Main Text
Mono- and binucleated polyploid hepatocytes (4n, 8n, 16n and higher) are found in all mammalian species, but the functional significance of this conserved phenomenon remains unknown 1-4. Polyploidization occurs through failed cytokinesis, begins at weaning in rodents and increases with age 2,5-7. Previously, we demonstrated that the opposite event, i.e. ploidy-reversal, also occurs in polyploid hepatocytes generated by artificial cell fusion 8-10. This suggested the intriguing possibility that somatic “reductive mitoses” can also happen in normal hepatocytes. Here we show that multipolar mitotic spindles form frequently in mouse polyploid hepatocytes and can result in one-step ploidy-reversal to generate offspring with halved chromosome content. Proliferating hepatocytes produce a highly diverse population of daughter cells with multiple numerical chromosome imbalances as well as uniparental origins. Our findings support a dynamic model of hepatocyte polyploidization, ploidy-reversal and aneuploidy, a phenomenon which we term the “ploidy-conveyor.” We propose that this mechanism evolved to generate genetic diversity and permits adaptation of hepatocytes to xenobiotic or nutritional injury.
We first tested whether normal polyploid hepatocytes can undergo ploidy-reversal in vivo. Highly pure (> 99%; Fig. 1a, Supplementary Fig. 1) octaploid hepatocytes isolated by fluorescence activated cell sorting (FACS) from male mice hemizygous for Rosa26 (lacZ) were transplanted into female Fah-/- mice, a model for selective liver replacement 11, and the frequency of donor-derived hepatocytes was assessed following extensive (>70%) liver repopulation (Supplementary Fig. 2). Hepatocytes from repopulated recipient mice were loaded with Hoechst and fluorescein di-ß-D-galactopyranoside (FDG), a fluorescent substrate of ß-galactosidase (ß-gal), and analyzed by FACS. Donor-derived ß-gal was expressed by octaploid (90 ± 2%), tetraploid (83 ± 5%) and diploid hepatocytes (59 ± 5%) (Fig. 1b). Donor-derived FAH expression was also detected in most octaploid (89 ± 3%), tetraploid (86 ± 4%) and diploid (67 ± 11%) hepatocytes (data not shown). The overall ploidy distribution of donor hepatocytes was the same as found in normal liver of an aged mouse. Diploid and octaploid hepatocytes proliferated at equivalent rates (Supplementary Fig. 3), thus eliminating the possibility the high percentage of near-diploid donor-derived hepatocytes resulted from overgrowth by rare contaminating diploids. Cytogenetics confirmed the presence of reduced-ploidy donor-derived cells (Fig. 1c). Surprisingly, most donor cells were aneuploid, i.e. had numerical chromosome gains and/or losses (Figs. 1d, 1e). In addition to analyzing single hepatocytes, liver sections from repopulated livers were stained for donor markers. While most Fah+ nodules contained both Y-chromosome (mChrY) and ß-gal activity, loss of either marker was found in ∼5% of FAH+ repopulation nodules (Supplementary Fig. 4). Together, these findings indicate that most normal polyploid hepatocytes undergo ploidy-reversal and marker segregation when forced to divide extensively.
Fig. 1. Purified octaploid hepatocytes generate reduced-ploidy daughters in vivo.
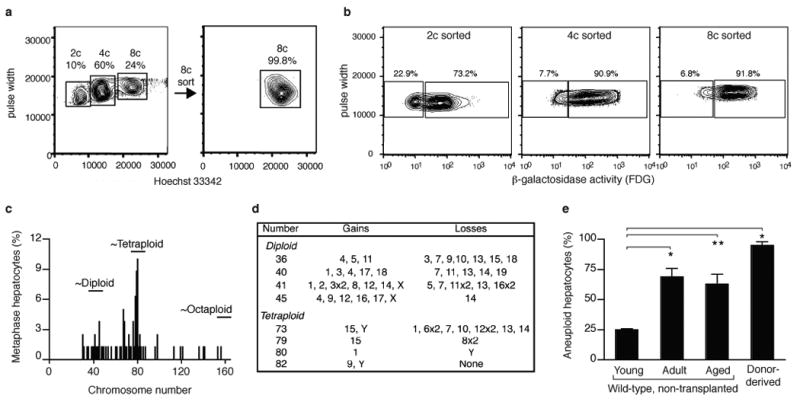
a, Hepatocytes were separated into ploidy populations by FACS with 2c, 4c and 8c DNA content, corresponding to diploid, tetraploid and octaploid hepatocytes, respectively. Highly pure viable octaploid hepatocytes were collected.
