Abstract
Accumulating evidence suggests that Raf kinase inhibitor protein (RKIP), which negatively regulates multiple signaling cascades including the Raf and nuclear factor κB (NF-κB) pathways, functions as a metastasis suppressor. However, the basis for this activity is not clear. We investigated this question in a panel of breast cancer, colon cancer and melanoma cell lines. We found that RKIP negatively regulated the invasion of the different cancer cells through three-dimensional extracellular matrix barriers by controlling the expression of matrix metalloproteinases (MMPs), particularly, MMP-1 and MMP-2. Silencing of RKIP expression resulted in a highly invasive phenotype and dramatically increased levels of MMP-1 and MMP-2 expression, while overexpression of RKIP decreased cancer cell invasion in vitro and metastasis in vivo of murine tumor allografts. Knockdown of MMP-1 or MMP-2 in RKIP-knockdown cells reverted their invasiveness to normal. In contrast, when examining migration of the different cancer cells in a two-dimensional, barrier-less environment, we found that RKIP had either a positive regulatory activity or no activity, but in no case a negative one (as would be expected if RKIP suppressed metastasis at the level of cell migration itself). Therefore, RKIP’s function as a metastasis suppressor appears to arise from its ability to negatively regulate expression of specific MMPs, and thus invasion through barriers, and not from a direct effect on the raw capacity of cells to move. The NF-κB pathway, but not the Raf pathway, appeared to positively control the invasion of breast cancer cells. A regulatory loop involving an opposing relationship between RKIP and the NF-κB pathway may control the level of MMP expression and cell invasion.
Keywords: Raf kinase inhibitor protein; Raf/MEK/ERK, NF-κB, matrix metalloproteinases; cancer cell migration, invasion and metastasis
1. Introduction
Raf kinase inhibitor protein (RKIP) is an endogenous modulator of the function of multiple proteins, including Raf-1 kinase [1,2] and kinases involved in the activation of the transcriptional regulator nuclear factor κB (NF-κB) [3,4]. RKIP thus negatively regulates the Raf/mitogen-activated protein kinase/extracellular signal-regulated kinase (ERK) kinase (MEK)/ERK pathway [1,2] and the NF-κB pathway [3,4]. RKIP has been implicated in a range of normal and disease processes, such as cell growth, apoptosis, cell migration, angiogenesis, reproduction, neurodegeneration and cancer metastasis (for reviews, see refs. [5,6]). RKIP was first discovered on the basis of its ability to bind phosphatidylethanolamine and is thus also known as phosphatidylethanolamine-binding protein (PEBP) [7,8]; however, this interaction may be non-specific [9]. RKIP/PEBP has been recognized to consist of an evolutionarily conserved multigene family, with the most widely expressed member in mammalian tissues being RKIP-1 or PEBP-1 (for reviews, see refs. [5,6]), the focus of this study and herein referred to simply as RKIP.
Evidence has emerged that RKIP can function as a suppressor of cancer metastasis. Its expression was originally found to be lower in metastatic human prostate cancer cells than non-metastatic ones and to correlate inversely with prostate cancer cell invasiveness but not proliferation in vitro [10,11]. Expression of RKIP is also inversely related to the metastatic potential of human melanoma [12], human breast cancer [13,14], human colorectal cancer [15–18], human ovarian cancer [19] and human nasopharyngeal cancer [20]. The expression levels of RKIP progressively decreases in human breast and prostate cancer cell lines of increasing metastatic capacity [21]. In contrast, in murine fibrosarcoma cells, RKIP expression positively correlates with metastasis [22].
Forced overexpression of RKIP in human prostate cancer cells reduces tumor angiogenesis and metastasis in vivo in an orthotopic murine xenograft model [10]. Overexpression of RKIP also results in decreased metastasis of human breast [14] and human ovarian [19] cancer cells in murine xenograft models. Furthermore, RKIP may constitute a useful prognostic marker for predicting the clinical outcome of certain cancers in human patients. It has been shown to be a prognostic marker for colorectal cancer [15–18], prostate cancer [23], gastrointestinal stromal tumors [24] and intestinal-type, though not diffuse-type, gastric adenocarcinoma [25].
The basis for RKIP’s metastasis-suppressing activity is not yet clear. It could involve a direct effect on cell movement itself, in which case one would expect that RKIP would negatively regulate cell migration. Alternatively, it could entail the ability of cells to degrade extracellular matrix (ECM) barriers, adhere in complex three-dimensional settings or some other process relevant to invasion and metastasis. While RKIP expression inversely correlates with cell movement in human hepatocellular carcinoma cells [26], RKIP instead appears to positively control the motility of Madin-Darby canine kidney (MDCK) epithelial cells and MCF7 human breast carcinoma cells [27–29], as well as rat hepatic stellate cells [30]. RKIP also regulates cell-substratum and cell-cell adhesion in MDCK cells [28,29]. Other processes that could affect the formation of viable metastases have been shown to be regulated by RKIP, including cell cycle progression [19,31,32], apoptosis [11,33] and angiogenesis [10]. It is therefore evident that there is a great deal of complexity in the cellular operations of RKIP and that we do not yet have a framework for understanding how RKIP acts to suppress tumor metastasis.
Here we report that RNA interference (RNAi)-based silencing of RKIP expression in a number of different cancer cell lines resulted in either no change in the rate of cell migration in a scratch-wound assay or a decreased rate of cell movement in a cell type-dependent manner. In contrast, silencing of RKIP expression resulted in an increased level of invasion of cells through a Matrigel barrier in four of the cancer cell lines tested (BT-20 and T47D human breast carcinoma cells, 168FARN murine breast carcinoma cells and LoVo human colon carcinoma cells) and no change in the degree of invasion in HCT-116 human colon carcinoma cells. Conversely, overexpression of RKIP in 4T1 murine breast carcinoma and MDA-MB-435 human melanoma cells led to decreased invasion without change in the rate of migration. Treatments over a range of concentrations with a small-molecule inhibitor of matrix metalloproteinases (MMPs), which constitute one of the major families of ECM-degrading enzymes, suggested that MMPs may be involved in the changes in invasion arising from modulation of RKIP expression. We looked at expression of MMPs and found that knockdown of RKIP correlated with increased expression of certain MMPs. MMP-1 and MMP-2, in particular, were dramatically upregulated in RKIP-knockdown cells. Most importantly, we found that knockdown of MMP-1 or MMP-2 in RKIP-knockdown T47D cells “rescued” the cells from their highly invasive phenotype, restoring a control level of invasion. Knockdown of RKIP and/or MMP-1 or MMP-2 had little or no effect on migration or proliferation of T47D cells. The NF-κB pathway appeared to be involved in the invasion of T47D cells, but not the Raf/MEK/ERK pathway. Our data suggest that RKIP functions as a metastasis suppressor not by negatively regulating cell migration but through negative regulation of MMP production and invasion through ECM barriers, ostensibly through a regulatory feedback loop involving RKIP and proinvasive factors such as NF-κB. These results help to define different roles for RKIP in cell migration, on the one hand, and invasion, on the other, and shed light on the mechanism by which RKIP acts as a metastasis suppressor through negative control of the expression of specific MMPs.
2. Materials and methods
2.1. Reagents
All cell culture media were from Invitrogen. Fetal bovine serum (FBS) was from Atlanta Biologicals, and newborn calf serum was from Lonza BioWhittaker. Cell culture plates, transwell chambers, Matrigel and fibronectin were from BD Biosciences. GM 1489, a general MMP inhibitor, was purchased from Calbiochem/EMD Chemicals. U0126, a MEK inhibitor, was from Tocris Bioscience. Dehydroxymethylepoxyquinomicin (DHMEQ), an NF-κB inhibitor, was a kind gift from Kazuo Umezawa (Keio University, Japan). Rabbit anti-RKIP antibody was from Invitrogen, and mouse monoclonal anti-α-tubulin antibody was from Sigma. Horseradish peroxidase secondary antibodies were from Santa Cruz Biotechnology.
2.2. Cell culture
BT-20 cells were cultured in minimum essential medium with 10% FBS. T47D, 4T1 and MDA-MB-435 cells were cultured in Dulbecco’s modified Eagle’s medium with 10% FBS. 168FARN cells were cultured in 5% FBS/5% newborn calf serum. HCT-116 cells were cultured in McCoy’s 5A medium with 10% FBS. LoVo cells were cultured in RPMI 1640 medium with 10% FBS. Cells were grown in a humidified tissue culture incubator at 37°C in 5% CO2.
2.3. Preparation of knockdown cells
For RNAi-based silencing of expression of RKIP, MMP-1 and MMP-2 in the human cell lines, we prepared knockdown cells expressing the corresponding small interfering RNAs (siRNAs) or a firefly luciferase-specific siRNA as control from the short hairpin RNA (shRNA) expression vectors pSUPER.retro-neo (OligoEngine) for the RKIP and firefly luciferase shRNAs and pSUPER.retro-puro (OligoEngine) for the MMP-1 and MMP-2 shRNAs, as previously reported [29,34]. The MMP-1 and MMP-2 shRNA expression constructs were kindly supplied by Joan Massagué (Memorial Sloan Kettering Cancer Center). The target sequences were: 5'-GATTCAGGGAAGCTCTACA-3' (RKIP-1/PEBP-1) [29], 5'-AGCGGAGAAATAGTGGCCC-3' (MMP-1) [34], 5'-GGACGGACTCCTGGCTCAT-3' (MMP-2) [34] and 5'-CGTACGCGGAATACTTCGA-3' (firefly luciferase control) [29]. Since the RKIP and firefly luciferase shRNAs were in an expression vector containing a neomycin-resistance gene and the MMP-1 and MMP-2 shRNAs were in an expression vector containing a puromycin-resistance gene, selection for double-knockdown cells was achieved in medium containing both G418 and puromycin. To silence expression of RKIP in the 168FARN murine cells, however, we infected cells with lentivirus encoding a murine RKIP-specific shRNA (with target sequence of 5'-GGTGTACGAGCAGGAACAG-3') or firefly luciferease as control (same target sequence as above) in the lentiviral shRNA expression vector pLL3.7, as described previously [35]. Knockdown was confirmed by Western blot analysis, as follows. Cells were lysed with 20 mM Tris, pH 7.4, 150mM NaCl, 2mM EDTA, and 1% Triton X-100. Samples (10–50 µg) were separated by sodium dodecyl sulfate-polyacrylamide eletrophoresis and then electrophoretically transferred from the gel to polyvinylidene fluoride membranes (Millipore). The primary antibody (anti-RKIP or anti-α-tubulin) were diluted in phosphate-buffered saline, pH 7.4, 0.2% Tween-20, 5% bovine serum albumin, 0.002% sodium azide. Following three washes, blots were incubated with the appropriate horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature. Proteins were visualized with enhanced chemiluminescence with the BioRad ChemiDoc EQ system.
