demonstrated that the expression of Cx30.3 starts at early embryonic stages and is strongly expressed in some adult tissues, including the inner ear, lateral line, the fins and the skin. Interestingly, its mammalian orthologs, Cx26 and Cx30, are similarly expressed and mutations in human Cx26 and Cx30 cause deafness and skin diseases (Richard, 2000, Gerido and White, 2004). In the human inner ear, Cx26 and Cx30 are present in the supporting epithelial cells of the cochlea and are thought to play an essential role in the recycling of potassium ions during auditory transduction (Kikuchi et al., 2000, Lefebvre and Van De Water, 2000). In human skin, Cx26 is expressed in hair follicles, eccrine sweat glands and in the basal keratinocytes of the palmar and plantar epidermis, while Cx30 is found in the upper, differentiated epidermal layers (Richard, 2000, Richard, 2005, Mese et al., 2007). As demonstrated in the present study, zebrafish Cx30.3 has similar expression patterns to its mammalian orthologs. If Cx30.3 plays a similar role in development and homeostasis in zebrafish, these animals could serve as a valuable research model for human hearing defects or skin diseases, as well as evaluating therapeutic entities to ameliorate such disorders.
Hair cells in the inner ear are involved in the sensory perception of sound and motion in mammals. Like other vertebrates, zebrafish also uses hair cells to detect sound and motion. Although the anatomy of the zebrafish inner ear differs significantly from that of the human inner ear, many of the same genetic pathways are utilized for the development and function of the inner ear in all vertebrates (Whitfield, 2002). For example, myosin VIIa is required and functions the same way in hair cells, whether in human or zebrafish (Hasson, 1999). In addition, fish have a lateral line system of hair cells, which are assembled to form the so-called “neuromasts”, the sensory organs of the lateral line. Hair cells in neuromasts are directly exposed to the surrounding water and sense its movement along the surface of the head and body, and play essential roles in a variety of behaviors, including prey detection, predator avoidance, school swimming, and sexual courtship (Ghysen and Dambly-Chaudiere, 2004). It is the neuromasts of the lateral line that make zebrafish an excellent model system for studying hair cell development and function, because fish hair cells are readily accessible on the surface of the animal (Ghysen and Dambly-Chaudiere, 2004, Goodrich, 2005). With the identification and characterization of zebrafish Cx30.3, this study has laid the groundwork for future studies on the biological function of Cx30.3 in hearing and skin development, for example, using morpholino knockdown approaches.
EXPERIMENTAL PROCEDURES
Fish care
Wild type zebrafish (Danio rerio) were purchased from a local pet store and maintained in an aquarium facility at 28.5 °C on a daily photoperiod of 14 h light and 10 h dark. The embryos were incubated in egg water (60 μg/ml Instant Ocean sea salts) in a 28.5 °C water bath, and staged using the time elapsed since fertilization. Embryos for in situ hybridization studies were treated with 0.003% 1-phenyl-2-thiourea (Sigma, Mississauga, ON, Canada) in egg water beginning at ~24 hpf (hours post fertilization) to prevent pigment formation. Zebrafish maintenance, breeding and embryo collection followed standard protocols (Westerfield, 1995). The use of experimental animals was approved by the University of Manitoba Institutional Animal Care Committee.
Cloning and sequence analysis
PAC clones positive for cx48.5 were previously isolated in this lab (Cheng et al., 2004). Cx48.5 positive PAC clones were digested with EcoRI and PstI, and the resulting fragments were subcloned into pBluescript II SK (Stratagene, La Jolla, CA). Subclones were screened with a 32P-labelled Cx48.5 probe (located in exon 2). One positive EcoRI subclone, carrying a ~12 kb fragment upstream of the translational start site, was selected for further analysis. This subclone was sent to the DNA Center at the University of Calgary for sequencing. The assembled sequence was used in a BLAST search of GenBank and resulted in the discovery of a new zebrafish connexin, Cx30.3. The predicted amino acid sequence was analyzed by tools on the ExPASy server (http://www.expasy.org/tools/protparam.html, Gasteiger et al., 2003), and transmembrane domains were predicted by software on the DAS Transmembrane Prediction server (http://www.sbc.su.se/~miklos/DAS/, Cserzo et al., 1997). Amino acid sequence similarity analysis was performed by aligning the predicted Cx30.3 amino acid sequence using MegAlign software (DNASTAR, Inc, Madison, WI).