b, Hepatocytes isolated from mice repopulated by 8c donors were loaded with Hoechst + FDG. All ploidy classes expressed ß-gal (n=6 recipients).
c, d, Chromosome number (c) and representative aneuploid karyotypes (d) of donor-derived mChrY+ hepatocytes in repopulated livers (n=3). Chromosomal gains/losses are described relative to the nearest ploidy.
e, Percentage of numerical aneuploidy in donor-derived hepatocytes from repopulated mice or wild-type hepatocytes from non-transplanted mice (average ± s.e.m.; see Supplementary Fig. 5 for details). * P<0.006; ** P=0.01.
The transplantation experiments demonstrating ploidy-reversal were done using the Fah knockout mouse. Therefore, the high degree of aneuploidy observed (Fig. 1d) and marker loss (Supplementary Fig. 4) could be attributed to toxic metabolites made by Fah deficient hepatocytes 12. To address this potential artifact, the karyotypes of hepatocytes from wild-type mice were determined. Although chromosome counts clustered around 40, 80 and 160 chromosomes, frequent chromosome gains and losses were detected in adult hepatocytes (Supplementary Fig. 5). At weaning, most hepatocytes were normal diploids, and by adulthood >60% of hepatocytes had numerical abnormalities (Fig. 1e). Thus, normal hepatocytes become aneuploid in adult mice, and this phenomenon is unrelated to Fah deficiency.
Due to the time required for liver repopulation (6-8 weeks) and the dynamic ability of hepatocytes to change ploidy, it was unclear whether the ploidy-reversal in vivo occurred in a single step or gradually over the course of multiple cell divisions. Therefore, we tracked ploidy changes using a short-term (1-2 mitoses) in vitro system. As expected, diploid hepatocytes became binucleated and polyploidized (Supplementary Figs. 6a, 6b). To investigate ploidy-reversal, we followed the fate of polyploid hepatocytes. Concurrent with DNA replication and mitosis (data not shown), the percentage of binucleated cells dropped dramatically, generating populations with >80% mononucleated cells (Supplementary Fig. 6a). After 5 days in culture pure tetraploid hepatocytes had produced daughters with 8c and 2c DNA content (18 ± 1% and 0.9 ± 0.1%, respectively) (Figs. 2a, 2b). Cultured octaploid hepatocytes showed similar ploidy redistribution (Supplementary Fig. 6c, Fig. 2b). The emergence of reduced-ploidy daughter cells over 1-2 cell cycles indicates a single-step mechanism for ploidy-reversal.
Fig. 2. Polyploid hepatocytes undergo ploidy-reversal and unequal marker segregation in vitro.
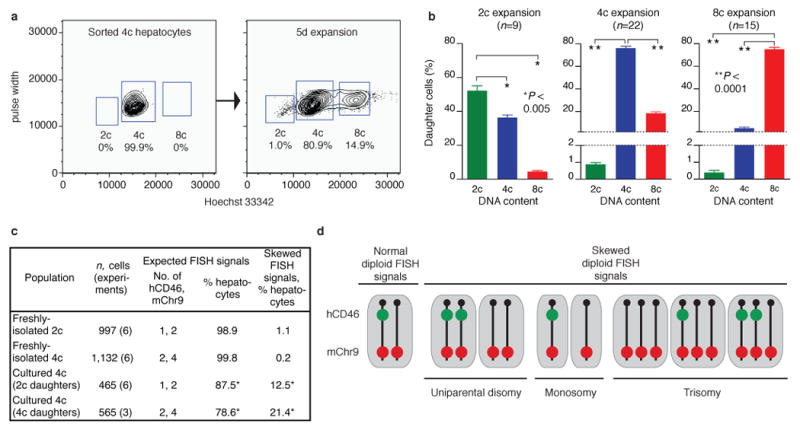
a, b, DNA content of cultured hepatocyte populations was determined by FACS. Representative plot is shown for freshly-sorted 4c hepatocytes and cells expanded for 5d (a). Results are summarized over multiple experiments (b). Data points represent average values ± s.e.m.