2.4. Preparation of RKIP-overexpressing cells
RKIP was overexpressed in 4T1 or MDA-MB-435 cells by retroviral-mediated gene transfer. Retroviral constructs encoding N-terminally FLAG-tagged rat RKIP in the pQCXIP (Clontech) vector or N-terminally hemagglutinin (HA)-tagged rat RKIP in the pWZL-blasticidin vector were used to generate RKIP-overexpressing 4T1 and MDA-MB-435 cells, respectively, as previously described [11]. Expression of the tagged RKIP fusions was confirmed by Western blot analysis (vide supra).
2.5. Wound closure
Cells were grown to confluence in 24-well tissue-culture-treated plates and then scratch wounded and analyzed by tracing the wound edge in digital images and determining the remaining open area as a function of time after wounding, as previously described [27]. Because MDA-MB-435 cells migrated more as individuals following wounding of confluent monolayer rather than as a more cohesive sheet like the other cells, the open area was determined by tracing the edge of the remaining intact cell monolayer, as with the other cells, but then subtracting the area occupied by cells migrating into the wound area as individuals to obtain the correct unoccupied area as a function of time. The data were graphed with GraphPad Prism software. Cell viability was confirmed at the end of each experiment by the trypan blue dye exclusion assay and observation of cell morphology, noting any signs of rounding up or detachment.
2.6. Cell invasion
The polycarbonate membrane (8-µm pore size) of transwell inserts (BD Biosciences) was coated with 25 µg of Matrigel (BD Biosciences). Chemoattractive medium was added to the lower chambers (24-well BD Falcon TC companion plates), consisting of 600 µL medium with 10% FBS for all the cell lines (supplemented with 10 µg of fibronectin for the T47D and HCT-116 cells). Cells were serum starved overnight. Cell suspensions containing 5 × 104 cells/mL were then prepared in serum-free cell culture medium. In experiments with compound treatments (GM 1489, U0126 or DHMEQ), compound was added to the serum-free medium prior to resuspension of the cells. 100 µL of the cell suspension were added to each of the Matrigel-coated inserts of the upper chambers. BT-20, T47D, HCT-116 and LoVo cells were incubated at 37°C with 5% CO2 for 48 h, while 168FARN, 4T1 and MDA-MB-435 cells were incubated at 37°C with 5% CO2 for 24 h (because of their higher rate of invasion through Matrigel).
Non-invaded cells were removed from the upper surface of the membrane by gently scrubbing with a cotton swab. The cells on the lower surface of the membrane (invaded cells) were fixed with methanol, stained with crystal violet solution, rinsed with water and air dried. The mean values for the number of control, RKIP-knockdown or RKIP-overexpressing cells invaded through the membrane were determined from digital images captured on an inverted microscope.
Initial “titration” experiments were performed with GM 1489 for each cell line to evaluate cytotoxicity of the compound by the trypan blue dye exclusion assay and counting viable cell numbers to ensure that all subsequent invasion experiments were performed at subtoxic concentrations. Once this was determined for each cell line, a suspension of cells treated with GM 1489 or dimethyl sulfoxide (DMSO) carrier solvent alone as the control for comparison was added to Matrigel-coated insert chambers (with the final concentration of DMSO being uniform for each treatment). The lower chamber was also treated with GM 1489 in the chemoattractive medium. Cells were then incubated at 37°C with 5% CO2 for 48 h in all cases, except for the MDA-MB-435 cells, which were incubated for 24 h, corresponding to the respective conditions of the invasion assays. Similar initial “titration” experiments were performed with U0126 and DHMEQ to establish subtoxic concentrations for treatment of T47D cells in the invasion assays.
2.7. Murine tumor allograft experiments
All animal work was performed in accordance with an IACUC (Institutional Animal Care and Use Committee) approved protocol. Female BALB/cJ mice (1.5–2 months old) were purchased from Jackson Laboratory. Approximately 1 × 106 control or RKIP-overexpressing 4T1 cells in 50 µl phosphate-buffered saline, pH 7.4, were injected into the mammary fat pad. To facilitate in vivo visualization of the disseminated cancer cells, we engineered the 4T1 cells to express a dual-function firefly luciferase-yellow fluorecent protein (YFP) fusion reporter gene by lentiviral infection, and mice were imaged 30 days later with a Xenogen IVIS Spectrum imaging system, as previously described [36]. Mice were euthanized shortly after imaging and lungs were removed for visual inspection under dissection microscope.
2.8. Quantitative real-time reverse transcriptase-polymerase chain reaction (RT-PCR)
Total cellular RNA was extracted from control and knockdown T47D cells with Trizol reagent (Invitrogen) and reverse transcribed with random hexamer primers (Applied Biosystems). The resulting cDNAs were used for PCR with the SYBR-Green Master PCR mix (Qiagen) in triplicate. PCR and data collection were performed on an ABI7500 real-time thermal cycler (Applied Biosystems). β-actin or glyceraldehyde-3-phosphate dehydrogenase was used as an internal standard, as indicated in the relevant figures. The final concentration of the primers used in the reaction was 0.64 µM. The gene-specific primers were:
5'-TGATATCGGGGCTTTGATGT-3' (MMP-1 forward),
5'-CACTTCTCCCCGAATCGTAG-3' (MMP-1 reverse),
5'-TCCCATTTTGATGACGATGA-3' (MMP-2 forward),
5'-CCGTACTTGCCATCCTTCTC-3' (MMP-2 reverse),
5'-AGTGGAGGAAAACCCACCTT-3' (MMP-3 forward),
5'-CCAGGTCCATCAAAAGGGTA-3' (MMP-3 reverse),
5'-CATCGTCATCCAGTTTGGTG-3' (MMP-9 forward),
5'-TCGAAGATGAAGGGGAAGTG-3' (MMP-9 reverse),
5'-TGGTCCAGGAGATGAAGACC-3' (MMP-13 forward),
5'-GGAAGTTCTGGCCAAAATGA-3' (MMP-13 reverse),
5'-GCTGAGATCAAGGCCAATGT-3' (MMP-14 forward),
5'-ATGTAGGCATAGGGCACCTC-3' (MMP-14 reverse),
5'-ATCTGGCACCAGACCTTCTACAATGAGCTGCG-3' (β-actin forward),
5'-CGTCATACTCCTGCTTGCTGATCCACATCTGC-3' (β-actin reverse),
5'-TGCACCACCAACTGCTTAGC-3' (glyceraldehyde-3-phosphate dehydrogenase forward),
5'-GGCATGGACTGTGGTCATGAG-3' (glyceraldehyde-3-phosphate dehydrogenase reverse).
2.9. Cell proliferation
T47D cells were serum starved for 24 h in a tissue culture incubator at 37°C with 5% CO2 and then plated onto 96-well tissue culture plates at 105 cells per well in 100 µL in serum-containing medium. Initial cell numbers were determined for control cells by adding the tetrazolium salt WST-8 as per the manufacturer's instructions (Cell Counting Kit-8, Dojindo Molecular Technologies), incubating for 3 h at 37°C and then measuring absorbance at 450 nm in a UV-visible absorbance plate reader (Molecular Devices Spectramax Plus 384). Experimental cell cultures were allowed to grow in the tissue culture incubator for 48 h, and then cell numbers were determined.
3. Results
3.1. RKIP positively regulates cell migration or has no role in cell migration, depending on cell type
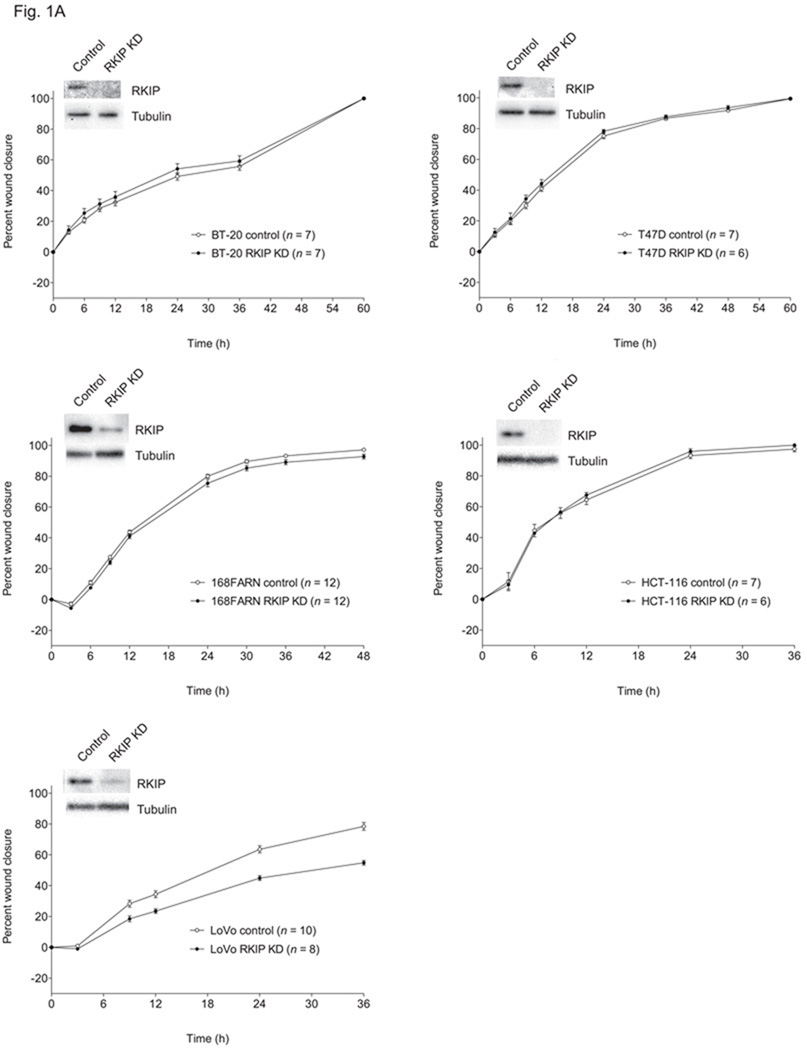
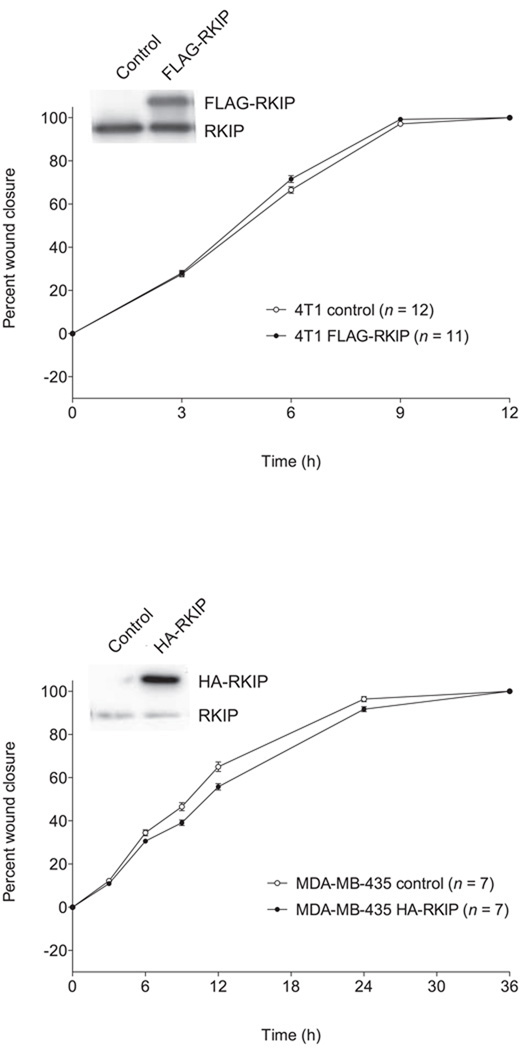
RKIP could function to suppress cancer metastasis in different ways, since there are a number of necessary but not individually sufficient conditions involved. One possibility is that invasion is reduced because of decreased rates of cancer cell migration. To test this, we evaluated cell migration in two-dimensional culture in the absence of any ECM barrier in classic scratch-wound assays. We found that RKIP had no motility-suppressing activity at all in any of the cancer cell lines tested, based on both RKIP knockdown (Fig. 1A) and RKIP overexpression (Fig. 1B). In fact, in LoVo cells, RNAi-based silencing of RKIP expression resulted in a reduced rate of cell migration (Fig. 1A), as it does in MDCK and MCF7 cells [28,29].
