Abstract
Background
NHAoc/NHA2 is highly and selectively expressed in osteoclasts and plays a role(s) in normal osteoclast differentiation, apoptosis and bone resorptive function in vitro. Extensive mutational analysis of a bacterial homologue, NhaA, has revealed a number of amino acid residues essential for its activity. Some of these residues are evolutionarily conserved and have been shown to be essential not only for activity of NhaA in bacteria, but also of NHAoc/NHA2 in eukaryotes.
Methods
The salt-sensitive Saccharomyces cerevisiae strain BW31a was used for heterologous expression of mutants of NHAoc/NHA2. Membrane expression of NHAoc/NHA2 was confirmed by confocal microscopy. Intracellular concentration of Na+ (a measure of Na+ antiporter activity) was estimated by atomic absorption spectroscopy. The growth phenotypes of cells expressing NHAoc/NHA2 mutants were studied on YNB agar supplemented with NaCl and by growth curves in YNB broth.
Results
Mutations in amino acid residues V161 and F357 reduced the ability of transfected BW31a cells to remove intracellular sodium and to grow in NaCl-containing medium. Yeast expressing the double mutant F357 F437 can not grow in 0.4 M NaCl, suggesting that these residues are also essential for antiporter activity.
Conclusions
Evolutionarily conserved amino acids are required for full antiporter function.
General Significance
Mutations in these amino acid residues may impact NHAoc activity and therefore osteoclast function in vitro and in vivo.
INTRODUCTION
The exchange of Na+ or K+ and H+ down their concentration gradients (antiport or exchange activity) occurs in cells in all phyla and kingdoms. This activity is essential to control intracellular pH, cell volume and reuptake of Na+, and cellular events such as migration, adhesion, proliferation and apoptosis [1–3].
We identified a mouse gene, nhaoc/NHA2, which is induced by RANKL stimulation of osteoclast precursors in vitro and in vivo [4,5]. Orthologues of nhaoc/NHA2 are found in all metazoans studied and define a newly recognized subfamily of metazoan proteins within the CPA2 family of antiporters [1,4,6,7]. Members of this family share a conserved N-terminus (~500 amino acids) predicted to have 10 to 12 transmembrane segments, and a short C-terminal tail (~50–100 amino acids). Together they form a novel family of antiporters which share a common ancestor with NhaA, the main antiporter of Escherichia coli [8].
NhaA has been extensively studied and is the only antiporter for which the 3D crystal structure is known. It is an electrogenic antiporter with a stoichiometry of 2H+/1Na+ whose activity is strongly pH-dependent [8]. Data from genetic-complementation, biochemical pull-down experiments, intermolecular cross-linking and cryo-electron microscopy of 2D crystals studies reveal that NhaA exists as a dimer in the native membrane.
Despite the evolutionary distance, NhaA and its eukaryotic orthologues show remarkable sequence similarity, suggesting that these proteins also have a similar structural architecture, characterized by 10 to 12 predicted transmembrane segments (TMS), depending on the software used [9]. Many amino acid residues are conserved, notably, a pair of adjacent aspartic acid residues, which are essential for antiporter activity in NhaA [10] as well as in HsNHA2 [7]. NhaA has been also subjected to extensive mutational studies. An analysis of the NhaA E241- F267 segment revealed several roles for this region of the protein: i.e. amino acids located in it participate in the “pH sensor”, have effects on the determination of the H+/Na+ stoichiometry, form part of the cytoplasmic funnel leading to the cation binding sites and contribute to the NhaA dimer interface. Finally, a F267C mutation reduces the H+/Na+ stoichiometry of NhaA and a double mutation F267C/F344C inactivates the antiporter activity [8].
NHAoc/NHA2 is unique in that it is predominantly expressed in osteoclasts and is required for osteoclast differentiation in vitro and in vivo [4,5]. In addition, nhaoc/NHA2 silencing inhibits osteoclast formation in vitro. Mutations in genes that affect osteoclast activity and function in mice usually result in osteopetrotic phenotypes in vivo. Likewise, based on our preliminary characterization of NHAoc/NHA2 expression and function, we predict that mutations in this gene that affect activity of the protein will also result in reduced osteoclast differentiation, activity and/or survival.
Nine isoforms of Na+/H+ antiporters have been described in humans. Most cells usually express several of them simultaneously [11], which hinders the proper biochemical and physiological characterization of a particular transporter, due to the presence of other molecules with similar transport velocity and ion specificity in the same cell. These difficulties can be overcome by expressing a given transporter in a host lacking ion transport systems. One such organism employed for heterologous expression is the yeast Saccharomyces cerevisiae.
Saccharomyces cerevisiae is one of the best characterized eukaryotic organisms. It is a unicellular fungus and yet yeast cells are very similar to higher eukaryotes with regards to cell structure and physiology. Because of this, yeasts have been utilized to study a variety of cell functions, including ion transport mechanisms [12–14]. Two different types of membrane transporters mediate Na+ efflux: Na+-ATPases and Na+/H+ antiporters. The ENA genes (ENA1 to ENA4, this number can vary according to the strain) encode Na+-ATPases. The NHA1 gene encodes the H+/cation antiporter.
The Saccharomyces cerevisiae BW31a mutant strain (ena1–4Δ nha1Δ) is very sensitive to salt because it lacks the main sodium and potassium extrusion systems. For this reason, phenotype complementation of BW31a cells (the ability of a heterologously expressed gene to rescue the salt-sensitive growth phenotype) is a powerful approach for the functional characterization of novel antiporters.
The goal of this study is to characterize the antiporter activity of NHAoc/NHA2 mutants in a yeast model. We hypothesize that evolutionarily conserved amino acids that are essential for NhaA antiporter activity will have a similar role in NHAoc/NHA2. We also hypothesize that mutations in those amino acid residues will impact NHAoc/NHA2 activity, and because of its restricted expression pattern, osteoclast function.