c, ∼ 99% of freshly-isolated 2c or 4c hepatocytes had the expected number of hCD46 and mChr9 signals. Cultured hepatocytes displayed chromosome missegregation. *, P < 0.0001 compared to freshly-isolated cells.
d, Schematic of the observed FISH signals in 2c daughters derived from cultured 4c hepatocytes.
To test whether polyploid hepatocyte mitosis could also produce aneuploidy, we analyzed the frequency and nature of chromosome missegregation in vitro. Mice hemizygous for a yeast artificial chromosome containing the human CD46 gene (hCD46) 13 on mChr9 were utilized (Supplementary Figs. 7a-7c). Using fluorescence in situ hybridization (FISH), chromosome signals in hepatocyte nuclei were quantified before and after proliferation (Supplementary Fig. 8a). After 5d expansion by 4c hepatocytes, the ploidy distribution shifted to include cells with 2c and 8c DNA content, as illustrated previously (Fig. 2a). Daughter cells with reduced ploidy (2c cells) and equal ploidy (4c cells) were hybridized with probes for hCD46 and mChr9. Approximately 99% of hepatocytes analyzed directly without prior culture (2c and 4c) contained the appropriate number of FISH signals (Fig. 2c). In contrast, FISH signals were skewed in 2c and 4c daughter cells (12.5% and 21.4%, respectively), indicating a diverse and aneuploid population of daughter cells (Figs. 2c, 2d and Supplementary Fig. 8b). Importantly, ∼1/3 of the 2c cells displayed uniparental disomy for mChr9. These data show that proliferating hepatocytes routinely generate a genetically diverse population of daughter cells.
The mechanism by which hepatocytes generated aneuploid or reduced-ploidy daughter cells was unknown. We rationalized that increased numbers of centrosomes in polyploid hepatocytes could lead to multipolar divisions and/or chromosome missegregation. To test this idea, cell divisions by polyploid hepatocytes were analyzed in vitro. In ∼half of tetraploid mitoses, bipolar spindles were established and maintained by centrosome clustering (Fig. 3a). The remaining tetraploid hepatocytes contained spindles oriented in a multipolar configuration, with centrosomes oriented on 3-4 distinct poles (reflecting either true multipolar spindles or alignment of prometaphase chromosomes from binucleated hepatocytes) (Figs. 3b, 3c). Octaploid hepatocytes established multipolar spindles with as many as 8 poles (Fig. 3d). However, only ∼1% of cells in anaphase or telophase were oriented with tripolar spindles (Fig. 3e). Additionally, hepatocytes with 2 discrete mitotic spindles synchronized in metaphase (Fig. 3f) and anaphase (Fig. 3g) were identified, an event we called “double mitosis.” Mitotic figures nearly identical to those seen in cultured hepatocytes were also observed during hepatocyte proliferation in vivo (Figs. 3i-3l).
Fig. 3. Polyploid hepatocyte mitoses with multipolar spindles and chromosome segregation defects.