Fig. 1.
Effects of altered RKIP expression on migration of different cancer cell lines. The values represent the means and standard errors of the mean (SEM) for the percent closure of wounds in cell monolayers as a function of time (n = indicated number of separately treated wounds on multiwell tissue culture-treated plates from three independent experiments). (A) BT-20, T47D and 168FARN breast cancer cells and HCT-116 and LoVo colon cancer cells stably expressing a control siRNA for firefly luciferase (control) and those expressing an RKIP-specific siRNA (RKIP KD). Above the graphs are representative Western blots showing expression of RKIP and α-tubulin in the corresponding cell lines. (B) Control and RKIP-overexpressing 4T1 breast cancer cells and MDA-MB-435 melanoma cells (stably expressing FLAG-RKIP and HA-RKIP, respectively). Above the graphs are representative Western blots showing expression of tagged and endogenous RKIP in the corresponding cell lines.
3.2. RKIP negatively controls cancer cell invasion
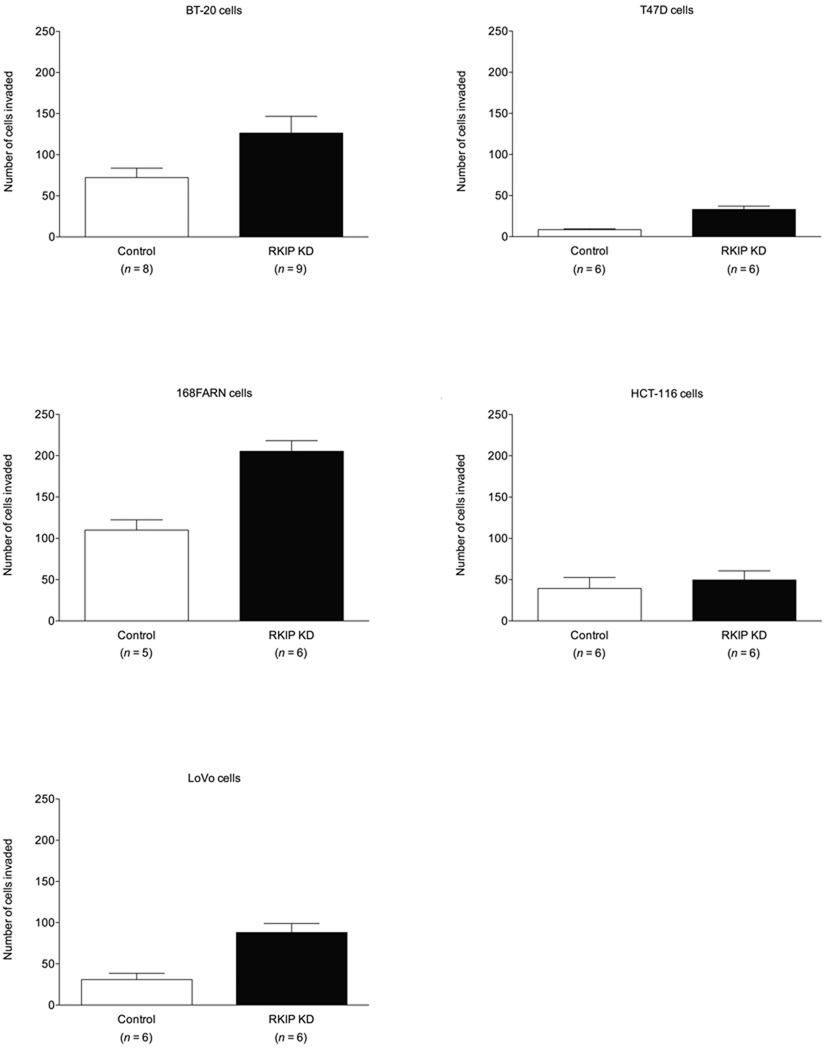
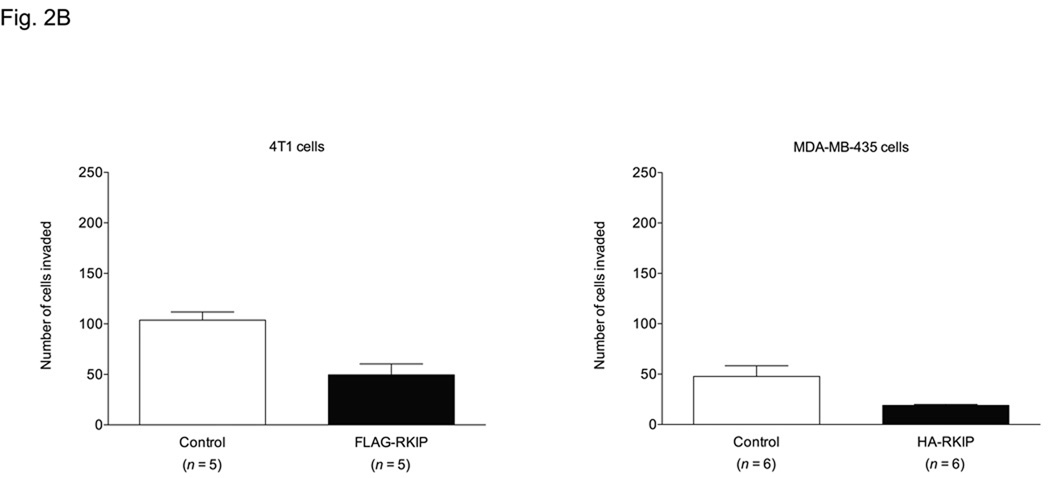
We examined the role of RKIP cell invasion in an in vitro invasion assay in the same cancer cells tested in the cell migration assay. Knockdown of RKIP in BT-20, T47D and 168FARN and LoVo cells resulted in increased cell invasion through Matrigel (Fig. 2A). In HCT-116 cells, however, there was no difference in invasion with RKIP knockdown (Fig. 2A). The converse manipulation, forced overexpression of RKIP, resulted in reduced invasiveness in 4T1 and MDA-MB-435 cells (Fig. 2B). The results with the MDA-MB-435 cells are consistent with those recently reported [37].
Fig. 2.
Effects of altered RKIP expression on invasion of different cancer cell lines through Matrigel. The values represent the means and SEM for the number of cells invading through Matrigel (n = indicated number of wells from three independent experiments) over 48 h for BT-20, T47D, HCT-116 and LoVo cells or over 24 h for 168FARN, 4T1 and MDA-MB-435 cells. (A) Control and RKIP-knockdown BT-20, T47D, 168FARN, HCT-116 and LoVo cells. The fold increase in invasion for each cell line for RKIP-knockdown cells over corresponding control cells was: T47D = 3.67; LoVo = 2.84; 168FARN = 1.86; BT-20 = 1.75; HCT-116 = 1.28. (B) Control and RKIP-overexpressing 4T1 and MDA-MB-435 cells. The fraction of invaded RKIP-overexpressing cells compared to corresponding control cells was: MDA-MB-435 = 0.40; 4T1 = 0.48.
It is pertinent to note that 168FARN cells are derived from the same mouse mammary tumor as 4T1 cells but have lower metastatic potential than 4T1 cells [38]. While formation of viable metastases, of course, involves much more than the ability of cells to invade ECM, in our experiments 168FARN control cells and 4T1 control cells were similarly invasive (Fig. 2). Another point worth noting is that while MDA-MB-435 cells have long been considered a breast cancer cell line and used as a model for metastatic breast carcinoma, evidence has emerged to suggest that they are instead of melanoma origin [39–45], although this question is still not settled [46–49]. MDA-MB-435 cells are nevertheless a valuable model system for highly metastatic cancer.
Comparing the fold increase in invasion relative to controls in the RKIP-knockdown cells, we found that invasion of RKIP-knockdown T47D cells increased by 3.67-fold, the highest as compared to the increase showed by RKIP-knockdown LoVo (2.84-fold), 168FARN (1.86-fold) and BT-20 (1.75-fold) cells. Conversely, the fraction of cells invading through Matrigel in the experiments with the RKIP-overexpressing cells compared to the controls was 0.40 for the MDA-MB-435 cells and 0.48 for the 4T1 cells. Knockdown of RKIP in T47D cells had little to no effect on cell proliferation (vide infra), preculding that possibility as an explanation for the difference in T47D cell numbers invading through Matrigel.