Phylogenetic analyses and multiple sequence alignment
To assess which sequences were most closely related to Cx30.3, a phylogenetic analysis was performed on protein sequences aligned using ClustalW2 with default parameters (Larkin et al., 2007). Connexin sequences were obtained from GenBank. A neighbor-joining tree was created by ClustalW2. Bootstrap analysis was based on 1000 neighboring replicates. The phylogram was generated using TreeView (Page, 1996).
RNA extraction
RNA was isolated from zebrafish embryos and adult fish tissues using Trizol reagent (Invitrogen Corp., Carlsbad, CA) according to the manufacturer’s instructions. RNA concentration was determined by UV spectroscopy. For regular RT-PCR (reverse transcription polymerase chain reaction), a DNase I treatment was performed to remove residual genomic DNA contamination, whereas for real time RT-PCR and RACE (rapid amplification of cDNA ends) experiments, no DNase treatments were performed in order to avoid any loss and degradation of RNA.
RACE and sequencing of UTRs
5′ and 3′ RACE were performed using a GeneRacer Kit (Invitrogen) according to the manufacturer’s recommended protocols. In brief, total RNA samples were treated with calf intestinal phosphatase (CIP) in order to de-phosphorylate the 5′ends of truncated mRNA and non-mRNA molecules. The dephosphorylated RNA samples were then treated with tobacco acid pyrophosphatase (TIP) to remove the 5′ cap structure from mature full-length mRNA, leaving a 5′ phosphate group available. Using T4 RNA ligase, a universal RNA oligonucleotide (provided by the manufacturer, 5′-CGA CUG GAG CAC GAG GAC ACU GAC AUG GAC UGA AGG AGU AGA AA -3′) was then ligated to the de-capped mRNA, providing a priming site for the following PCR reactions. Total RNA samples for both 5′ and 3′ RACE were prepared from adult zebrafish retina tissue. Reverse transcription was carried out with SuperScript III reverse transcriptase and the GeneRacer oligo dT primer (5′-GCT GTC AAC GAT ACG CTA CGT AAC GGC ATG ACA GTG (T)24 -3′). For 5′RACE, the gene-specific reverse primer used was 5′-TCC CAT GAT GGC GGA GTT GAA G-3′ for the first round of PCR and 5′-GCT GCT ATG ACC AGG ATG CAA ATG-3′ for the nested PCR. For 3′RACE, the gene-specific forward primers used was 5′-ATC TCA CTC TGA AGA CCT CAG-3′ for the first round of PCR and 5′-GAG AAA ATG TCC ACA GAA AAG GCG-3′ for the nested PCR.
The nested PCR products were cloned into the pCR4-TOPO vector (Invitrogen). The ligation mixture was then used to transform One Shot® TOP10 chemically competent E. coli cells (Invitrogen). Several positive colonies were randomly picked up and grown in LB medium at 37°C with shaking (250 rpm) overnight. Plasmids were extracted from 3 clones for each of the 5′ and 3′RACE, and sent out for sequencing (DNA Centre, University of Calgary).
RT-PCR
Two separate sets of total RNA samples from embryos at different developmental stages (0.5, 1, 2, 3, 4, 5 dpf) were isolated for RT-PCR experiments. For each sample, 3 μg of total RNA was treated with DNase I and then reverse transcribed using Superscript II reverse transcriptase (Invitrogen) with oligo-dT(18) primers (Invitrogen). PCR primers were designed to span the intron of Cx30.3 in order to discriminate RT-PCR products from genomic contamination. A forward primer (5′-ATC TCA CTC TGA AGA CCT CAG-3′) that anneals in the first exon and a reverse primer (5′-GCT GCT ATG ACC AGG ATG CAA ATG-3′) that anneals in the second exon amplified a 252 bp fragment from the cDNA template. The gene encoding elongation factor 1α (EF1α) was also amplified as a positive control to assess the presence and integrity of the cDNA in the samples. Accordingly, EF1α primers (forward: 5′-CAA GGG CTC CTT CAA GTA CGC CTG-3′, and reverse: 5′-GGC AGA ATG GCA TCA AGG GCA-3′) were designed to span an intron, and this primer pair amplifies a 569 bp fragment from a cDNA template or a 733 bp fragment from genomic DNA (Cheng et al., 2004, Christie et al., 2004).