To test these hypotheses, we expressed mutants of NHAoc/NHA2 in Saccharomyces cerevisiae BW31a cells and assessed the ability of the expressed mutants to rescue the salt sensitive phenotype. We have introduced several mutations in the NHAoc/NHA2 molecule: these are a) mutations in Aspartic Acid residues 278 and 279 to Cysteine individually. A double D278C-D279C mutant, which has previously been shown to abolish activity [7,18] was used as negative control b) three point mutations of hydrophobic amino-acid residues: Isoleucine 159, Valine 161 and Phenylalanine 357, which are amino acids that can be substituted in humans as a result of single nucleotide polymorphisms (SNP) (I159T, V161A and F357C) and whose conservation throughout evolution suggests that each may be an important determinant of NHAoc/NHA2 activity, and c) a mutation of F437 to Cysteine alone or F357C-F437C double mutant. F437 is homologous to NhaA F344.
MATERIALS AND METHODS
Yeast strains, media and growth conditions
For heterologous expression of NHAoc/NHA2 we used the salt-sensitive Saccharomyces cerevisiae strain BW31a (ena1-4Δ, nha1Δ)[12]. Yeast cells were cultured in YPD (2% peptone, 1% yeast extract, 2% glucose) or YNB media (0.67% yeast nitrogen base w/o amino acids, 2% glucose or galactose, and appropriate supplements).
DNA manipulations
DNA manipulation, including the isolation of plasmids and cell transformations, were performed according to standard protocols.
Plasmid construction
For expression of NHAoc/NHA2 we used the plasmid pYES2.1-NHAoc/NHA2, which harbors the ORF of human NHAoc/NHA2 in the pYES2.1/V5-His-TOPO vector (Invitrogen) for galactose-inducible expression in yeast. The human NHAoc/NHA2 cassette was generated by Taq PCR amplification from plasmid SC128249 (OriGene Technologies, Inc, Rockville, MD) using the following primers:
| Sense: | 5′-GCCATGGGGGATGAAGATAAAAG - 3′ |
| Antisense: | 5′-AACTTGCACAGAAGTCTCTCC - 3′ |
Mutations of pYES2.1-NHAoc were done using the NEB Phusion Site-directed Mutagenesis Kit (F-541S), according to the manufacturer’s instructions, using the following primers. All mutations were confirmed by sequencing.
D278C single mutation
Sense: Phos-TGCATTCTGGCCATCACTGGCTTC, Antisense: Phos-ATCGAAGCTGCCAGCTGCCATGAG
D279C single mutation
Sense: Phos - GACATTCTGGCCATCACTGGCTTC, Antisense: Phos – ACAGAAGCTGCCAGCTGCCATGAG
D278C D279C double mutation
Sense: Phos-TGCATTCTGGCCATCACTGGCTTC, Antisense: Phos – ACAGAAGCTGCCAGCTGCCATGAG
F357C single mutation
Sense: Phos-TGTGGTTTCCCTGGATCAGGAGGA, Antisense: Phos-ATGCACACTGCTGAACACAGCTAG
F437C single mutation
Sense: Phos-TGTCTGATGGTGTGTTTTGCTGG, Antisense: Phos-TGTAGTCAAAATTCGTATCAATAC
F357 F437 - C357 F437 double mutation
Using F357C mutated vector as template, introduce the second (F437C) mutation
I159T
Sense: Phos-TTTCTCATCAGAAATACCCCAGTCATCAACGAT, Antisense: Phos-CCCTGCAAGCAGCATGCCAAGAAGAGA
V161A
Sense: Phos-GCCATCAACGATAATGTGCAGATC, Antisense: Phos-TGGGATATTTCTGATGAGAAACCC
Yeast Electroporation
BW31a cells were prepared for electroporation as follows: One vial of BW31a cells was inoculated into 50 ml YPD and cultured overnight at 30°C with shaking. Cells were harvested in a 50 ml sterile conical tube, centrifuged at 1500 rpm (4°C) for 5 minutes and kept on ice. Cells were next serially washed with 40 ml then 20 ml ice-cold sterile water, resuspended in 5 ml 1M ice-cold Sorbitol and pelleted at 2,000 rpm for 5 minutes at 4°C. Finally, the cells were resuspended in 200 μl ice-cold 1M Sorbitol. 40 μl of yeast suspension were mixed with 1 μl (~0.5μg) DNA in a pre-chilled electroporation cuvette (Cell Projects Ltd. Cat #: EP-102) and electroporated with one pulse: V= 1.5kV, 25 μF, 200 Ohms (Time constant ~4.8 sec). 200ul 1M Sorbitol was immediately added to the cells and 100 μl of electroporated cells were spread onto 2% glucose YNB plates. The plates were incubated at 30°C. Colonies appeared usually after 3 days. Individual colonies were picked up, tested for expression of NHAoc/NHA2 by western blot analysis, and selected for phenotypic tests.
Western blot analysis
For expression studies, yeast protein extracts were subjected to western blot analysis. Briefly, cultures were grown for 48 hours at 30°C in YNB medium supplemented with 2% Galactose to induce expression. Cells (100 μl aliquot) were centrifuged, washed with water, resuspended in 12.5 μl lysis buffer (ZYMO RESEARCH, Orange, CA) plus 0.5 μl Zymolase and incubated for 1 hour at 37°C. The ly sates were further diluted with 25 μl 2X SDS sample buffer and used for western blot analysis using anti V5 antibody (Invitrogen dilution: 1/2000) as the primary antibody and an HRP-conjugated secondary antibody (Cell Signaling Technology, Inc., 1:1000). Blots were washed in TBST to remove unbound antibodies, incubated with 10 ml LumiGLO with gentle agitation for 1 minute at room temperature and exposed to X-ray film.