3.3. RKIP overexpression results in decreased metastasis in vivo in a murine allograft model
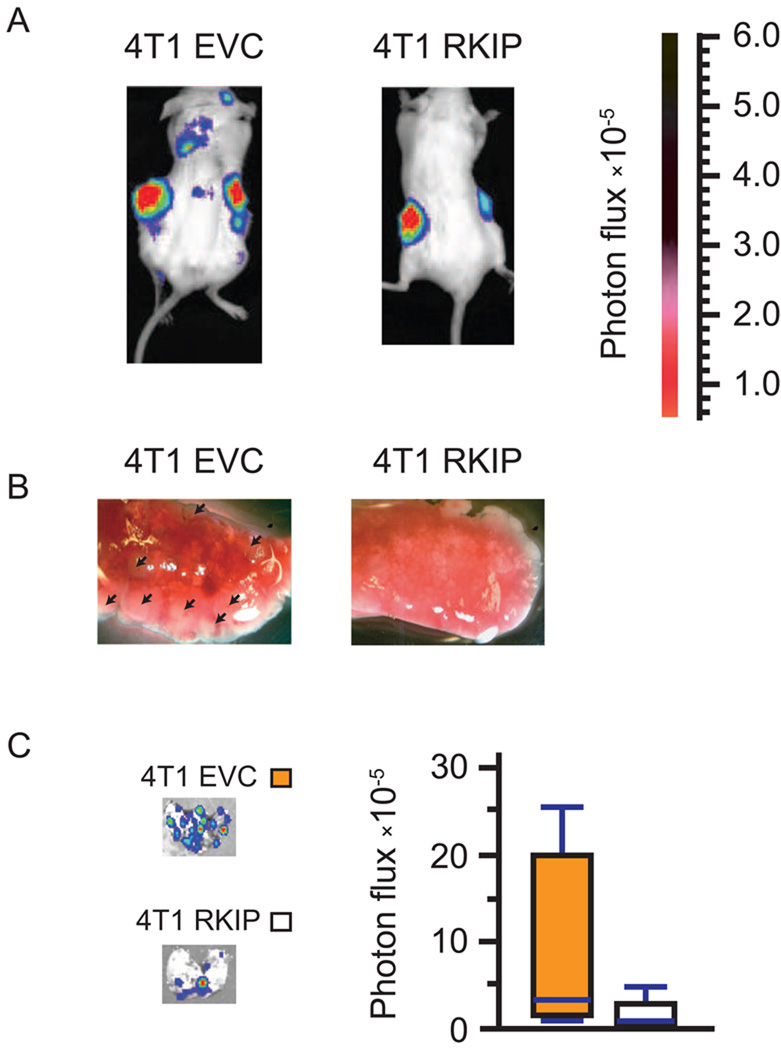
To examine the effects of RKIP on cancer cell invasion at the organismal level, we engineered 4T1 cells to express a dual-function firefly luciferase-YFP fusion reporter gene. YFP-positive 4T1 cells expressing RKIP or empty vector control were orthotopically allografted into mammary fat pads of Balb/cJ mice. Four weeks after implantation, the luciferase-tagged cancer cells were imaged by non-invasive bioluminescence imaging. Significantly increased foci of luciferase signal were observed in mice bearing RKIP-expressing 4T1 allografts (Fig. 3A). The mice were then sacrificed and the lungs were removed for visual inspection (Fig. 3B) and quantification by bioluminescence imaging (Fig. 3C). Consistent with the results from whole-animal imaging, RKIP-expressing tumors formed more macroscopically visible lung metastases with increased luciferase signal when compared with their control counterparts.
Fig. 3.
(A) Xenogen images of mice orthotopically injected with empty vector control (EVC) or RKIP-expressing 4T1 cells. Both control and RKIP-expressing cells were also expressing a firefly luciferase-YFP fusion reporter gene for bioluminescent detection. (B) Representative photos of the lungs of mice bearing control (left panel) or RKIP-expressing (right panel) 4T1 allografts 30 days after implantation. The arrows point to the metastatic nodules in the lung. (C) Xenogen images of mice orthotopically injected with control or RKIP-expressing cells, along with a standard box plot of the data.
3.4. RKIP regulates the expression of MMPs
The degree of invasion through Matrigel, but not the raw ability of cells to move in the absence of an ECM barrier, was dependent on the level of RKIP expression. Therefore, the differences in invasive potential between control cells and those with altered levels of RKIP expression must be due an invasion-relevant process other than cell motility itself, the most obvious such process being the capacity to proteolytically digest the ECM. Since MMPs are ECM-degrading proteases with well-documented roles in cancer cell invasion and metastasis (for reviews, see refs. [50,51]), we investigated whether the function of RKIP in the control of invasiveness was dependent on MMP expression and activity.
As a preliminary experiment to confirm the importance of MMPs, we treated BT-20, T47D and LoVo control and RKIP-knockdown cells, as well as MDA-MB-435 normal and RKIP-overexpressing cells, with GM 1489, a general MMP inhibitor [52]. We evaluated the effect of treatment with this MMP inhibitor on cell invasion. (We verified prior to these experiments that GM 1489 at the concentrations used in these cells was not cytotoxic and had no effect by itself on cell numbers. In addition, to confirm selectivity of GM 1489 against cell invasion over raw migration, we also tested the compound in the wound closure assay and found no inhibition of cell migration in a barrier-less environment.)
As shown in the Table 1, control “wild-type” cells were more sensitive to GM1489, based on minimum inhibitory concentration and/or half-maximal inhibitory concentration (IC50), than RKIP-knockdown cells but less sensitive than RKIP-overexpressing cells (Table 1). If RKIP negatively regulates MMP expression, cells expressing lower levels of RKIP would be expected to produce higher levels of MMP and thus require higher concentrations (higher IC50) of GM 1489 to inhibit invasion, whereas the converse would be true for the RKIP-overexpressing cells (lower IC50). The data are consistent with this hypothesis (Table 1). Interestingly, control LoVo cells were not significantly inhibited at all by GM 1489 at subtoxic concentrations of the inhibitor, yet the RKIP-knockdown LoVo cells were inhibited by GM 1489 (Table 1). This implies that while invasion may not be MMP-limited in control LoVo cells but that the increased invasion in RKIP-knockdown LoVo cells nevertheless results from increased MMP activity above the normal level.
Table 1.
Sensitivity of RKIP-knockdown and RKIP-overexpressing cells to pharmacological inhibition of MMPs.
| Cell type | MICa (µM) | IC50b (µM) | MLCc (µM) |
|---|---|---|---|
| BT-20 control | 0.5 | 1.6 | 20 |
| BT-20 RKIP KD | 1 | 3.2 | 20 |
| T47D control | 0.5 | 1.8 | 20 |
| T47D RKIP KD | 0.5 | 3.0 | 20 |
| LoVo control | No subtoxic activity | NA | 20 |
| LoVo RKIP KD | 1 | 5.4 | 20 |
| MDA-MB-435 control | 5 | 7.3 | 20 |
| MDA-MB-435 HA-RKIP (RKIP-overexpressing) |
No subtoxic activity | NA | 20 |
Minimum inhibitory concentration (MIC) represents the lowest concentration at which GM 1489, a general MMP inhibitor, exhibited a statistically significant antiinvasive effect by Student’s t tests for each cell type. Data are derived from three independent experiments.
The half-maximal inhibitor concentration (IC50) for inhibition of invasion by GM 1489. Only subtoxic concentrations, for which there was no sign of cytotoxicity in “titration” experiments done initially, were used for the IC50 determinations with GraphPad Prism software.
Minimum lethal concentration (MLC) represents the lowest concentration at which signs of cytototoxicity were first observed based on the trypan blue dye exclusion assay in preliminary “titration” experiments.
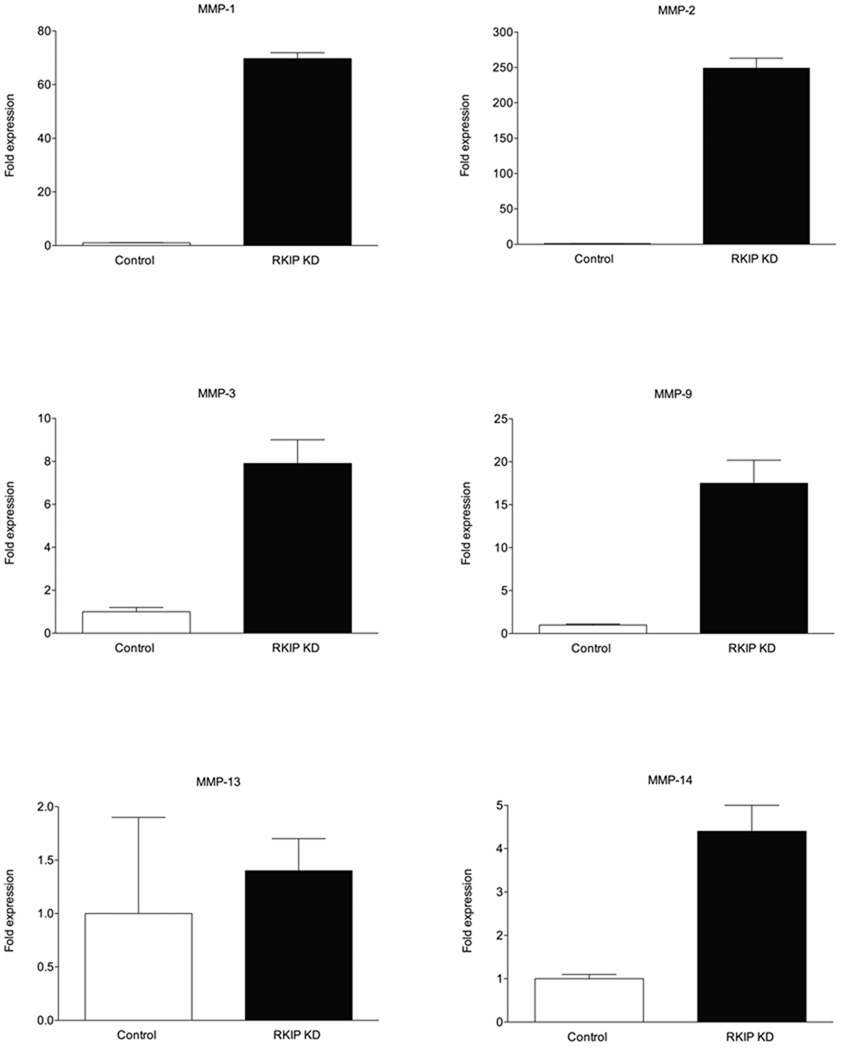
We next examined whether changes in the level of RKIP expression correlated with changes in the expression of specific MMPs already implicated in cancer cell invasion and metastasis (for reviews, see refs. [50,51]). Because the fold increase in invasion with RKIP knockdown was greatest in the T47D cells, we quantitated the expression level of various MMPs in control and RKIP-knockdown T47D cells. Silencing of RKIP expression resulted in dramatically increased expression of MMP-1 and MMP-2 of 69.7-fold and 249.0-fold over the control cells, respectively, by quantitative real-time RT-PCR (Fig. 4). MMP-3 expression was 7.9-fold, MMP-9 was 17.5-fold, MMP-13 was 1.4-fold and MMP-14 was 4.4-fold above the level in control cells (Fig. 4).
Fig. 4.
MMP-1 and MMP-2 are highly expressed in RKIP-knockdown cells. The values represent the means and standard deviation from three independent experiments for fold expression of different MMPs in RKIP-knockdown T47D cells over control cells, as determined by quantitative real-time RT-PCR (with β-actin, whose expression was unaffected by RKIP knockdown, as an internal standard). The fold expression values for the different MMPs from the graphs are: MMP-1 = 69.7; MMP-2 = 249.0; MMP-3 = 7.9; MMP-9 = 17.5; MMP-13 = 1.4; MMP-14 = 4.4.