Real-time RT-PCR
The tissue distribution pattern of cx30.3 mRNA expression was assessed by real-time PCR, using the iQ5 Real-Time PCR detection system (Bio-Rad). 3 μg of total RNA were reverse transcribed by Superscript III (Invitrogen), with a mix of random hexamers and oligo-dT(18). The cx30.3 primers used for real-time PCR were the same as those for RT-PCR (described above, the predicted amplicon is 252 bp in size). Zebrafish ef1α was used as a reference cDNA, using primers designed to span the first intron splicing boundary of the ef1α gene, so as to avoid amplifying genomic sequences in real-time PCR reactions. The forward primer was 5′-GAG GAG TGA TCT CTC AAT CTT GAA AC-3′, and the reverse primer was 5′-CCA CCG CAT TTG TAG ATC AGA TGG-3′ and the amplicon was 133 bp in length. The specificity and efficiency of the cx30.3 and ef1α primers were validated by standard-curve methods according to the manufacturer’s specifications (Bio-Rad).
cDNA samples (1 μl per well) were mixed with 250 nM of each primer and 10 μl of SYBR Green Supermix (Bio-Rad) reagent in a final volume of 20 μl. PCR conditions were as follows: 95°C for 3 min followed by 40 cycles at 95°C for 10 sec, 62°C for 10 sec, and 72°C for 10 sec. After amplification, melting curve analysis was employed to check PCR specificity, and/or monitor genomic DNA contamination. In addition, a reverse transcriptase negative control and a template-negative control were also tested in parallel. Each sample was tested in duplicate reactions and all PCR runs were performed twice with cDNAs prepared from independent batches of RNA samples. The relative expression of cx30.3 gene transcripts was normalized to the ef1α reference gene, using the comparative Ct method.
In situ hybridization
Digoxigenin-labelled anti-sense and sense RNA probes for cx30.3 were synthesized by in vitro transcription with DIG-11 UTP (Roche, Mississauga, ON). The DNA template was from one of the 3′RACE products of cx30.3 and contained the entire 3′UTR region plus a 94 bp C-terminal fragment. The resulting anti-sense and sense RNA probes were about 1.4 kb in size. The in situ hybridization (ISH) was carried out with various developmental stages of embryos between 1 and 5 dpf (days post fertilization), following Thisse’s protocol with slight modifications (Thisse et al., 1993, Christie et al., 2004). Embryos were mounted in glycerol for whole mount observations. To obtain plastic sections, the embryos were embedded in JB-4 resin (Polysciences; Warrington, PA) and 5 μm sections were cut with a microtome. The embryos were observed, and all images were recorded using a Zeiss Axioskop FS microscope equipped with a Sony DXC-950 CCD camera and Northern Eclipse software (Empix, Mississauga, ON).
Functional analysis/Electrophysiology
The cx30.3 coding sequence was PCR amplified using Platinum Taq DNA polymerase High Fidelity (Invitrogen) with primers carrying BamHI and EcoRI restriction sites (forward primer 5′-GGT TAA TTG GAT CCG ACA GGA TGA GTT GGG GAG CAC-3′, and reverse primer 5′-CCA TGA TGG CGG AAT TCA AGA TCA TC-3′). The PCR products were cut with BamHI and EcoRI and cloned into a pCS2+ expression vector (Turner and Weintraub, 1994). The accuracy of the DNA primary sequence and its open reading frame (ORF) were confirmed by sequencing.
The aforementioned cx30.3 plasmid was linearized at the NotI restriction site of pCS2+, and transcribed using the SP6 mMessage mMachine (Ambion). Adult Xenopus females were anesthetized, the ovaries were removed and Stage V–VI oocytes were collected after the ovarian lobes were de-folliculated in a solution containing 50mg/ml collagenase B, and 50mg/ml hyaluronidase in modified Barth’s medium (MB) without Ca2+. Cells were first injected with 10ng of antisense Xenopus Cx38 oligonucleotide to eliminate coupling caused by endogenous intercellular channels, and cultured overnight. Oligonucleotide injected oocytes were then injected with Cx30.3 cRNA transcripts (5ng/cell) or H2O as a negative control. The vitelline membranes were then removed in a hypertonic solution (200mM aspartic acid, 10mM HEPES, 1mMMgCl2, 10mM EGTA, and 20mM KCl at pH 7.4), and the oocytes were manually paired with the vegetal poles apposed in MB media.