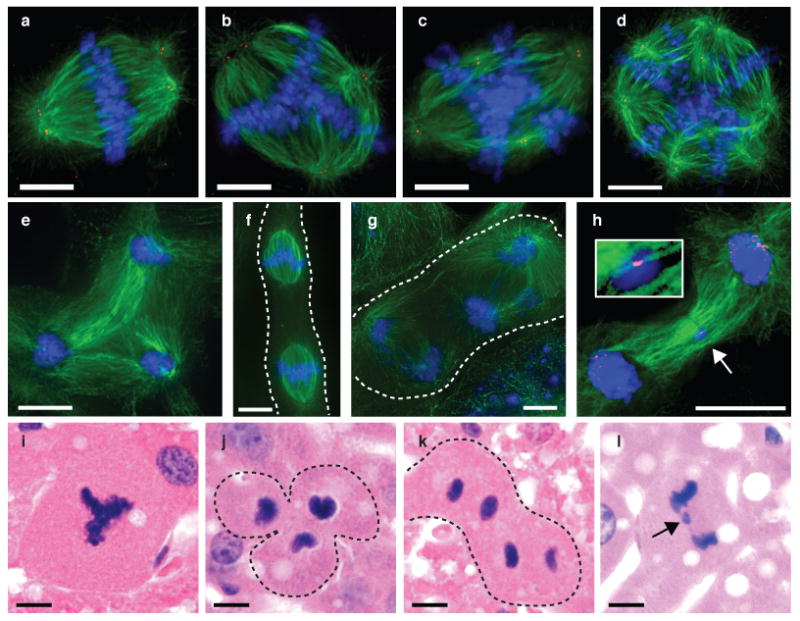
a-h, Mitotic figures were detected in cultured hepatocytes by visualizing DNA (blue) and microtubules (green). Centrioles (red) were detected in a-d; centromeres (red) were detected in h. (n=5 experiments). 4c hepatocytes contained bipolar (a) or multipolar spindles (b, c). Multipolar spindles were also seen in 8c hepatocytes (d). Three daughter nuclei emerged from a tripolar telophase (e). Double mitosis was detected in metaphase (f) and anaphase (g). Lagging chromosomes (arrow and inset) were also seen (h). i-l, Hepatocytes dividing in vivo formed similar mitotic figures: multipolar spindles (i), tripolar division (j), double mitosis (k) and lagging chromosomes (l) (n=4 mice). Scale bars are 10 μm.
The high percentage of multipolar metaphases and much lower frequency of multipolar anaphases/telophases was surprising. We hypothesized multipolar spindles could represent a temporary step in mitosis, and are then reorganized to be bipolar. Similar spindle dynamics were recently documented in cancer cells containing supernumerary centrosomes 14. A consequence of multipolar spindle reorientation is chromosome missegregation. Microtubules from different poles can attach to a single kinetochore, and failure to repair such merotelic attachments can lead to incomplete chromosome segregation 14. Consistent with spindle reorganization, we identified lagging chromosomes in 25-50% of tetraploid hepatocytes undergoing bipolar anaphase (Fig. 3h), suggesting that merotely contributes to marker loss (i.e., aneuploidy and loss-of-heterozygosity) observed in proliferating polyploid hepatocytes.
To determine whether multipolar mitoses produced viable offspring, we monitored hepatocyte divisions by time-lapse microscopy. As expected, diploids completed bipolar cell division with successful (89%) or failed cytokinesis (7%) (Fig. 4a). For analysis of polyploid hepatocytes, we focused mostly on tetraploids, but similar findings were seen with octaploids and non-fractionated hepatocytes (i.e., a mixture of all ploidy classes that were never exposed to Hoechst or subjected to FACS). Nearly 90% of tetraploid hepatocytes divided in a bipolar manner (Fig. 4a, Supplementary Fig. 9, Supplementary Movie 1), and in many time-lapse sequences their daughters (14%) divided again. Approximately 7% of tetraploid mitoses failed to complete cytokinesis (Fig. 4a, Supplementary Fig. 10, Supplementary Movie 2). Half of mononucleated and binucleated tetraploids transitioned from an early multipolar intermediate (as seen by chromosome alignment along multiple axes) to a standard bipolar configuration (Fig. 4c). Mitotic arrest and apoptosis were never seen.
Fig. 4. Live cell imaging of multipolar mitoses in hepatocytes.