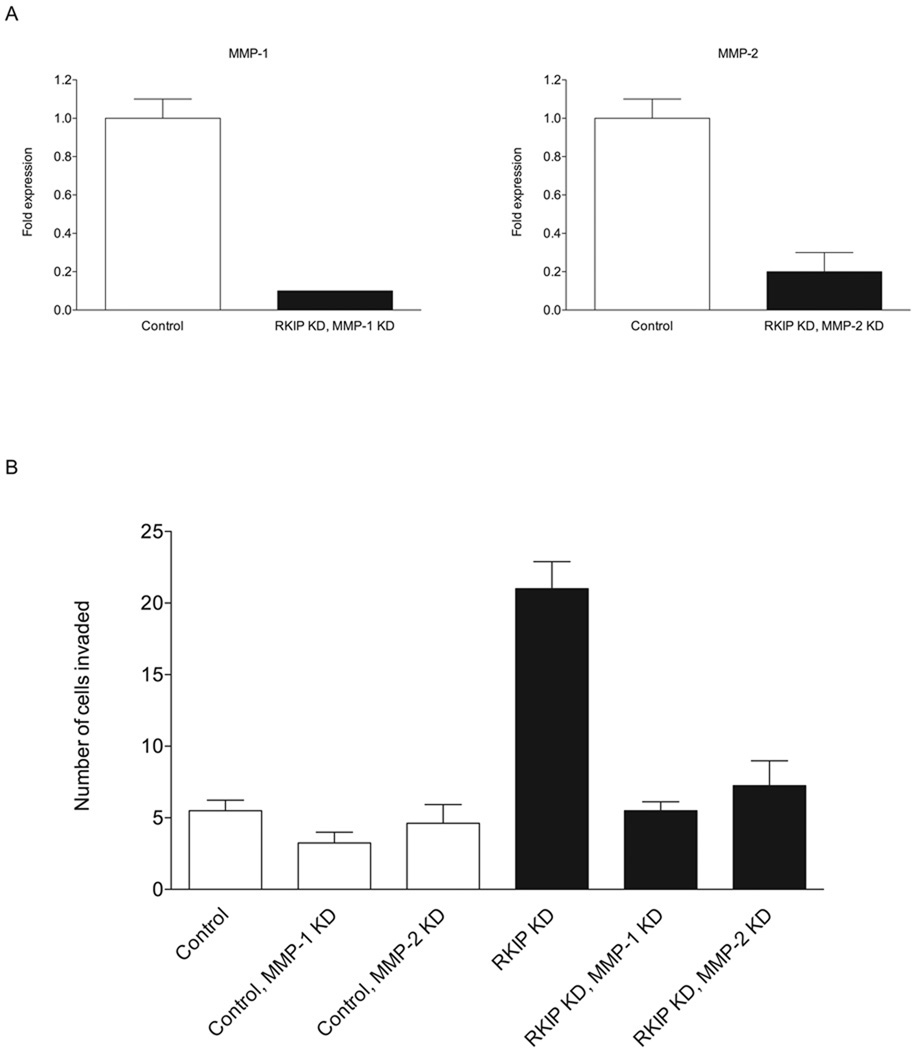
3.5. Knockdown of MMP-1 or MMP-2 in RKIP-knockdown cells reduces the degree of cell invasion to control levels
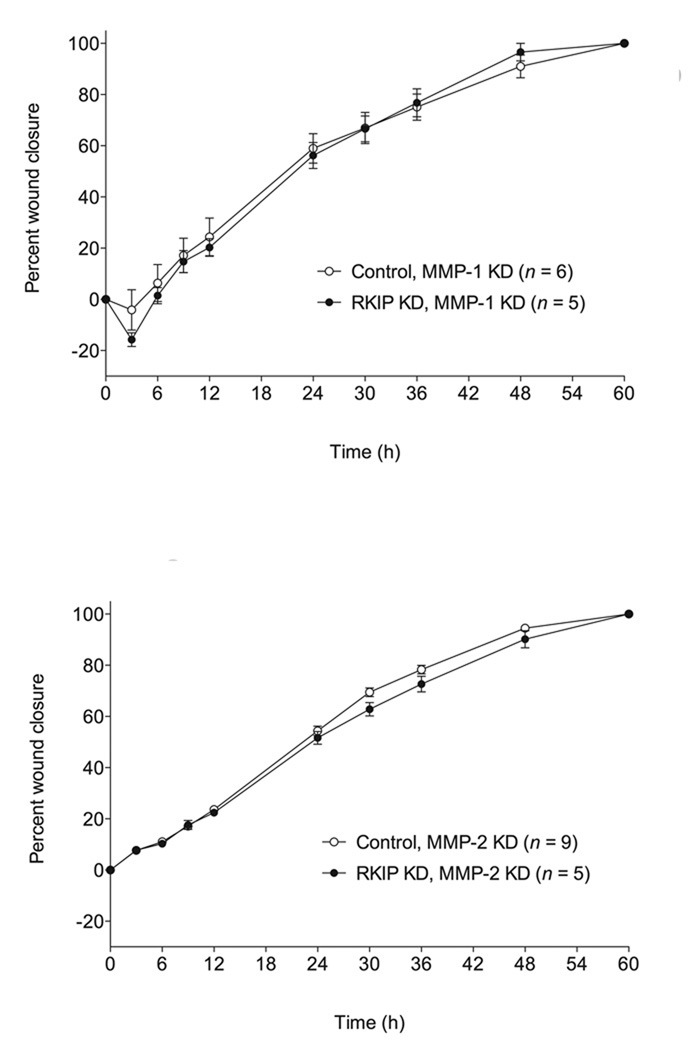
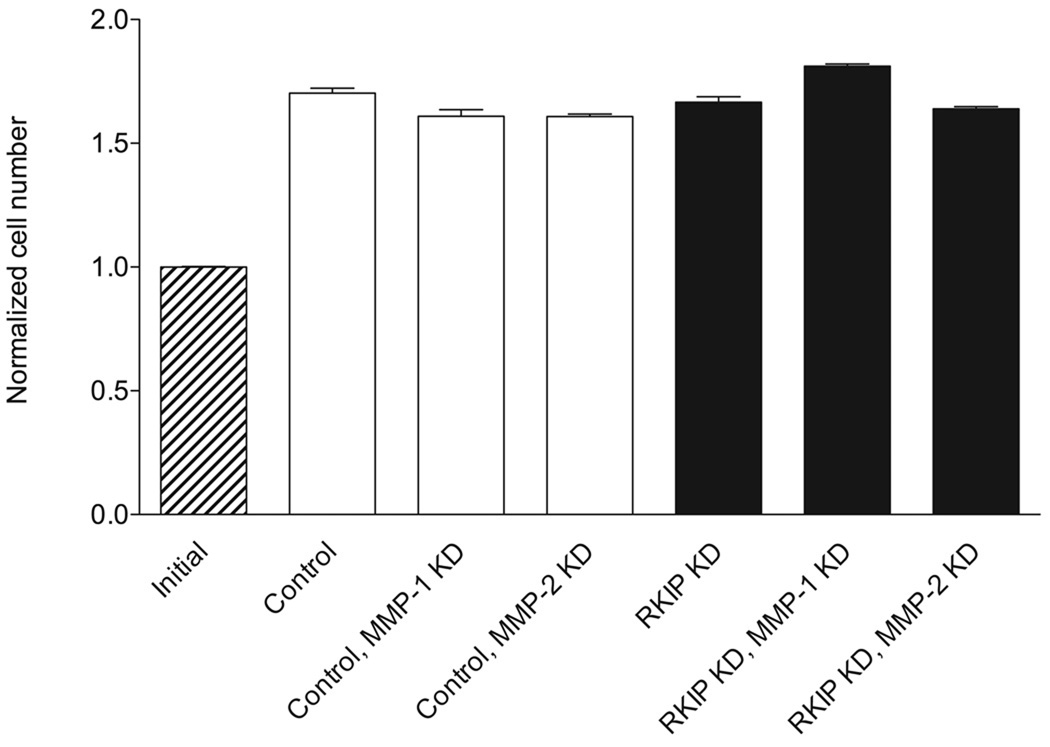
Since MMP-1 and MMP-2 expression were very strongly negatively correlated with RKIP expression in T47D cells, we prepared RKIP/MMP-1-double-knockdown and RKIP/MMP-2-double-knockdown T47D cells. When we tested these cells in the invasion assay, we found that knockdown of either MMP-1 or MMP-2 resulted in a reversion of RKIP-knockdown cells to a control level of cell invasion (Fig. 5), with no effect on the rate of cell migration in the wound closure assay (Fig. 6). As a control experiment to determine whether differences in cell numbers in the invasion experiment could be due to differential rates of cell growth, we investigated the rate of proliferation of the different cells and found that growth rates were similar for all the cells (Fig. 7).
Fig. 5.
Knockdown of MMP-1 or MMP-2 “rescues” the highly invasive phenotype resulting from RKIP knockdown. (A) Expression of MMP-1 and MMP-2 in the RKIP/MMP-1 and RKIP/MMP-2 double-knockdown T47D cells, as determined by quantitative real-time RT-PCR (with glyceraldehyde-3-phosphate dehydrogenase, whose expression was unaffected by knockdown of RKIP and the MMPs, as an internal standard). “Control” refers to cells expressing a control siRNA for firefly luciferase alone. (B) Double knockdown of RKIP and MMP-1 or MMP-2 restores a normal rate of invasion of T47D cells. The values represent the means and SEM for the number of cells invading through Matrigel for nine wells from three independent experiments. “Control” refers to cells expressing a control siRNA for firefly luciferase, either alone or in combination with expression of siRNAs for MMP-1 or MMP-2, as indicated. Invasion of control and RKIP-single-knockdown cells was evaluated in experiments independent from those in Fig. 2 and in parallel with the double-knockdown cells.
Fig. 6.
Knockdown of MMP-1 or MMP-2 in RKIP-knockdown cells has no effect on the rate of cell migration in the absence of an ECM barrier. The values represent the means and SEM for the percent closure of wounds as a function of time in T47D cell monolayers (n = indicated number of separately treated wounds on multiwell tissue culture-treated plates from three independent experiments). “Control” refers to cells expressing a control siRNA for firefly luciferase in combination with expression of siRNAs for MMP-1 or MMP-2, as indicated.
Fig. 7.
Knockdown of RKIP and/or MMP-1 or MMP-2 has little or no effect on the rate of cell proliferation. T47D cells were serum starved for 24 h, then plated at a uniform density onto multiwell tissue culture-treated plates in serum-containing medium. After 48 h, cell numbers were determined. The values represent the means and SEM for cell number normalized to the mean initial control cell number for 15 wells from three independent experiments.