Gap junctional coupling between oocyte pairs was measured using the dual whole-cell voltage clamp technique (DeRosa et al., 2007). Current and voltage electrodes (1.2mm diameter, omega dot; Glass Company of America, Millville, NJ, USA) were pulled to a resistance of 1–2MΩ with a horizontal puller (Narishige, Tokyo, Japan) and filled with solution containing 3M KCl, 10mM EGTA, and 10mM HEPES, pH 7.4. Voltage clamp experiments were performed using two GeneClamp 500 amplifiers (Axon Instruments, Foster City Calif., USA) controlled by a PC-compatible computer through a Digidata 1320A interface (Axon Instruments). The pCLAMP 8.0 software (Axon Instruments) was used to program stimulus and data collection paradigms.
For measurements of junctional conductance, cell pairs were first clamped at −40mV to eliminate any transjunctional potentials. Following the −40mV clamp, a single cell was subjected to alternating pulses of ± 20mV, while the current produced by the change in voltage was recorded in the second cell. The current delivered to the second cell was equal in magnitude to the junctional current. Junctional conductance was calculated by dividing the measured current by the voltage difference, Gj = Ij/(V1−V2).
To determine voltage gating properties, transjunctional potentials (Vj) of opposite polarity were generated by hyperpolarizing or depolarizing one cell in 20mV (range = ± 120mV) steps while clamping the second cell at −40mV. Currents were measured at the end of the voltage pulse, at which time they approached steady state (Ijss). Macroscopic conductance (Gjss) was calculated by dividing Ijss by Vj, normalized to the values determined at ± 20mV, and plotted against Vj. Data describing the relationship of Gjss as a function of Vj were analyzed using Origin 6.1 (Microcal Software, Northampton, Mass., USA) and fit to a Boltzmann relation of the form: Gjss = (Gjmax −Gjmin)/1 + exp[A(Vj−V0)] = Gjmin, where Gjss is the steady state junctional conductance, Gjmax (normalized to unity) is the maximum conductance, Gjmin is the residual conductance at large values of Vj, and V0 is the transjunctional voltage at which Gjss = (Gjmax − Gjmin)/2. The constant A = nq/kT represents the voltage sensitivity in terms of gating charge as the equivalent number (n) of electron charges (q) moving through the membrane, k is the Boltzmann constant, and T is the absolute temperature.
Acknowledgments
We would like to dedicate this paper to Dr. Gunnar Valdimarsson, who passed away on May 9, 2010. Gunnar was a mentor, valued collaborator and friend. We shall all miss him.
This work was supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) grant and a Manitoba Health Research Council (MHRC) grant awarded to Dr. G. Valdimarsson, and National Institutes of Health Grants R01 EY013163 & R01 AR059505 awarded to Dr. T. W. White. Liang Tao was financially supported by a MHRC fellowship, and his contributions to the work were originally part of a thesis at the University of Manitoba.
Grant Sponsor: Natural Sciences and Engineering Research Council of Canada; Grant Sponsor: Manitoba Health Research Council; Grant Sponsor: National Institutes of Health; Grant numbers: R01 EY013163 & R01 AR059505
References
- Barrio LC, Suchyna T, Bargiello T, Xu LX, Roginski RS, Bennett MV, Nicholson BJ. Gap junctions formed by connexins 26 and 32 alone and in combination are differently affected by applied voltage. Proc Natl Acad Sci U S A. 1991;88:8410–8414. doi: 10.1073/pnas.88.19.8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer EC, Paul DL, Goodenough DA. Connexin family of gap junction proteins. J Membr Biol. 1990;116:187–194. doi: 10.1007/BF01868459. [DOI] [PubMed] [Google Scholar]
- Bruzzone R, White TW, Goodenough DA. The cellular internet: On-line with connexins. Bioessays. 1996;18:709–718. doi: 10.1002/bies.950180906. [DOI] [PubMed] [Google Scholar]
- Cason N, White TW, Cheng S, Goodenough DA, Valdimarsson G. Molecular cloning, expression analysis, and functional characterization of connexin44.1: A zebrafish lens gap junction protein. Dev Dyn. 2001;221:238–247. doi: 10.1002/dvdy.1133. [DOI] [PubMed] [Google Scholar]