Confocal Microscopy
BW31a cells transformed with the indicated plasmids were grown in YNB supplemented with Glucose or Galactose (to induce expression of NHAoc/NHA2) as indicated. Cells were fixed by adding 37% formaldehyde to the culture followed by incubation at 30°C for 30 minutes. Cells were then pelleted, washed in 0.1M potassium phosphate pH 6.5 and treated with 2ul Zymolase (5 unit, Zymo Research, E1004) and 5ul of B-mercaptoethanol for 60 minutes at 37°C to digest the cell wall. Cover slips (No. 1.5 thickness - Fisher 12-545-81) were then coated with poly-lysine solution (Sigma P8920) for several minutes, washed once with water, and allowed to air dry. Cells (25ul) were then placed on the cover slips and allowed to attach for several minutes. The excess was aspirated after several minutes and the slides were immersed successively in ice-cold methanol for 6 minutes, then ice-cold acetone for 30 seconds, and allowed to air dry. Cells were blocked once in PBS-BSA (5mg/ml powdered bovine serum albumin in 1X PBS) and incubated in primary antibody (anti V5 from Invitrogen, R960-25) (diluted in PBS-BSA) in a moist chamber overnight at 4°C. Cells were then washed four times in the wells with PBS-BSA and incubated in secondary antibody (Alexa Fluor® 488, Invitrogen A-11017 diluted in PBS-BSA) in a moist chamber for 2 hours. Cells were washed three times with PBS-BSA, once with PBS and then incubated in 1X PBS-DAPI (SIGMA D9542, final concentration 0.1ug/ml) for at least 30 seconds. Cells were then washed in PBS and mounted onto microscope slides. Images were captured with a Leica SP5X confocal microscope.
Phenotypic Tests: Solid and Liquid Media
The growth phenotypes of cells expressing NHAoc/NHA2 mutants were estimated by drop test technique on YNB agar supplemented with NaCl. Briefly, freshly grown cells of each tested strain were resuspended in water and adjusted to the same initial OD600 (about 1). 3 μl aliquots of ten-fold serial dilutions of yeast suspensions were spotted on agar plates. Plates were incubated at 30°C (usually for 4 days) and images were obtained using a digital camera. To estimate the growth of cells in liquid YNB media with or without NaCl, we prepared a pre-culture by growing the previously electroporated yeast in 2 ml YPD plus adenine (20 μg/ml) overnight at 30°C. The cells were centrifuged, washed once in YNB and resuspended in YNB/2% galactose at OD600≈0.05. The growth was followed by measuring OD600 every 12 hours for 72 hours. YNB was supplemented with varying NaCl concentrations (indicated in the figures).
Determination of Cell Na+ Content
Fresh BW31a cells expressing NHAoc/NHA2 wild-type antiporter or its mutant versions were inoculated in 50 ml of YNB media and grown overnight (30°C; 160 rpm) to OD600 ~ 0.6. Fresh YNB medium with 1 M NaCl was added to reach a final concentration of 100 mM NaCl in the cell suspensions. Forty ml of suspension were incubated at 30 °C for 30 minutes, then 5-ml aliquots (three for each strain) were withdrawn, washed with cold deionized water and resuspended in 5 ml of 10 mM TRIS, 0.1 mM MgCl pH 4.5 (adjusted with solid Ca(OH)2). Immediately, cells were collected on Millipore membrane filters, washed, acid extracted, and the intracellular concentration of Na+ was estimated by atomic absorption spectroscopy [15]. The experiment was repeated three times, i.e. the sodium content measured in 9 samples for each strain. The intracellular content of Na+ was expressed in nmol of Na+ per mg of cell dry weight. The intracellular Na+ content in cells growing in the absence of NaCl was below 15 nmol/mg of dry weight. Cells expressing ScNHA1 were used as positive controls and cells with the empty vector were used as negative controls.
RESULTS
Bacterial NhaA and mammalian NHAoc/NHA2 have extensive sequence similarity
SIM [16,17] is a program which finds a user-defined number of best non-intersecting alignments between two protein sequences or within a sequence. The alignments are reported in order of decreasing similarity score and share no aligned pairs. We aligned NhaA and NHAoc/NHA2 using the BLOSUM30 comparison matrix with the following parameters: Number of alignments to be computed: 20; Gap open penalty: 12, Gap extension penalty: 4 (Figure 1). Two of the returned alignments indicate several potentially conserved amino acids, which are indicated with an asterisk “*”. Also highlighted (with a red asterisk) are: NHAoc/NHA2 D278-D279, I159, V161, F357 and F437 as well as their matching NhaA residues. D278-D279 are homologous to D163-D164 of bacterial NhaA. These two amino acids have been found to be essential for antiport activity in bacterial, yeast and mammalian antiporters. I159, V161 and F357 are amino acids that are substituted as a result of human SNPs. F357 and F437 are homologous to F267 and F344 of NhaA, respectively. F267 regulates the stoichiometry of NhaA and a double F267C F344C mutation can abolish NhaA antiporter activity [8].
Figure 1. NhaA - NHAoc/NHA2 sequence alignment.