3.6. The NF-κB pathway, but not the Raf/MEK/ERK pathway, appears to be involved in invasion of T47D cells
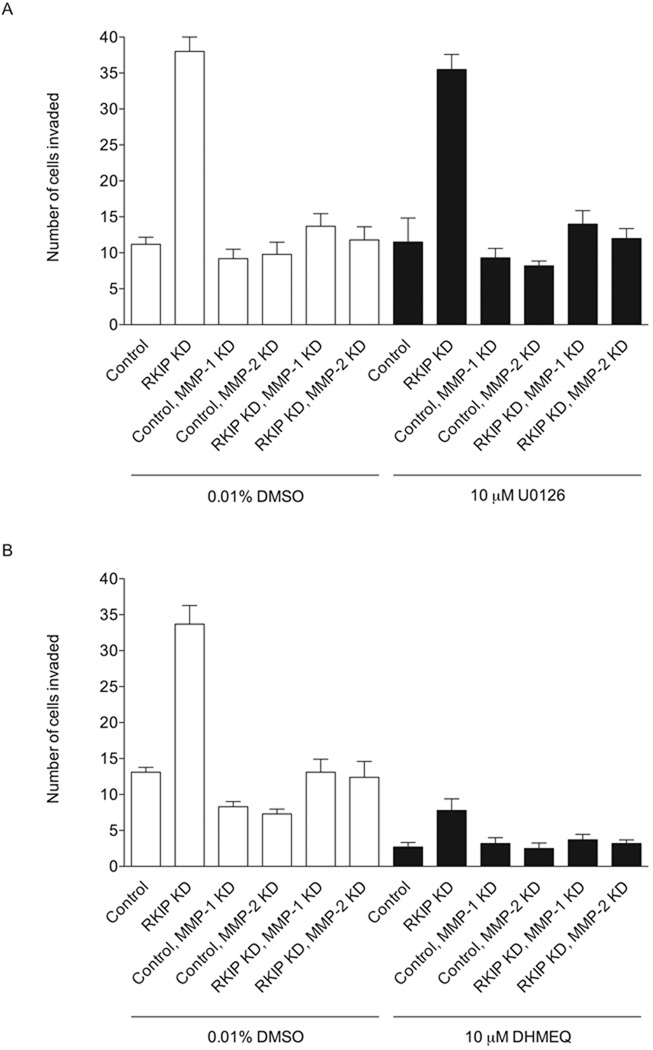
In order to determine whether the Raf/MEK/ERK or NF-κB pathways may be involved in invasion of T47D cells through Matrigel, we conduced invasion experiments in the presence of U0126, a MEK inhibitor [53], or DHMEQ, an NF-κB inhibitor [54]. DHMEQ reduced the rate of invasion of control, RKIP-knockdown and RKIP/MMP-1 and RKIP/MMP-2 double-knockdown T47D cells, while U0126 had no significant effect on invasion, as determined by Student’s t test, for any of the cells (Fig. 9). The degree of invasion of RKIP-knockdown cells in the presence of DHMEQ is 23% that of the untreated RKIP-knockdown cells.
Fig. 9.
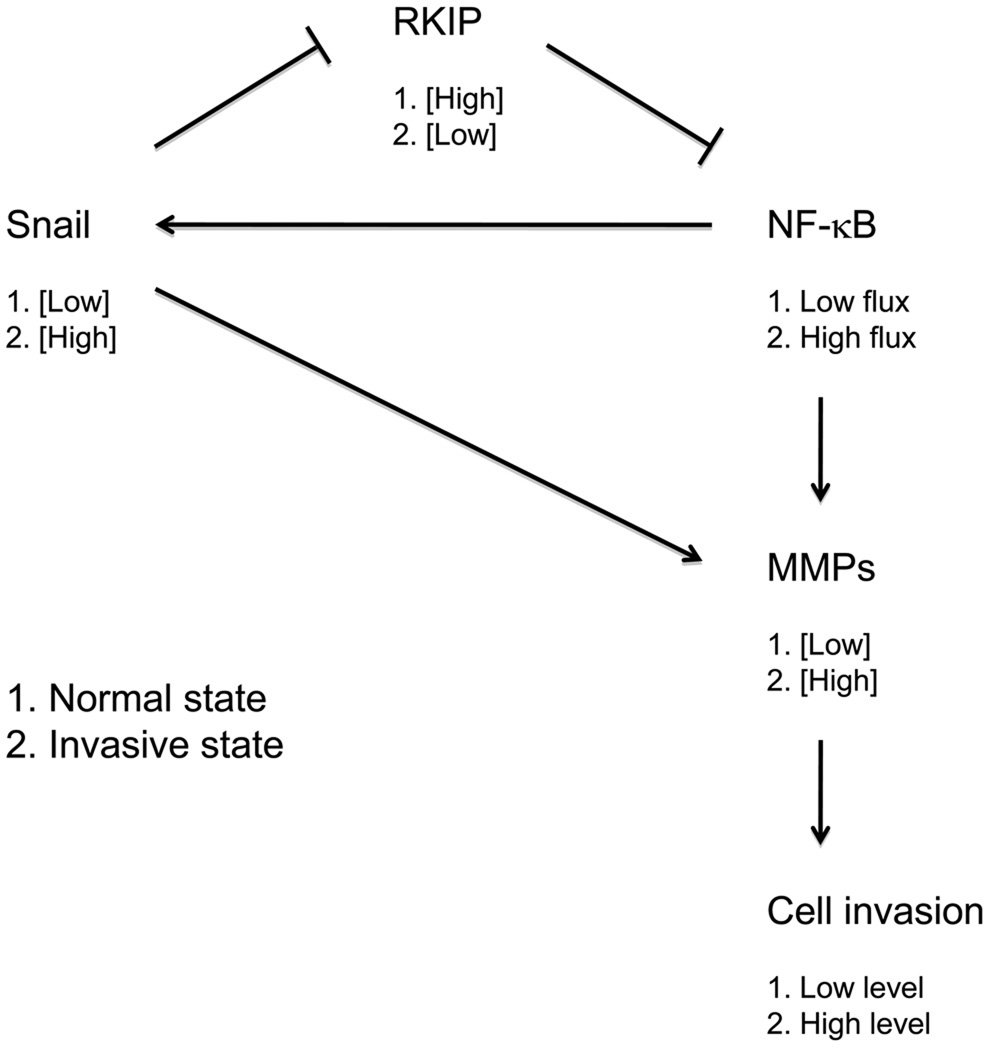
Schematic diagram of a plausible regulatory loop controlling RKIP-limited cell invasion. See text for details.
4. Discussion
We sought to better understand the basis of RKIP’s function as a suppressor of cancer metastasis. To do so, we silenced expression of RKIP in cancer cell lines generally considered to have low metastatic potential and, conversely, overexpressed RKIP in more metastatic cancer cell lines. We examined the effect of these manipulations on the migration and invasion of these cells, as well as metastasis in vivo in one case, and investigated the regulation of MMP expression by RKIP and roles of the RKIP-regulated Raf and NF-κB pathways in cancer cell invasion.
In all cell lines we have tested to date, silencing of RKIP expression either reduces or has no effect on the rate of cell migration. RKIP appears to have a positive function in the control of cell migration in MDCK and MCF7 cells [27–29] and in LoVo cells (Fig. 1). RKIP also appears to play a positive role in the migration of hepatic stellate cells [30]. In contrast, another study suggests that RKIP may play a negative role in the migration of hepatocellular carcinoma cells [26].
The present study thus reveals that RKIP has either a positive role or no observable role in cell migration in different cancer cell lines. The metastasis suppressor function of RKIP does not appear to be mediated at the level of cell migration through any migration-suppressing activity. Instead, we found for all but one of the cancer cell lines tested that knockdown of RKIP resulted in increased invasion through Matrigel (Fig. 2A). Conversely, overexpression of RKIP results in decreased invasion in vitro (Fig. 2B) and in markedly decreased formation of metastases in vivo from murine tumor allografts (Fig. 3), demonstrating a major suppressive function of RKIP in invasion and metastasis.
Our results demonstrate the divergence of functions of RKIP in two-dimensional cell migration in the absence of an ECM barrier compared to three-dimensional cell migration in the presence of a barrier. The resolution to these contrasting findings is that RKIP has an important negative role in the control of the expression of the ECM-degrading proteases MMP-1 and MMP-2, as determined by quantitative real-time RT-PCR (Fig. 4). This is in general agreement with other studies showing that overexpression of RKIP reduces the levels of expression of certain MMPs in C-28/I2 human chondrocytes [55], MDA-MB-231 human breast carcinoma cells [56] and SNB-19 human brain cancer cells [57] (though not in PC-3 human prostate cancer cells [57], NCI-H460 human lung cancer cells [57] and SK-OV-3 human ovarian cancer cells [57]). The invasion-suppressing activity of RKIP could be explained by regulation of MMP-1 and MMP-2 expression, since double knockdown of RKIP and MMP-1 or MMP-2 completely or almost completely restores a normal level of invasiveness in T47D cells (Fig. 5), while not affecting migration in the absence of an ECM barrier (Fig. 6) or the rate of cell proliferation (Fig. 7). The invasive phenotype of cells with silenced RKIP expression appears to be the result of elevated expression of specific MMPs and the consequent increased degradation of ECM barriers to invasion, not the result of any change in the basic locomotive potential of the cells.
RKIP therefore modulates the activity of factors that control the expression of specific MMPs, particularly MMP-1 and MMP-2, in T47D cells. This regulation could take the form of negative control of activators or positive control of repressors of MMP transcription. It could also involve post-transcriptional regulation such as modulation of mRNA splicing or stability. While these questions remain to be answered, a plausible outline of how RKIP might control MMP expression can be fleshed out.
The mechanism by which RKIP suppresses cancer cell invasion and metastasis appears to involve negative control of MMP expression through inhibition of the NF-κB pathway but not the Raf/MEK/ERK pathway (Fig. 8). RKIP is known to modulate the function of a number of protein kinases, including Raf-1 [1,2], B-Raf [35] (although B-Raf activation may not be as tightly controlled by RKIP as is Raf-1 activation [58]) and kinases that control activation of NF-κB [3,4]. RKIP consequently negatively regulates the Raf/MEK/ERK [1,2] and the NF-κB pathways [3,4]. Activated NF-κB complexes regulate MMP transcription (for a review, see ref. [59]). In addition, while the Raf/MEK/ERK pathway also controls expression of MMPs through a number of transcription factors, including CCAAT/enhancer-binding protein β, Ets, Fos and Jun (for a review, see ref. [59]), it may be of lesser importance in T47D cell invasion (Fig. 8). Furthermore, the expression of the proinvasive transcriptional regulator Snail is partly controlled by the NF-κB [60,61] and Raf/MEK/ERK [14,62] pathways, and RKIP negatively regulates Snail expression [14,63,64]. Snail, in turn, promotes expression of a number of MMPs, including MMP-1 and MMP-2, although the mechanism is unclear and likely to be indirect (for a review, see ref. [65]).
Fig. 8.
The NF-κB pathway but not the Raf/MEK/ERK pathway is involved in cell invasion. Invasion of control, RKIP-knockdown and RKIP/MMP-1 and RKIP/MMP-2 double-knockdown T47D cells through Matrigel in the presence or absence of 10 µM U0126, a MEK inhibitor (A), or 10 µM DHMEQ, an NF-κB inhibitor (B), was evaluated. The values represent the means and SEM for the number of cells invading for six wells from three independent experiments. The experiments were conducted with the same cells and in the same manner as in Fig. 5B, with untreated cells assayed again in parallel with compound-treated cells.
Interestingly, expression of RKIP is itself negatively controlled by Snail to ostensibly form a regulatory feedback loop [21,63,64]. This network may by default reinforce “low” Snail and “high” RKIP expression (Fig. 9). However, the opposite metastable state (high Snail and low RKIP expression) may be favored in response to normal embryonic invasion signals or when the system becomes dysregulated during the progression of RKIP-limited metastatic cancers (Fig. 9). Implicit in the findings reported here, the major output of this regulatory network limiting invasiveness of cells is control of the expression level of specific MMPs. The contribution of Snail to the regulation of MMP expression could be through repression of RKIP expression exclusively, thus affecting flux through the NF-κB pathway, or also through additional RKIP-independent pathways branching from the central regulatory loop. In both cases, the feedback system between RKIP and Snail would control output to MMP expression and cell invasion.