- Cheng S, Shakespeare T, Mui R, White TW, Valdimarsson G. Connexin 48.5 is required for normal cardiovascular function and lens development in zebrafish embryos. J Biol Chem. 2004;279:36993–37003. doi: 10.1074/jbc.M401355200. [DOI] [PubMed] [Google Scholar]
- Christie TL, Mui R, White TW, Valdimarsson G. Molecular cloning, functional analysis, and RNA expression analysis of connexin45.6: A zebrafish cardiovascular connexin. Am J Physiol Heart Circ Physiol. 2004;286:H1623–32. doi: 10.1152/ajpheart.00800.2003. [DOI] [PubMed] [Google Scholar]
- Cruciani V, Mikalsen SO. The vertebrate connexin family. Cell Mol Life Sci. 2006;63:1125–1140. doi: 10.1007/s00018-005-5571-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruciani V, Mikalsen SO. Evolutionary selection pressure and family relationships among connexin genes. Biol Chem. 2007;388:253–264. doi: 10.1515/BC.2007.028. [DOI] [PubMed] [Google Scholar]
- Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: The dense alignment surface method. Protein Eng. 1997;10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- Dahl G, Werner R, Levine E, Rabadan-Diehl C. Mutational analysis of gap junction formation. Biophys J. 1992;62:172–180. doi: 10.1016/S0006-3495(92)81803-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl G, Miller T, Paul D, Voellmy R, Werner R. Expression of functional cell-cell channels from cloned rat liver gap junction complementary DNA. Science. 1987;236:1290–1293. doi: 10.1126/science.3035715. [DOI] [PubMed] [Google Scholar]
- Dahl E, Manthey D, Chen Y, Schwarz HJ, Chang YS, Lalley PA, Nicholson BJ, Willecke K. Molecular cloning and functional expression of mouse connexin-30, a gap junction gene highly expressed in adult brain and skin. J Biol Chem. 1996;271:17903–17910. doi: 10.1074/jbc.271.30.17903. [DOI] [PubMed] [Google Scholar]
- DeRosa AM, Xia CH, Gong X, White TW. The cataract-inducing S50P mutation in Cx50 dominantly alters the channel gating of wild-type lens connexins. J Cell Sci. 2007;120:4107–4116. doi: 10.1242/jcs.012237. [DOI] [PubMed] [Google Scholar]
- Dobrowolski R, Willecke K. Connexin-caused genetic diseases and corresponding mouse models. Antioxid Redox Signal. 2009;11:283–295. doi: 10.1089/ars.2008.2128. [DOI] [PubMed] [Google Scholar]
- Eastman SD, Chen TH, Falk MM, Mendelson TC, Iovine MK. Phylogenetic analysis of three complete gap junction gene families reveals lineage-specific duplications and highly supported gene classes. Genomics. 2006;87:265–274. doi: 10.1016/j.ygeno.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerido DA, White TW. Connexin disorders of the ear, skin, and lens. Biochim Biophys Acta. 2004;1662:159–170. doi: 10.1016/j.bbamem.2003.10.017. [DOI] [PubMed] [Google Scholar]
- Ghysen A, Dambly-Chaudiere C. Development of the zebrafish lateral line. Curr Opin Neurobiol. 2004;14:67–73. doi: 10.1016/j.conb.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Goodenough DA. Bulk isolation of mouse hepatocyte gap junctions. characterization of the principal protein, connexin. J Cell Biol. 1974;61:557–563. doi: 10.1083/jcb.61.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich LV. Hear, hear for the zebrafish. Neuron. 2005;45:3–5. doi: 10.1016/j.neuron.2004.12.027. [DOI] [PubMed] [Google Scholar]
- Hasson T. Molecular motors: Sensing a function for myosin-VIIa. Curr Biol. 1999;9:R838–41. doi: 10.1016/s0960-9822(00)80040-8. [DOI] [PubMed] [Google Scholar]
- John SA, Revel JP. Connexon integrity is maintained by non-covalent bonds: Intramolecular disulfide bonds link the extracellular domains in rat connexin-43. Biochem Biophys Res Commun. 1991;178:1312–1318. doi: 10.1016/0006-291x(91)91037-d. [DOI] [PubMed] [Google Scholar]
- Kikuchi T, Kimura RS, Paul DL, Takasaka T, Adams JC. Gap junction systems in the mammalian cochlea. Brain Res Brain Res Rev. 2000;32:163–166. doi: 10.1016/s0165-0173(99)00076-4. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. ClustalW and ClustalX version 2. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lefebvre PP, Van De Water TR. Connexins, hearing and deafness: Clinical aspects of mutations in the connexin 26 gene. Brain Res Brain Res Rev. 2000;32:159–162. doi: 10.1016/s0165-0173(99)00075-2. [DOI] [PubMed] [Google Scholar]