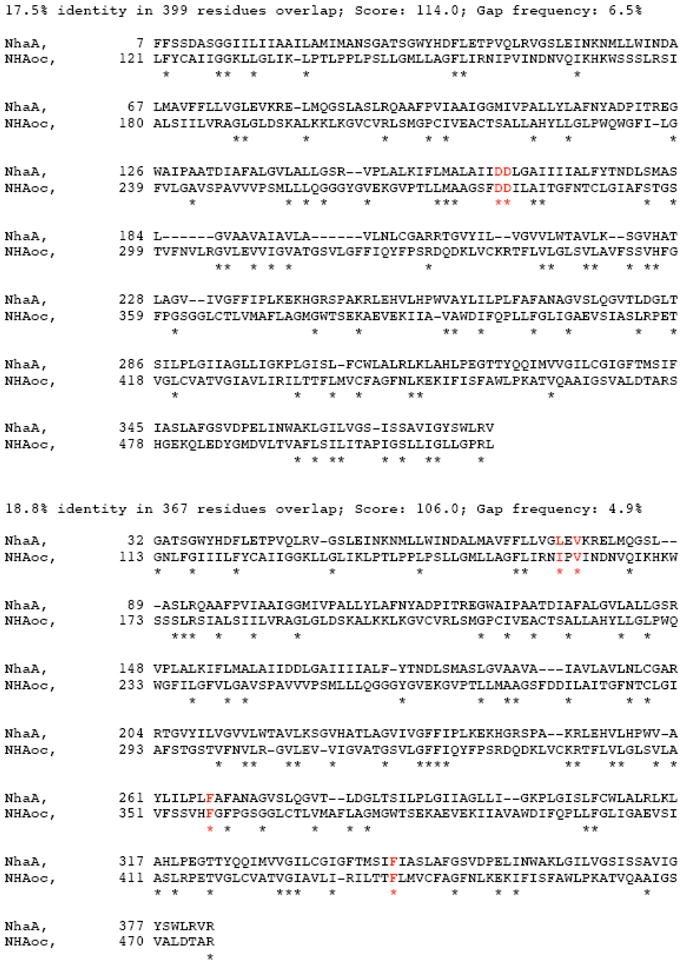
The alignments are reported in order of decreasing similarity score and share no aligned pairs. We aligned NhaA and NHAoc/NHA2 using the BLOSUM30 comparison matrix with the following parameters: Number of alignments to be computed: 20; Gap open penalty:12, Gap extension penalty:4. Above are two of the returned alignments indicating with a “*” the conserved amino acids. Also highlighted (in red): NHAoc/NHA2 Asp278-Asp279, I159, V161, F357 and F437 as well as their matching NhaA residues. Asp278-Asp279 are homologous to Asp163 - Asp164 of NhaA, which are essential for antiport activity. I159, V161 and F357 are amino acids that are substituted as a result of human SNPs. F357 is homologous to F267 of NhaA and amino acid that regulates the stoichiometry of NhaA.
Heterologous expression of human NHAoc/NHA2 can rescue the salt-sensitive phenotype of BW31a yeast
For expression in yeast, we cloned the cDNA of NHAoc/NHA2 into pYES2.1/V5-His TOPO TA under the control of the inducible GAL1 promoter. This plasmid allows for the production of a V5 tag/fusion protein that can be detected using an anti V5 antibody. We used the vector pNHA1-985GFP (expressing Saccharomyces cerevisiae’s own antiporter, NHA1) [12] and the empty pYES2.1/V5-His TOPO TA vector as positive and negative controls, respectively. Plasmids were electroporated into the salt-sensitive BW31a (ena1– 4Δ nha1Δ) yeast strain. Western blot analysis of independent transformants (Figure 2, bottom) shows galactose-inducible expression of a ~60 KDa protein in lanes 2 – 7, corresponding to the NHAoc/NHA2 – V5 fusion protein. To study NHAoc/NHA2 function, we tested the ability of high-expressing transformants 2 – 5 to grow on YNB plates supplemented with varying concentrations of NaCl (0 – 0.4 M) and galactose (Figure 2, top). Glucose containing plates were used as a control for galactose inducible expression. All strains grew equally well on YNB plates without any added salt, plus glucose or galactose, indicating that over-expressed NHAoc/NHA2 was not toxic to cells (0 M NaCl). When plated on YNB media supplemented with 0.4 M NaCl, cells expressing the NHAoc/NHA2 exchanger grew significantly better than the negative control cells, confirming that NHAoc/NHA2 has antiporter activity and can rescue the salt sensitivity of BW31a cells, as previously reported [1,4,6,7]. NHAoc/NHA2 expression did not result in the same sodium tolerance as that achieved by expression of the yeast Nha1 antiporter (lane +). NHAoc/NHA2 expressing cells can grow in the presence of NaCl only in galactose-containing plates, confirming that the salt tolerance is only improved when the heterologous protein is expressed and that cells grown on glucose remain salt-sensitive.
Figure 2. Solid agar growth of the alkali-metal-cation sensitive BW31a yeast strain expressing human NHAoc/NHA2.