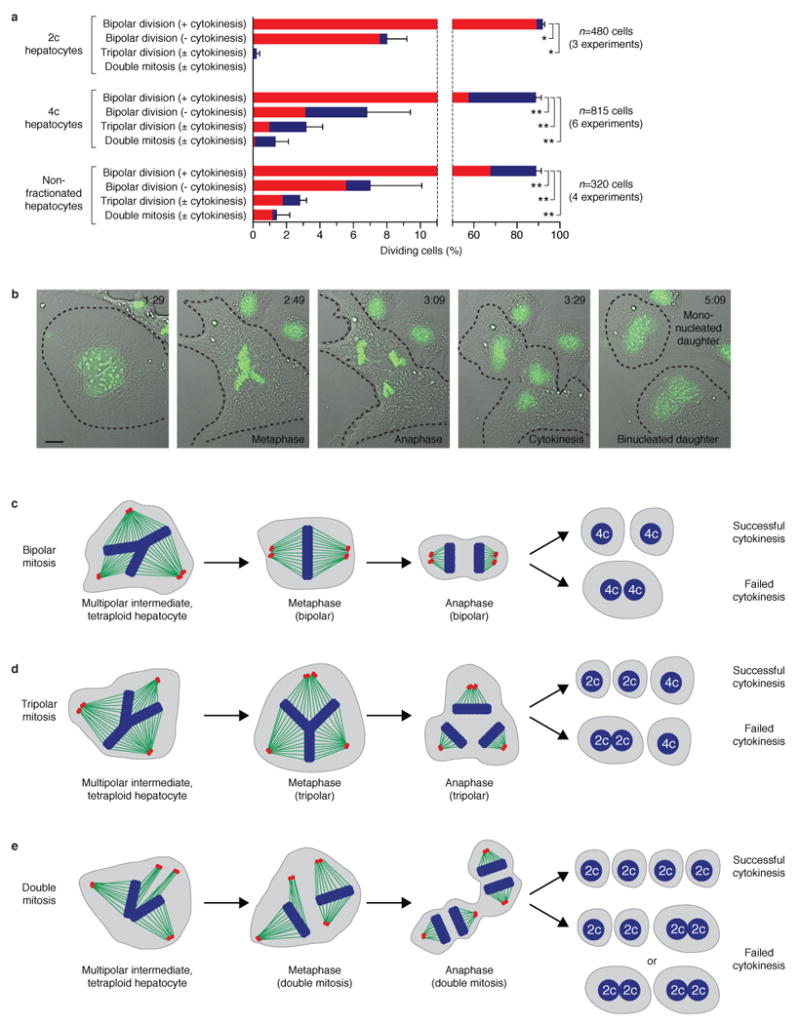
a, Summary of mitotic events captured by time-lapse microscopy. The percentage (average ± s.e.m.) of dividing mononucleated (red) and binucleated (blue) hepatocytes is shown. * P<0.001; ** P<0.0001.
b, Images (related to Supplementary Movie 3) of a binucleated tetraploid hepatocyte undergoing tripolar division. Time (hours : minutes) is indicated. Mitosis was tracked with DIC and Hoechst (pseudocolored green). Cell boundaries are marked. Scale bar is 10 μm.
c-e, Schematic depiction of divisions completed by polyploid (mono- or binucleated) hepatocytes. The outcome of each division depends on spindle reorganization and completion/failure of cytokinesis. Metaphase chromosomes (blue), mitotic spindles (green) and centrosomes (red) are indicated.
In addition to standard divisions producing 2 daughters, mitosis along multiple axes was seen. For example, 3.2% of tetraploid hepatocytes (mononucleated and binucleated) were captured undergoing tripolar mitosis (Figs. 4a, 4b, Supplementary Movie 3). All of the daughter cells were viable for the duration of the imaging session (up to 16h), and in some cases (15% of daughters) we were able to film subsequent mitoses (Supplementary Fig. 11, Supplementary Movie 4). While ∼10% of tripolar divisions completed 3-way cytokinesis, most divisions (∼90%) ended in partial failed cytokinesis (Fig. 4d). Nuclear content frequently segregated in a 4:2:2 ratio, which is consistent with 1 tetraploid daughter nucleus and 2 reduced-ploidy diploids (Supplementary Fig. 12). Furthermore, 1.2% of tetraploid hepatocytes completed double mitotic events (Fig. 4a). Double mitoses by either binucleated (Supplementary Fig. 13, Supplementary Movie 5) or mononucleated tetraploids (Supplementary Fig. 14, Supplementary Movie 6) generated 4 distinct nuclei via 2 synchronized mitoses (Fig. 4e). By definition, birth of 4 mononucleated cells from a parental cell represents a ploidy-reversal event. Daughters were viable and appeared healthy throughout imaging (as long as 10h).