- Liu X, Gorovsky MA. Mapping the 5′ and 3′ ends of tetrahymena thermophila mRNAs using RNA ligase mediated amplification of cDNA ends (RLM-RACE) Nucleic Acids Res. 1993;21:4954–4960. doi: 10.1093/nar/21.21.4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan E, White TW, Ugonabo C, Olson C, Nagy JI, Valdimarsson G. Zebrafish Cx35: Cloning and characterization of a gap junction gene highly expressed in the retina. J Neurosci Res. 2003;73:753–764. doi: 10.1002/jnr.10712. [DOI] [PubMed] [Google Scholar]
- Meşe G, Londin E, Mui R, Brink PR, White TW. Altered gating properties of functional Cx26 mutants associated with recessive non-syndromic hearing loss. Hum Genet. 2004;115:191–199. doi: 10.1007/s00439-004-1142-6. [DOI] [PubMed] [Google Scholar]
- Meşe G, Richard G, White TW. Gap junctions: basic structure and function. J Invest Dermatol. 2007;127:2516–2524. doi: 10.1038/sj.jid.5700770. [DOI] [PubMed] [Google Scholar]
- Nag A, Narsinh K, Kazerouninia A, Martinson HG. The conserved AAUAAA hexamer of the poly(A) signal can act alone to trigger a stable decrease in RNA polymerase II transcription velocity. RNA. 2006;12:1534–1544. doi: 10.1261/rna.103206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyamada M, Oyamada Y, Takamatsu T. Regulation of connexin expression. Biochim Biophys Acta. 2005;1719:6–23. doi: 10.1016/j.bbamem.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Page RDM. TREEVIEW: An application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Rahman S, Evans WH. Topography of connexin32 in rat liver gap junctions. evidence for an intramolecular disulphide linkage connecting the two extracellular peptide loops. J Cell Sci. 1991;100:567–578. doi: 10.1242/jcs.100.3.567. [DOI] [PubMed] [Google Scholar]
- Richard G. Connexins: A connection with the skin. Exp Dermatol. 2000;9:77–96. doi: 10.1034/j.1600-0625.2000.009002077.x. [DOI] [PubMed] [Google Scholar]
- Richard G. Connexin disorders of the skin. Clin Dermatol. 2005;23:23–32. doi: 10.1016/j.clindermatol.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Schaefer BC. Revolutions in rapid amplification of cDNA ends: New strategies for polymerase chain reaction cloning of full-length cDNA ends. Anal Biochem. 1995;227:255–273. doi: 10.1006/abio.1995.1279. [DOI] [PubMed] [Google Scholar]
- Sohl G, Willecke K. Gap junctions and the connexin protein family. Cardiovasc Res. 2004;62:228–232. doi: 10.1016/j.cardiores.2003.11.013. [DOI] [PubMed] [Google Scholar]
- Spray DC, Harris AL, Bennett MV. Equilibrium properties of a voltage-dependent junctional conductance. J Gen Physiol. 1981;77:77–93. doi: 10.1085/jgp.77.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang R, Dodd A, Lai D, McNabb WC, Love DR. Validation of zebrafish (danio rerio) reference genes for quantitative real-time RT-PCR normalization. Acta Biochim Biophys Sin (Shanghai) 2007;39:384–390. doi: 10.1111/j.1745-7270.2007.00283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C, Thisse B, Schilling TF, Postlethwait JH. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development. 1993;119:1203–1215. doi: 10.1242/dev.119.4.1203. [DOI] [PubMed] [Google Scholar]
- Turner DL, Weintraub H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- Unger VM, Kumar NM, Gilula NB, Yeager M. Three-dimensional structure of a recombinant gap junction membrane channel. Science. 1999;283:1176–1180. doi: 10.1126/science.283.5405.1176. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book. University of Oregon Press; Eugene: 1995. [Google Scholar]
- White TW. Unique and redundant connexin contributions to lens development. Science. 2002;295:319–320. doi: 10.1126/science.1067582. [DOI] [PubMed] [Google Scholar]
- White TW. Nonredundant gap junction functions. News Physiol Sci. 2003;18:95–99. doi: 10.1152/nips.01430.2002. [DOI] [PubMed] [Google Scholar]
- White TW, Paul DL. Genetic diseases and gene knockouts reveal diverse connexin functions. Annu Rev Physiol. 1999;61:283–310. doi: 10.1146/annurev.physiol.61.1.283. [DOI] [PubMed] [Google Scholar]
- Whitfield TT. Zebrafish as a model for hearing and deafness. J Neurobiol. 2002;53:157–171. doi: 10.1002/neu.10123. [DOI] [PubMed] [Google Scholar]