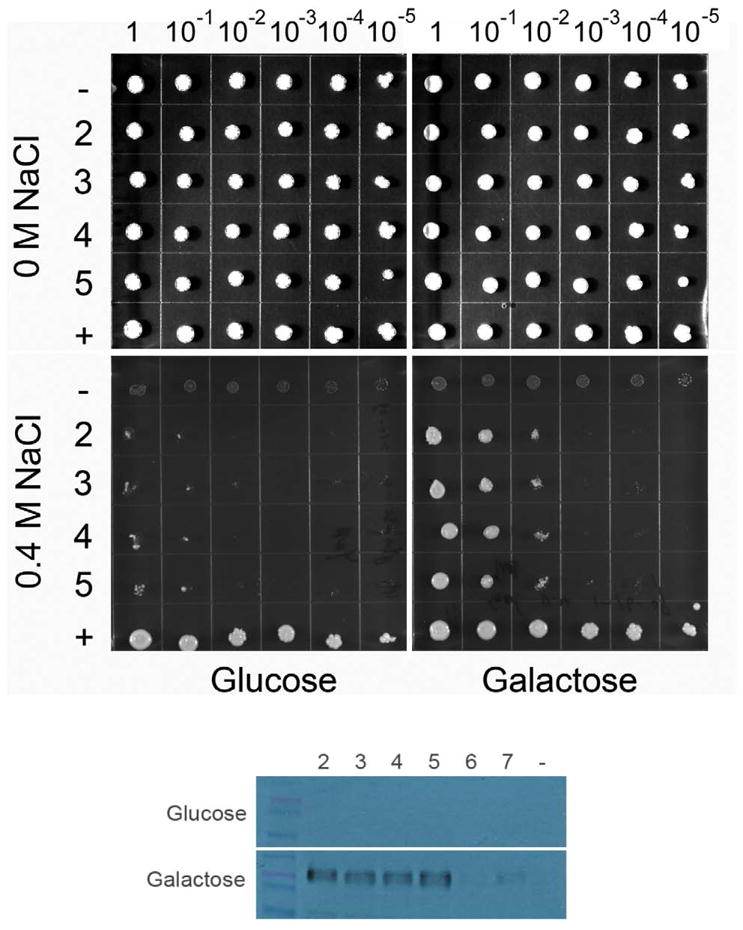
(top) Ten-fold serial dilutions of yeast suspensions (1 – 10−5) were spotted on YNB plates supplemented with NaCl and 2% glucose or 2% galactose, as indicated. Cells transformed with the four independent plasmids that were high expressers of NHAoc/NHA2 from the GAL1 promoters were tested (rows 2, 3, 4 and 5). Cells transformed with empty vector (row -) and cells expressing their own NHA1 antiporter (pNHA1–985GFP, row +) were used as negative and positive controls, respectively. (bottom) Western Blot analysis of BW31a cells transformed with NHAoc/NHA2 expression plasmids (lanes 2 – 7). Blots were incubated with a V5 antibody. Clones 2–5 were high expressers of NHAoc/NHA2 and were therefore used in the subsequent experiment seen in the top panel of this figure. Cells transformed with an empty vector (lane -) were used as negative control. A ~60 KDa protein, corresponding to NHAoc/NHA2 – V5 fusion protein, can be detected in lanes 2–5.
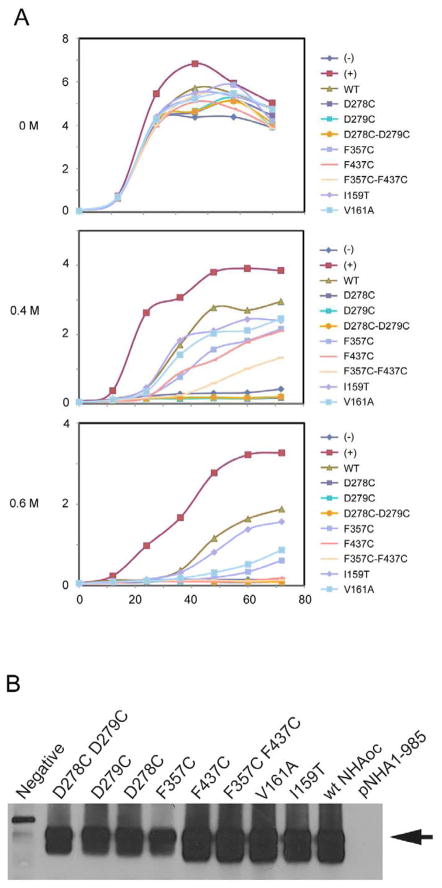
Phenotypes of mutants of NHAoc/NHA2: Growth curves in liquid media
To characterize the growth and NaCl tolerance of cells expressing mutants of NHAoc/NHA2 in detail, we performed growth tests in liquid media. All mutants of NHAoc/NHA2 expressed at equivalent levels (Figure 3B). We did not observe any difference in growth rates between cells over expressing any version of NHAoc/NHA2 exchanger and control cells in liquid YNB media without any sodium added, confirming that the expression of the NHAoc/NHA2 exchanger (wild type or mutant) was not toxic to the cells. In medium containing 0.4 and 0.6 M NaCl the growth rates changed significantly (Figure 3A). Cells transfected with the empty vector (negative control) grew very slowly. Cells expressing wild type NHAoc/NHA2 exchanger showed improved ability to grow, though they grew more slowly than cells expressing the yeast NHA1 antiporter. These results confirmed the observation from drop tests in solid agar and indicated that NHAoc is functional in BW31a cells.
Figure 3. (A, B) Growth curves of BW31a cells in liquid media.
Cells were transformed with a plasmid expressing wild type or different mutants of NHAoc/NHA2 and grown in YNB broth and 2% galactose and without NaCl or with 0.4 M and 0.6 M NaCl. Cells transformed with an empty vector (−) and cells transformed with Saccharomyces cerevisiae NHA1 antiporter (+) were used as negative and positive controls, respectively.
(B) Western Blot analysis of BW31a cells transformed with a plasmid expressing wild type or different mutants of NHAoc/NHA2 using a V5-specific antibody, show equivalent protein expression of NHAoc/NHA2 (60KD band, black arrow).
Mutations in aspartic acid residues in positions 278 and 279 completely abolished the ability of BW31a cells to grow in NaCl, in agreement with previous results [7,18]. In addition, we also mutated D278 and D279 separately and observed that each residue is essential for activity. Mutation I159T did not affect the ability of BW31a cells to grow in NaCl, whereas mutations V161A and F357C resulted in reduced growth (~50% inhibition compared to WT NHAoc/NHA2). Finally, yeast expressing the double mutant F357 F437 can not grow in 0.6 M NaCl (93 % inhibition of growth), suggesting that these residues are indeed homologues of NhaA F267 F344 which are essential for antiporter activity [8].