Our data demonstrate that hepatocytes can increase (failed cytokinesis) and reduce (multipolar mitosis) their ploidy, thus resulting in the concept of a “ploidy-conveyor.” This dynamic mechanism not only generates numerical chromosome “abnormalities,” but also uniparental chromosome sets. Given that 5-10% of all genes are thought to be monoallelically expressed 15, this segregation pattern produces tremendous genetic heterogeneity.
The pervasive presence of aneuploid genotypes in the liver raises the question of whether this phenomenon serves a physiological purpose. Elegant studies in yeast showed aneuploidy can provide a strong selective advantage in response to multiple environmental stressors 16. Our findings suggest the provocative possibility that hepatocyte polyploidization evolved precisely to result in subsequent ploidy-reversal, aneuploidy and genetic diversity. Therefore, hepatic injury, which produces liver regeneration, could result in selection of hepatocytes that are genetically most resistant to the injury from a pre-existing pool of diverse genotypes. Genetic analysis of hepatocytes following liver injury may reveal favorable genotypes that differ from the germ line. Indeed, our own group has already observed an example of “favorable loss-of-heterozygosity” followed by selection in the Fah deficient mouse 17.
Methods Summary
Mouse surgery
The Institutional Animal Care and Use Committee of Oregon Health & Science University approved all mouse experiments. Hepatocytes from 3-6 month old donors were transplanted intrasplenically 9,12. 2/3 partial hepatectomy was described previously 18. Livers were harvested after 44h, a time point when the maximum number of hepatocyte mitotic figures are found 19.
Cell isolation and flow cytometry
Primary hepatocytes were isolated by two-step collagenase perfusion 11 and cultured hepatocytes were isolated by trypsinization. Detailed protocols are described for isolation/analysis of hepatocyte ploidy populations by flow cytometry in Methods.
Hepatocyte expansion in vitro
FACS-purified hepatocytes from 3-6 month old mice were seeded at 1000 to 2000 cells/cm2 on either Primaria tissue culture plastic (Beckton Dickinson), collagen-coated Lab-Tek II, CC2-treated chamber slides (Nunc) or collagen-coated ibi-Treat 8-well μ-slides (Ibidi). Cells were initially incubated in hepatocyte culture medium containing DMEM with 4.5 g/l glucose (HyClone), 10% FBS (HyClone), nonessential amino acids (Cellgro) and antibiotic-antimycotic (Cellgro). After 24h, the culture medium was replaced with SUM3 medium supplemented with 0.5% FBS. Cells were cultured for defined intervals. Detailed protocols for in vitro assays are described in Methods: karyotype analysis; interphase FISH 20; high resolution imaging of mitotic figures; and time-lapse microscopy.
Histology and immunocytochemistry
Histological analyses 21 and FAH immunocytochemistry 8 were previously reported. The frequency of FACS-isolated hepatocytes in G2/M 22 was determined by Cyclin A (Santa Cruz) staining.
Statistical significance
Statistical significance was determined using 2-sided Student's t-test (Figs. 1e, 2b, 4a) or Fisher's exact test (Fig. 2c). P values less than 0.05 were considered significant.