In summary, the metastasis-suppressing activity of RKIP does not result from a negative function of RKIP on the raw ability of cells to move in the absence of an ECM barrier. In fact, RKIP either has a positive function or no function in the control of cell movement, depending on the cell type. RKIP’s metastasis-suppressing activity instead appears to arise from a negative role in the modulation of NF-κB-mediated expression of specific MMPs and thus degradation of ECM and cell invasion.
Acknowledgements
We thank Eric Tsung for technical assistance, Dr. Robert Negrin for the firefly luciferase-YFP expression construct, Dr. Joan Massagué for the MMP-1 and MMP-2 shRNA expression constructs and Dr. Kazuo Umezawa for the sample of DHMEQ. This work was supported by National Institutes of Health grants GM077622 (G. Fenteany) and CA133479 (K.C. Yeung).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
None declared.
References
- 1.Yeung K, Seitz T, Li S, Janosch P, McFerran B, Kaiser C, Fee F, Katsanakis KD, Rose DW, Mischak H, Sedivy JM, Kolch W. Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature. 1999;401:173–177. doi: 10.1038/43686. [DOI] [PubMed] [Google Scholar]
- 2.Yeung K, Janosch P, McFerran B, Rose DW, Mischak H, Sedivy JM, Kolch W. Mechanism of suppression of the Raf/MEK/extracellular signal-regulated kinase pathway by the Raf kinase inhibitor protein. Mol. Cell. Biol. 2000;20:3079–3085. doi: 10.1128/mcb.20.9.3079-3085.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yeung KC, Rose DW, Dhillon AS, Yaros D, Gustafsson M, Chatterjee D, McFerran B, Wyche J, Kolch W, Sedivy JM. Raf kinase inhibitor protein interacts with NF-κB-inducing kinase and TAK1 and inhibits NF-κB activation. Mol. Cell. Biol. 2001;21:7207–7217. doi: 10.1128/MCB.21.21.7207-7217.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang H, Park S, Sun SC, Trumbly R, Ren G, Tsung E, Yeung KC. RKIP inhibits NF-κB in cancer cells by regulating upstream signaling components of the IκB kinase complex. FEBS Lett. 2010;584:662–668. doi: 10.1016/j.febslet.2009.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klysik J, Theroux SJ, Sedivy JM, Moffit JS, Boekelheide K. Signaling crossroads: the function of Raf kinase inhibitory protein in cancer, the central nervous system and reproduction. Cell Signal. 2008;20:1–9. doi: 10.1016/j.cellsig.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeng L, Imamoto A, Rosner MR. Raf kinase inhibitory protein (RKIP): a physiological regulator and future therapeutic target. Expert Opin. Ther. Targets. 2008;12:1275–1287. doi: 10.1517/14728222.12.10.1275. [DOI] [PubMed] [Google Scholar]
- 7.Bernier I, Tresca JP, Jolles P. Ligand-binding studies with a 23 kDa protein purified from bovine brain cytosol. Biochim. Biophys. Acta. 1986;871:19–23. doi: 10.1016/0167-4838(86)90128-7. [DOI] [PubMed] [Google Scholar]
- 8.Schoentgen F, Saccoccio F, Jolles J, Bernier I, Jolles P. Complete amino acid sequence of a basic 21-kDa protein from bovine brain cytosol. Eur. J. Biochem. 1987;166:333–338. doi: 10.1111/j.1432-1033.1987.tb13519.x. [DOI] [PubMed] [Google Scholar]
- 9.Atmanene C, Laux A, Glattard E, Muller A, Schoentgen F, Metz-Boutigue MH, Aunis D, Van Dorsselaer A, Stefano GB, Sanglier-Cianferani S, Goumon Y. Characterization of human and bovine phosphatidylethanolamine-binding protein (PEBP/RKIP) interactions with morphine and morphine-glucuronides determined by noncovalent mass spectrometry. Med. Sci. Monit. 2009;15:BR178–BR187. [PubMed] [Google Scholar]
- 10.Fu Z, Smith PC, Zhang L, Rubin MA, Dunn RL, Yao Z, Keller ET. Effects of Raf kinase inhibitor protein expression on suppression of prostate cancer metastasis. J. Natl. Cancer Inst. 2003;95:878–889. doi: 10.1093/jnci/95.12.878. [DOI] [PubMed] [Google Scholar]
- 11.Chatterjee D, Bai Y, Wang Z, Beach S, Mott S, Roy R, Braastad C, Sun Y, Mukhopadhyay A, Aggarwal BB, Darnowski J, Pantazis P, Wyche J, Fu Z, Kitagwa Y, Keller ET, Sedivy JM, Yeung KC. RKIP sensitizes prostate and breast cancer cells to drug-induced apoptosis. J. Biol. Chem. 2004;279:17515–17523. doi: 10.1074/jbc.M313816200. [DOI] [PubMed] [Google Scholar]
- 12.Schuierer MM, Bataille F, Hagan S, Kolch W, Bosserhoff AK. Reduction in Raf kinase inhibitor protein expression is associated with increased Ras-extracellular signal-regulated kinase signaling in melanoma cell lines. Cancer Res. 2004;64:5186–5192. doi: 10.1158/0008-5472.CAN-03-3861. [DOI] [PubMed] [Google Scholar]
- 13.Hagan S, Al-Mulla F, Mallon E, Oien K, Ferrier R, Gusterson B, Garcia JJ, Kolch W. Reduction of Raf-1 kinase inhibitor protein expression correlates with breast cancer metastasis. Clin. Cancer Res. 2005;11:7392–7397. doi: 10.1158/1078-0432.CCR-05-0283. [DOI] [PubMed] [Google Scholar]
- 14.Dangi-Garimella S, Yun J, Eves EM, Newman M, Erkeland SJ, Hammond SM, Minn AJ, Rosner MR. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J. 2009;28:347–358. doi: 10.1038/emboj.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Mulla F, Hagan S, Behbehani AI, Bitar MS, George SS, Going JJ, Garcia JJ, Scott L, Fyfe N, Murray GI, Kolch W. Raf kinase inhibitor protein expression in a survival analysis of colorectal cancer patients. J. Clin. Oncol. 2006;24:5672–5679. doi: 10.1200/JCO.2006.07.5499. [DOI] [PubMed] [Google Scholar]
- 16.Minoo P, Zlobec I, Baker K, Tornillo L, Terracciano L, Jass JR, Lugli A. Loss of Raf-1 kinase inhibitor protein expression is associated with tumor progression and metastasis in colorectal cancer. Am. J. Clin. Pathol. 2007;127:820–827. doi: 10.1309/5D7MM22DAVGDT1R8. [DOI] [PubMed] [Google Scholar]
- 17.Zlobec I, Baker K, Minoo P, Jass JR, Terracciano L, Lugli A. Node-negative colorectal cancer at high risk of distant metastasis identified by combined analysis of lymph node status, vascular invasion, and Raf-1 kinase inhibitor protein expression. Clin. Cancer Res. 2008;14:143–148. doi: 10.1158/1078-0432.CCR-07-1380. [DOI] [PubMed] [Google Scholar]
- 18.Zlobec I, Baker K, Terracciano L, Peter S, Degen L, Beglinger C, Lugli A. Two-marker protein profile predicts poor prognosis in patients with early rectal cancer. Br. J. Cancer. 2008;99:1712–1717. doi: 10.1038/sj.bjc.6604729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li HZ, Wang Y, Gao Y, Shao J, Zhao XL, Deng WM, Liu YX, Yang J, Yao Z. Effects of Raf kinase inhibitor protein expression on metastasis and progression of human epithelial ovarian cancer. Mol. Cancer Res. 2008;6:917–928. doi: 10.1158/1541-7786.MCR-08-0093. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y, Ouyang GL, Yi H, Li MY, Zhang PF, Li C, Li JL, Liu YF, Chen ZC, Xiao ZQ. Identification of RKIP as an invasion suppressor protein in nasopharyngeal carcinoma by proteomic analysis. J. Proteome Res. 2008;7:5254–5262. doi: 10.1021/pr800602c. [DOI] [PubMed] [Google Scholar]
- 21.Beach S, Tang H, Park S, Dhillon AS, Keller ET, Kolch W, Yeung KC. Snail is a repressor of RKIP transcription in metastatic prostate cancer cells. Oncogene. 2008;27:2243–2248. doi: 10.1038/sj.onc.1210860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayashi E, Kuramitsu Y, Okada F, Fujimoto M, Zhang X, Kobayashi M, Iizuka N, Ueyama Y, Nakamura K. Proteomic profiling for cancer progression: Differential display analysis for the expression of intracellular proteins between regressive and progressive cancer cell lines. Proteomics. 2005;5:1024–1032. doi: 10.1002/pmic.200401132. [DOI] [PubMed] [Google Scholar]
- 23.Fu Z, Kitagawa Y, Shen R, Shah R, Mehra R, Rhodes D, Keller PJ, Mizokami A, Dunn R, Chinnaiyan AM, Yao Z, Keller ET. Metastasis suppressor gene Raf kinase inhibitor protein (RKIP) is a novel prognostic marker in prostate cancer. Prostate. 2006;66:248–256. doi: 10.1002/pros.20319. [DOI] [PubMed] [Google Scholar]
- 24.Martinho O, Gouveia A, Silva P, Pimenta A, Reis RM, Lopes JM. Loss of RKIP expression is associated with poor survival in GISTs. Virchows Arch. 2009;455:277–284. doi: 10.1007/s00428-009-0821-z. [DOI] [PubMed] [Google Scholar]
- 25.Chatterjee D, Sabo E, Tavares R, Resnick MB. Inverse association between Raf kinase inhibitory protein and signal transducers and activators of transcription 3 expression in gastric adenocarcinoma patients: implications for clinical outcome. Clin. Cancer Res. 2008;14:2994–3001. doi: 10.1158/1078-0432.CCR-07-4496. [DOI] [PubMed] [Google Scholar]
- 26.Lee HC, Tian B, Sedivy JM, Wands JR, Kim M. Loss of Raf kinase inhibitor protein promotes cell proliferation and migration of human hepatoma cells. Gastroenterology. 2006;131:1208–1217. doi: 10.1053/j.gastro.2006.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mc Henry KT, Ankala SV, Ghosh AK, Fenteany G. A non-antibacterial oxazolidinone derivative that inhibits epithelial cell sheet migration. ChemBioChem. 2002;3:1105–1111. doi: 10.1002/1439-7633(20021104)3:11<1105::AID-CBIC1105>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 28.Zhu S, Mc Henry KT, Lane WS, Fenteany G. A chemical inhibitor reveals the role of Raf kinase inhibitor protein in cell migration. Chem. Biol. 2005;12:981–991. doi: 10.1016/j.chembiol.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 29.Mc Henry KT, Montesano R, Zhu S, Beshir AB, Tang HH, Yeung KC, Fenteany G. Raf kinase inhibitor protein positively regulates cell-substratum adhesion while negatively regulating cell-cell adhesion. J. Cell. Biochem. 2008;103:972–985. doi: 10.1002/jcb.21470. [DOI] [PubMed] [Google Scholar]