NHAoc/NHA2 is expressed in the membrane of BW31a cells
The results shown in the previous section indicate that mutations V161A, F357C, F357C-F437C as well as D278C and D279C diminished the ability of BW31a cells to grow in NaCl to different extents, or failed to complement the salt-sensitive phenotype. However, inhibition of growth in the presence of NaCl could also be due to mis-localization of the mutant NHAoc/NHA2 rather than a loss of activity. To determine whether the mutations affected membrane localization of NHAoc/NHA2 in BW31a cells, we performed confocal image analysis of BW31a cells expressing wild-type antiporter or mutant versions. The results (Figure 4) indicate that all mutants tested localized to the plasma membrane. These results demonstrate that the mutations did not interfere with membrane localization of NHAoc/NHA2 and suggest that the mutations affected antiporter activity.
Figure 4. WT and mutant versions of NHAoc/NHA2 localize to the plasma membrane.
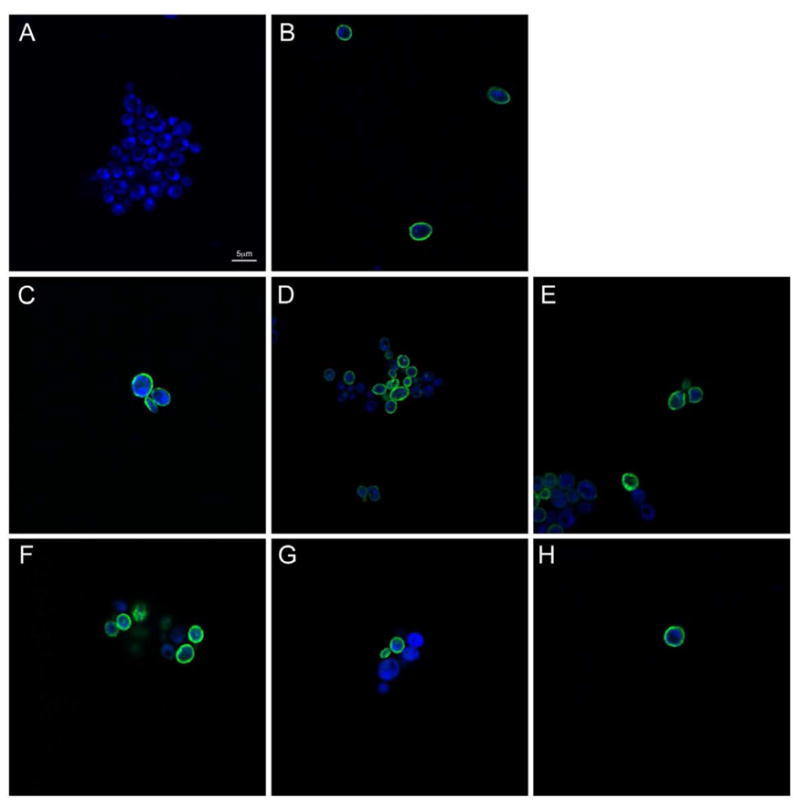
BW31a cells expressing different versions of NHAoc/NHA2 were fixed, permeabilized and stained using a primary anti-V5 antibody and a Alexa 488-conjugated fluorescent secondary antibody to detect the NHAoc/NHA2-V5 fusion protein. Confocal imaging of the cells shows anti-V5 reactivity in the membrane, indicating that none of these mutations interfere with protein localization. A) WT NHAoc/NHA2 in medium supplemented with glucose (no expression), B) WT NHAoc/NHA2 in medium supplemented with galactose (Induction), C) I159T, D) V161A, E) F357C, F) D278C, G) D279C, H) F357C-F437C.
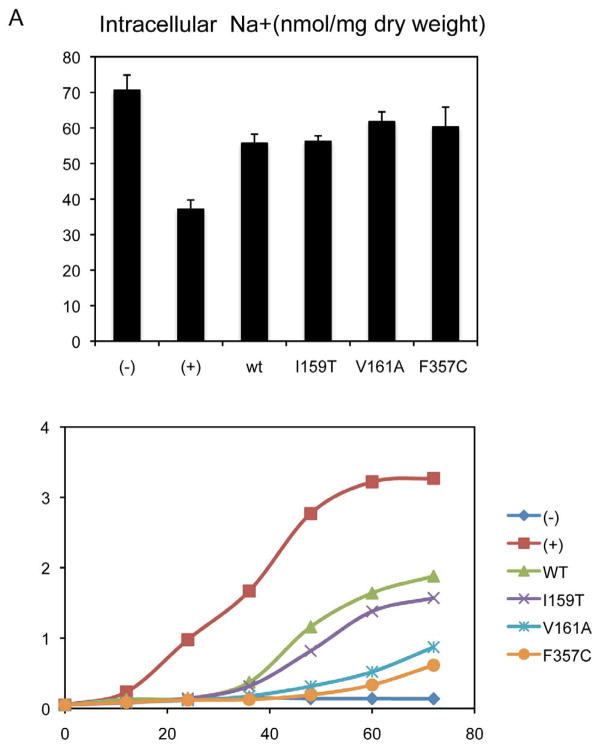
Mutations V161A and F357C inhibit Na+ antiport activity of NHAoc/NHA2
In order to test the ability of WT and mutant versions of NHAoc/NHA2 to mediate Na+ exchange, we performed and analysis of cell Na+ content. The results (Figure 5) indicate that there is no Na+/H+ exchange activity in negative control cells (−) as these cells have neither antiporter nor Na+/ATPase to pump Na+ out of the cells. Therefore, they contain more intracellular Na+ (relative intracellular content of Na+ is 70.58 nmol/mg dry weight or 100%). When BW31a cells express an active exchanger (e.g. cells expressing the yeast ScNHA1, (+)), the antiporter transports Na+ out of the cells, resulting in a lower intracellular Na+ content (37.13 nmol/mg dry weight or 52.6% of the negative control). In cells expressing the WT NHAoc/NHA2, the intracellular Na+ content is 55.69 nmol/mg dry weight or 78.9% of the negative control. These results conclusively show that NHAoc/NHA2 has Na+ antiporter activity. These findings are in complete agreement with the phenotypic complementation results since cells expressing wild type NHAoc/NHA2 exchanger showed improved Na+ antiporter activity compared to cells expressing no antiporter, though the antiport activity is less than that of the yeast ScNHA1 antiporter (Figure 5).