Methods
Mouse strains and surgery
The Institutional Animal Care and Use Committee of Oregon Health & Science University approved all mouse experiments. The following inbred mouse strains were used: wild-type (C57Bl, 129 and BALBc), transgenic Rosa26-lacZ (C57Bl and 129) 23, transgenic hCD46 13, Fah-/- (C57Bl and 129) 24. F1 hybrid mice (C57Bl × SJL) were also utilized. All mice were obtained from The Jackson Laboratories or bred at Oregon Health & Science University. Hepatocytes from 3-6 month old donors were transplanted intrasplenically 9,25. 2/3 partial hepatectomy was described previously 18. Livers were harvested after 44h, a time point when the maximum number of hepatocyte mitotic figures are found 19.
Histology and immunocytochemistry
Histological analyses 21 and FAH immunocytochemistry 8 were previously reported. The frequency of FACS-isolated hepatocytes in G2/M 22 was determined by Cyclin A (Santa Cruz) staining.
Isolation/analysis of defined hepatocyte ploidy populations
Primary hepatocytes were isolated by two-step collagenase perfusion 11 and cultured hepatocytes were isolated by trypsinization. For detection of hepatocyte ploidy populations, hepatocytes (2×106/ml) were incubated with 15 μg/ml Hoechst 33342 (Sigma) and 5 μM reserpine (Invitrogen) for 30 min at 37°. Cells were analyzed and/or sorted with an InFlux flow cytometer (Beckton Dickinson) using a 150 μm nozzle. Dead cells were excluded on the basis of 5 μg/ml propidium iodide (Invitrogen) incorporation. Cells adhering to each other (i.e., doublets) were eliminated on the basis on pulse width. Blood cells were excluded based on expression of CD45 using an APC-conjugated CD45 antibody (eBioscience). Ploidy populations were identified by DNA content using an ultraviolet 355 nm laser and 425-40 nm bandpass filter. Gating strategy is described (Supplementary Fig. 1b). Sorted hepatocytes were collected in DMEM with 4.5 g/l glucose (HyClone) containing 50% fetal bovine serum (FBS) (HyClone). The purity of sorted populations was determined at the end of each sort, and only highly purified populations (>99% pure) were used for subsequent assays. ß-gal activity in Hoechst-stained hepatocytes was detected using FDG reagent (Invitrogen) per manufacturer's instructions. Expression of hCD46 was detected with PE-conjugated hCD46 antibody (eBioscience).
Hepatocyte expansion in vitro
FACS-purified hepatocytes from 3-6 month old mice were seeded at 1000 to 2000 cells/cm2 on either Primaria tissue culture plastic (Beckton Dickinson), collagen-coated Lab-Tek II, CC2-treated chamber slides (Nunc) or collagen-coated ibi-Treat 8-well μ-slides (Ibidi). Cells were initially incubated in hepatocyte culture medium containing DMEM with 4.5 g/l glucose (HyClone), 10% FBS (HyClone), nonessential amino acids (Cellgro) and antibiotic-antimycotic (Cellgro). After 24h, the culture medium was replaced with SUM3 medium supplemented with 0.5% FBS. Cells were cultured for defined intervals.
SUM3 medium
SUM3 was prepared with the following reagents 26: 75% DMEM with 4.5 g/l glucose (HyClone), 25% Waymouth's MB 752/1 (Invitrogen), 2 mM L-glutamine (Invitrogen), antibiotic-antimycotic (Cellgro), 10 mM HEPES (Fisher), 50 ng/ml human epidermal growth factor (Invitrogen), 1.0 μg/ml insulin (Sigma), 30 nM sodium selenite (Sigma), 10 μg/ml transferrin (Sigma), 50 ng/ml Somatotropin (Sigma) and 1.0 μM 3,3′,5-Triiodo-L-thyronine sodium salt (T3) (Sigma).