- 30.Ma J, Li F, Liu L, Cui D, Wu X, Jiang X, Jiang H. Raf kinase inhibitor protein inhibits cell proliferation but promotes cell migration in rat hepatic stellate cells. Liver Int. 2009;29:567–574. doi: 10.1111/j.1478-3231.2009.01981.x. [DOI] [PubMed] [Google Scholar]
- 31.Zhang L, Fu Z, Binkley C, Giordano T, Burant CF, Logsdon CD, Simeone DM. Raf kinase inhibitory protein inhibits β-cell proliferation. Surgery. 2004;136:708–715. doi: 10.1016/j.surg.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 32.Eves EM, Shapiro P, Naik K, Klein UR, Trakul N, Rosner MR. Raf kinase inhibitory protein regulates Aurora B kinase and the spindle checkpoint. Mol. Cell. 2006;23:561–574. doi: 10.1016/j.molcel.2006.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woods Ignatoski KM, Grewal NK, Markwart SM, Vellaichamy A, Chinnaiyan AM, Yeung K, Ray ME, Keller ET. Loss of Raf kinase inhibitory protein induces radioresistance in prostate cancer. Int. J. Radiat. Oncol. Biol. Phys. 2008;72:153–160. doi: 10.1016/j.ijrobp.2008.04.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gupta GP, Nguyen DX, Chiang AC, Bos PD, Kim JY, Nadal C, Gomis RR, Manova-Todorova K, Massagué J. Mediators of vascular remodelling co-opted for sequential steps in lung metastasis. Nature. 2007;446:765–770. doi: 10.1038/nature05760. [DOI] [PubMed] [Google Scholar]
- 35.Park S, Yeung ML, Beach S, Shields JM, Yeung KC. RKIP downregulates B-Raf kinase activity in melanoma cancer cells. Oncogene. 2005;24:3535–3540. doi: 10.1038/sj.onc.1208435. [DOI] [PubMed] [Google Scholar]
- 36.Edinger M, Cao YA, Verneris MR, Bachmann MH, Contag CH, Negrin RS. Revealing lymphoma growth and the efficacy of immune cell therapies using in vivo bioluminescence imaging. Blood. 2003;101:640–648. doi: 10.1182/blood-2002-06-1751. [DOI] [PubMed] [Google Scholar]
- 37.Li HZ, Gao Y, Zhao XL, Liu YX, Sun BC, Yang J, Yao Z. Effects of Raf kinase inhibitor protein expression on metastasis and progression of human breast cancer. Mol. Cancer Res. 2009;7:832–840. doi: 10.1158/1541-7786.MCR-08-0403. [DOI] [PubMed] [Google Scholar]
- 38.Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992;52:1399–1405. [PubMed] [Google Scholar]
- 39.Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, Iyer V, Jeffrey SS, Van de Rijn M, Waltham M, Pergamenschikov A, Lee JC, Lashkari D, Shalon D, Myers TG, Weinstein JN, Botstein D, Brown PO. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 40.Ellison G, Klinowska T, Westwood RF, Docter E, French T, Fox JC. Further evidence to support the melanocytic origin of MDA-MB-435. Mol. Pathol. 2002;55:294–299. doi: 10.1136/mp.55.5.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J, Lee C, Wagner SN, Li C, Golub TR, Rimm DL, Meyerson ML, Fisher DE, Sellers WR. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 42.Christgen M, Lehmann U. MDA-MB-435: the questionable use of a melanoma cell line as a model for human breast cancer is ongoing. Cancer Biol. Ther. 2007;6:1355–1357. doi: 10.4161/cbt.6.9.4624. [DOI] [PubMed] [Google Scholar]
- 43.Rae JM, Creighton CJ, Meck JM, Haddad BR, Johnson MD. MDA-MB-435 cells are derived from M14 melanoma cells–a loss for breast cancer, but a boon for melanoma research. Breast Cancer Res. Treat. 2007;104:13–19. doi: 10.1007/s10549-006-9392-8. [DOI] [PubMed] [Google Scholar]
- 44.Lacroix M. Persistent use of "false" cell lines. Int. J. Cancer. 2008;122:1–4. doi: 10.1002/ijc.23233. [DOI] [PubMed] [Google Scholar]
- 45.Lacroix M. MDA-MB-435 cells are from melanoma, not from breast cancer. Cancer Chemother. Pharmacol. 2009;63:567. doi: 10.1007/s00280-008-0776-9. [DOI] [PubMed] [Google Scholar]
- 46.Sellappan S, Grijalva R, Zhou X, Yang W, Eli MB, Mills GB, Yu D. Lineage infidelity of MDA-MB-435 cells: expression of melanocyte proteins in a breast cancer cell line. Cancer Res. 2004;64:3479–3485. doi: 10.1158/0008-5472.CAN-3299-2. [DOI] [PubMed] [Google Scholar]
- 47.Chambers AF. MDA-MB-435 and M14 cell lines: identical but not M14 melanoma? Cancer Res. 2009;69:5292–5293. doi: 10.1158/0008-5472.CAN-09-1528. [DOI] [PubMed] [Google Scholar]
- 48.Hollestelle A, Schutte M. Comment Re: MDA-MB-435 and M14 Cell Lines: Identical but not M14 Melanoma? Cancer Res. 2009;69:7893. doi: 10.1158/0008-5472.CAN-09-2396. [DOI] [PubMed] [Google Scholar]
- 49.Montel V, Suzuki M, Galloy C, Mose ES, Tarin D. Expression of melanocyte-related genes in human breast cancer and its implications. Differentiation. 2009;78:283–291. doi: 10.1016/j.diff.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 50.Deryugina EI, Quigley JP. Matrix metalloproteinases and tumor metastasis. Cancer Metastasis Rev. 2006;25:9–34. doi: 10.1007/s10555-006-7886-9. [DOI] [PubMed] [Google Scholar]
- 51.Fingleton B. Matrix metalloproteinases: roles in cancer and metastasis. Front. Biosci. 2006;11:479–491. doi: 10.2741/1811. [DOI] [PubMed] [Google Scholar]
- 52.Holleran WM, Galardy RE, Gao WN, Levy D, Tang PC, Elias PM. Matrix metalloproteinase inhibitors reduce phorbol ester-induced cutaneous inflammation and hyperplasia. Arch. Dermatol. Res. 1997;289:138–144. doi: 10.1007/s004030050169. [DOI] [PubMed] [Google Scholar]
- 53.Favata MF, Horiuchi KY, Manos EJ, Daulerio AJ, Stradley DA, Feeser WS, Van Dyk DE, Pitts WJ, Earl RA, Hobbs F, Copeland RA, Magolda RL, Scherle PA, Trzaskos JM. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J. Biol. Chem. 1998;273:18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- 54.Matsumoto N, Ariga A, To-e S, Nakamura H, Agata N, Hirano S, Inoue J, Umezawa K. Synthesis of NF-κB activation inhibitors derived from epoxyquinomicin C. Bioorg. Med. Chem. Lett. 2000;10:865–869. doi: 10.1016/s0960-894x(00)00114-1. [DOI] [PubMed] [Google Scholar]
- 55.Xu L, Peng H, Glasson S, Lee PL, Hu K, Ijiri K, Olsen BR, Goldring MB, Li Y. Increased expression of the collagen receptor discoidin domain receptor 2 in articular cartilage as a key event in the pathogenesis of osteoarthritis. Arthritis Rheum. 2007;56:2663–2673. doi: 10.1002/art.22761. [DOI] [PubMed] [Google Scholar]
- 56.Delassus GS, Cho H, Park J, Eliceiri GL. New pathway links from cancer-progression determinants to gene expression of matrix metalloproteinases in breast cancer cells. J. Cell Physiol. 2008;217:739–744. doi: 10.1002/jcp.21548. [DOI] [PubMed] [Google Scholar]
- 57.Delassus GS, Cho H, Hoang S, Eliceiri GL. Many new down- and up-regulatory signaling pathways, from known cancer progression suppressors to matrix metalloproteinases, differ widely in cells of various cancers. J. Cell Physiol. 2010;224:549–558. doi: 10.1002/jcp.22157. [DOI] [PubMed] [Google Scholar]
- 58.Trakul N, Menard RE, Schade GR, Qian Z, Rosner MR. Raf kinase inhibitory protein regulates Raf-1 but not B-Raf kinase activation. J. Biol. Chem. 2005;280:24931–24940. doi: 10.1074/jbc.M413929200. [DOI] [PubMed] [Google Scholar]
- 59.Vincenti MP, Brinckerhoff CE. Signal transduction and cell-type specific regulation of matrix metalloproteinase gene expression: can MMPs be good for you? J. Cell. Physiol. 2007;213:355–364. doi: 10.1002/jcp.21208. [DOI] [PubMed] [Google Scholar]
- 60.Barbera MJ, Puig I, Dominguez D, Julien-Grille S, Guaita-Esteruelas S, Peiro S, Baulida J, Franci C, Dedhar S, Larue L, Garcia de Herreros A. Regulation of Snail transcription during epithelial to mesenchymal transition of tumor cells. Oncogene. 2004;23:7345–7354. doi: 10.1038/sj.onc.1207990. [DOI] [PubMed] [Google Scholar]
- 61.Julien S, Puig I, Caretti E, Bonaventure J, Nelles L, van Roy F, Dargemont C, de Herreros AG, Bellacosa A, Larue L. Activation of NF-κB by Akt upregulates Snail expression and induces epithelium mesenchyme transition. Oncogene. 2007;26:7445–7456. doi: 10.1038/sj.onc.1210546. [DOI] [PubMed] [Google Scholar]
- 62.Lan M, Kojima T, Murata M, Osanai M, Takano K, Chiba H, Sawada N. Phosphorylation of ezrin enhances microvillus length via a p38 MAP-kinase pathway in an immortalized mouse hepatic cell line. Exp. Cell Res. 2006;312:111–120. doi: 10.1016/j.yexcr.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 63.Baritaki S, Chapman A, Yeung K, Spandidos DA, Palladino M, Bonavida B. Inhibition of epithelial to mesenchymal transition in metastatic prostate cancer cells by the novel proteasome inhibitor, NPI-0052: pivotal roles of Snail repression and RKIP induction. Oncogene. 2009;28:3573–3585. doi: 10.1038/onc.2009.214. [DOI] [PubMed] [Google Scholar]
- 64.Baritaki S, Yeung K, Palladino M, Berenson J, Bonavida B. Pivotal roles of snail inhibition and RKIP induction by the proteasome inhibitor NPI-0052 in tumor cell chemoimmunosensitization. Cancer Res. 2009;69:8376–8385. doi: 10.1158/0008-5472.CAN-09-1069. [DOI] [PubMed] [Google Scholar]
- 65.Munshi HG, Stack MS. Reciprocal interactions between adhesion receptor signaling and MMP regulation. Cancer Metastasis Rev. 2006;25:45–56. doi: 10.1007/s10555-006-7888-7. [DOI] [PubMed] [Google Scholar]