Figure 5. Mutations V161A and F357C inhibit NHAoc/NHA2 Na+ antiport activity and fail to complement the salt-sensitive phenotype of BW31a cells.
(−) cells transfected with an empty vector, (+) yeast ScNha1, (WT) WT NHAoc/NHA2, I159T, V161A and F357C: three mutants of NHAoc/NHA2. A) Intracellular Na+ content after culturing BW31a cells for 30 minutes in 100 mM NaCl. B) Growth curves in YNB/0.6 M NaCl of BW31a cells expressing different mutants of NHAoc/NHA2.
We also tested mutants I159T, V161A and F357C. In cells expressing I159T the intracellular Na+ content was 56.18 nmol/mg dry weight or 79.6% of the negative control. This value is statistically undistinguishable from the WT NHAoc/NHA2, which indicates that this mutation has no effect on antiporter activity. V161A and F357C, on the other hand, resulted in Na+ contents of 61.75 (or 87.5 % of the negative control) and 60.29 nmol/mg dry weight (85.4 % of the negative control), respectively. These values show a 10 % (V161A) and 8 % (F357C) increase in the intracellular Na+, compared to the WT NHAoc/NHA2 which demonstrates that these mutations have an inhibitory effect on antiporter activity.
DISCUSSION
We have performed a mutational analysis of the osteoclast-specific Sodium-Proton antiporter NHAoc/NHA2 by phenotypic complementation in yeasts and found that evolutionarily conserved amino acids are required for full antiporter function.
Na+/H+ antiporters are ubiquitous throughout the biological kingdom. The main Na+/H+ antiporter of E. coli, NhaA, has eukaryotic orthologues. Among these are the murine NHAoc/NHA2 [4] and the human HsNHA2 [7], which are part of a newly recognized family of metazoan CPA2 antiporters [1]. To date, the structure of these metazoan CPA2 antiporters have not been solved, however the primary sequence predicts a structure with 10 to 12 transmembrane segments (TMS) comparable to that of NhaA [10], suggesting that these proteins have a similar structural architecture. In addition to their structural similarity and despite the enormous evolutionary distance, NhaA and its eukaryotic homologues also have some sequence similarity. Many amino acid residues are conserved, notably, a pair of adjacent Aspartic acid residues, which are essential for antiporter activity in NhaA [10] as well as in NHAoc/NHA2 and HsNHA2 [7] and in antiporters from other organisms. In this report we have confirmed those findings and we have described new mutations that affect NHAoc/NHA2 antiporter activity: Valine 161 to Alanine, Phenylalanine 357 to Cysteine and the double mutant Phenylalanine 357–437 to Cysteine. These mutations correspond to amino acid residues that are conserved between NhaA and NHAoc/NHA2 and are required for proper function.
The pathophysiology of many skeletal diseases is associated with either increased (osteoporosis, metastatic bone disease and Paget’s disease) or decreased (various types of osteopetrosis) bone resorption by osteoclasts. In normal osteoclasts, all resorption activity may be experimentally abolished by combined pharmacological inhibition of proton pumping and sodium/proton antiport [19]. This observation, together with our previous reports [4] suggest that a molecule responsible for Sodium/Proton Antiport, like NHAoc/NHA2, is likely to be intimately involved in the regulation of osteoclast function and bone resorption, which will ultimately have an impact in bone homeostasis.
Single nucleotide polymorphisms have recently been recognized as an important factor in the development of human osteoporosis. Review of the online GeneCards database (Genecards.org, Weizmann Institute of Science, Israel) reveals the existence of three human SNPs (non-synonymous) in the coding region of NHAoc/NHA2 result in three amino acid substitutions: I159T, V161A and F357C. These amino acid residues are evolutionarily conserved. In addition, one of them (F357) is a putative homolog to NhaA F267, an amino acid essential for stoichiometry of NhaA [8]. Our data supports a role for two of these amino acids (V161 and F357) in the activity of NHAoc/NHA2. Measured as the ability to sustain yeast growth in 0.6 M NaCl, the V161A and F357C mutants result in loss of antiporter activity of 54% and 68%, respectively. Measured as the ability to extrude intracellular Na+, V161A and F357C cause a loss of activity of 10 % and 8 %, respectively. Finally, we found that the double mutant F357C-F437C abolishes 93% of the antiporter activity. These results are in agreement with the phenotypic complementation results as mutation I159T did not inhibit cell growth in NaCl, whereas mutations V161A and F357C did inhibit cell growth in NaCl, reflecting a reduced capacity of those mutants to remove Na+ from cells. Our findings suggest that the three human NHAoc/NHA2 SNPs may impact bone health and alter fracture risk for those carrying these mutations due to altered NHAoc/NHA2 activity.
The functional characterization of NHAoc/NHA2 will provide a better understanding of the mechanisms responsible for altered bone resorption that characterizes many bone diseases. Knowing which domains of NHAoc/NHA2 are critical for antiporter function will aid us in the design of appropriate new therapies, based on pharmacological agents that may interfere with NHAoc/NHA2 activity and ultimately help maintain bone mass. These studies provide the groundwork and context of our efforts to understand the molecular mechanisms of osteoclast differentiation and function as they affect bone mass in health and disease.
Acknowledgments
This work was supported by the following grants: NIH-NIAMS R21AR057915-01A1 (RAB) GA AS CR IAA500110801 (HS), MSMT COST OC10012 (HS), and AV0Z 50110509 (HS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brett CL, Donowitz M, Rao R. Evolutionary origins of eukaryotic sodium/proton exchangers. Am J Physiol Cell Physiol. 2005;288:C223–C239. doi: 10.1152/ajpcell.00360.2004. [DOI] [PubMed] [Google Scholar]
- 2.Ritter M, Fuerst J, Woll E, Chwatal S, Gschwentner M, Lang F, Deetjen P, Paulmichl M. Na(+)/H(+)exchangers: linking osmotic dysequilibrium to modified cell function. Cell Physiol Biochem. 2001;11:1. doi: 10.1159/000047787. [DOI] [PubMed] [Google Scholar]
- 3.Slepkov ER, Rainey JK, Sykes BD, Fliegel L. Structural and functional analysis of the Na+/H+ exchanger. Biochem J. 2007;401:623. doi: 10.1042/BJ20061062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Battaglino RA, Pham L, Morse LR, Vokes M, Sharma A, Odgren PR, Yang M, Sasaki H, Stashenko P. NHA-oc/NHA2: a mitochondrial cation-proton antiporter selectively expressed in osteoclasts. Bone. 2008;42:180. doi: 10.1016/j.bone.2007.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pham L, Purcell P, Morse L, Stashenko P, Battaglino RA. Expression analysis of nha-oc/NHA2: a novel gene selectively expressed in osteoclasts. Gene Expr Patterns. 2007;7:846. doi: 10.1016/j.modgep.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rheault MR, Okech BA, Keen SB, Miller MM, Meleshkevitch EA, Linser PJ, Boudko DY, Harvey WR. Molecular cloning, phylogeny and localization of AgNHA1: the first Na+/H+ antiporter (NHA) from a metazoan, Anopheles gambiae. J Exp Biol. 2007;210:3848. doi: 10.1242/jeb.007872. [DOI] [PubMed] [Google Scholar]
- 7.Xiang M, Feng M, Muend S, Rao R. A human Na+/H+ antiporter sharing evolutionary origins with bacterial NhaA may be a candidate gene for essential hypertension. Proc Natl Acad Sci U S A. 2007;104:18677. doi: 10.1073/pnas.0707120104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tzubery T, Rimon A, Padan E. Structure-based functional study reveals multiple roles of TMS IX and loop VIII-IX in NhaA NA+/H+ antiporter of Escherichia coli at physiological pH. J Biol Chem. 2008 doi: 10.1074/jbc.M800482200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pribylova L, Papouskova K, Zavrel M, Souciet JL, Sychrova H. Exploration of yeast alkali metal cation/H+ antiporters: sequence and structure comparison. Folia Microbiol (Praha) 2006;51:413. doi: 10.1007/BF02931585. [DOI] [PubMed] [Google Scholar]
- 10.Hunte C, Screpanti E, Venturi M, Rimon A, Padan E, Michel H. Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature. 2005;435:1197. doi: 10.1038/nature03692. [DOI] [PubMed] [Google Scholar]
- 11.Orlowski J, Grinstein S. Diversity of the mammalian sodium/proton exchanger SLC9 gene family. Pflugers Arch. 2004;447:549. doi: 10.1007/s00424-003-1110-3. [DOI] [PubMed] [Google Scholar]
- 12.Flegelova H, Haguenauer-Tsapis R, Sychrova H. Heterologous expression of mammalian Na/H antiporters in Saccharomyces cerevisiae. Biochim Biophys Acta. 2006;1760:504. doi: 10.1016/j.bbagen.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 13.Kinclova-Zimmermannova O, Zavrel M, Sychrova H. Identification of conserved prolyl residue important for transport activity and the substrate specificity range of yeast plasma membrane Na+/H+ antiporters. J Biol Chem. 2005;280:30638. doi: 10.1074/jbc.M506341200. [DOI] [PubMed] [Google Scholar]
- 14.Sychrova H. Yeast as a model organism to study transport and homeostasis of alkali metal cations. Physiol Res. 2004;53(Suppl 1):S91–S98. [PubMed] [Google Scholar]
- 15.Kinclova O, Ramos J, Potier S, Sychrova H. Functional study of the Saccharomyces cerevisiae Nha1p C-terminus. Mol Microbiol. 2001;40:656. doi: 10.1046/j.1365-2958.2001.02412.x. [DOI] [PubMed] [Google Scholar]
- 16.Duret L, Gasteiger E, Perriere G. LALNVIEW: a graphical viewer for pairwise sequence alignments. Comput Appl Biosci. 1996;12:507. doi: 10.1093/bioinformatics/12.6.507. [DOI] [PubMed] [Google Scholar]
- 17.Huang XQ, Hardison RC, Miller W. A space-efficient algorithm for local similarities. Comput Appl Biosci. 1990;6:373. doi: 10.1093/bioinformatics/6.4.373. [DOI] [PubMed] [Google Scholar]
- 18.Fuster DG, Zhang J, Shi M, Bobulescu IA, Andersson S, Moe OW. Characterization of the sodium/hydrogen exchanger NHA2. J Am Soc Nephrol. 2008;19:1547. doi: 10.1681/ASN.2007111245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Del Fattore A, Peruzzi B, Rucci N, Recchia I, Cappariello A, Longo M, Fortunati D, Ballanti P, Iacobini M, Luciani M, Devito R, Pinto R, Caniglia M, Lanino E, Messina C, Cesaro S, Letizia C, Bianchini G, Fryssira H, Grabowski P, Shaw N, Bishop N, Hughes D, Kapur RP, Datta HK, Taranta A, Fornari R, Migliaccio S, Teti A. Clinical, genetic, and cellular analysis of 49 osteopetrotic patients: implications for diagnosis and treatment 3. J Med Genet. 2006;43:315. doi: 10.1136/jmg.2005.036673. [DOI] [PMC free article] [PubMed] [Google Scholar]