Cytogenetics: karyotypes and FISH
Karyotypes
First, freshly-isolated primary hepatocytes were seeded on Primaria tissue culture plastic (Beckton Dickinson) and incubated in medium containing DMEM with 4.5 g/l glucose (HyClone), 10% FBS (HyClone), nonessential amino acids (Cellgro) and antibiotic-antimycotic (Cellgro). Nonadherent cells were removed after 4h, and cultures were switched to SUM3 medium supplemented with 0.5% FBS. Secondly, after ∼40h, hepatocytes were treated with 150 mg/ml colcemid (Sigma) for 2-4 hr and harvested by trypsinization. Third, after extensive washing, cells were incubated for 10 min in 56 mM KCl with 5% FBS and fixed with methanol:acetic acid (3:1 ratio). Finally, chromosomes from ∼20 metaphase-arrested hepatocytes per sample were G-banded with a standard trypsin/Wright's stain protocol. Photographs were taken using CytoVision software from Applied Imaging.
FISH Probe Generation
FISH point probes for mChrY, mChr9 and hCD46 were developed from bacterial artificial chromosomes RP24-225i24 (CHORI), RP23-346E22 (Invitrogen) and RP11-454L1 (Invitrogen), respectively. Bacterial artificial chromosomes were nick translated with the CGH Nick Translation Kit (Abbott Molecular) using SpectrumOrange dUTP and SpectrumGreen dUTP (Abbott Molecular). Each probe was validated using beta-inverse calculation to establish its normal cut-off value.
FISH analysis
FACS-sorted cells were dropped onto slides, coated with 3:1 methanol:acetic acid, and allowed to dry. Further fixation involved incubation in 3:1 methanol:acetic acid for 5 min at room temperature. For aging, slides were baked for 5 min at 95°. Slides were incubated in 2XSSC for 30 min at 37°, followed by 0.005% pepsin (Sigma)/0.01N HCl (Sigma) for 13 min at 37°. Slides were rinsed in 1XPBS (Sigma) for 5 min at room temperature and then fixed in 1% formaldehyde/0.45% MgCl2 (Sigma)/PBS for 5 min at room temperature. Slides were again rinsed in 1XPBS for 5 min at room temperature and then dehydrated by 2 min washes in 70%, 80%, and 90% ethanol. Once slides dried, probes were added and coverslips sealed with rubber cement. Slides were incubated at 72° for 2 min and 37° overnight. Following incubation, slides were washed in 0.4XSSC+0.3% NP-40 (Sigma) at 72° for 2 min, placed in 2XSSC+0.15% NP-40 for 5s to 1 min at room temperature, and mounted with DAPI II (Abbott). Samples were analyzed and scored under a Nikon Eclipse E800 photoscope. For analysis of interphase nuclei, signals were counted for as many nuclei available, up to 200 for each sample. Signals were scored as separate if they were further than a signal's diameter apart in distance, according to established clinical practices of FISH signal interpretation and as described 20. Photographs were captured using CytoVision software from Applied Imaging.
High resolution imaging of dividing hepatocytes
Mitotic figures identified in vitro were detected in populations of hepatocytes from 2-4 month old F1 hybrid mice (SJL × C57Bl) growing on collagen-coated ibi-Treat 8-well μ-slides (Ibidi). After expansion for 2-4d, wells were fixed and immunostained as described 14. Briefly, cells were fixed with methanol and incubated with primary antibodies for alpha-Tubulin (clone DM1A, Sigma), Centrin-2 (N-17, Santa Cruz) or Centromere proteins (15-235, Antibodies, Incorporated). Primary antibodies were detected with species-specific secondaries conjugated to Alexafluor 488 or Alexafluor 555 (Invitrogen). Nuclei were stained with 40 μg/ml Hoechst 33342 (Invitrogen). The images were acquired on a high resolution wide field Core DV system (Applied Precision) equipped with a Nikon Coolsnap ES2 HQ camera. This system is an Olympus IX71 inverted microscope with a proprietary XYZ stage enclosed in a controlled environment chamber, DIC transmitted light and a short arc 250W Xenon lamp for fluorescence. Each image was acquired as Z-stacks (every 0.2 μm from the bottom to top of each mitotic spindle). Images were deconvolved with the appropriate optical transfer function using an algorithm of 10 iterations using SoftWoRx Image Restoration Software (Applied Precision).
Live cell imaging
Live cell imaging was performed with a high resolution wide field Core DV system with controlled environment chamber (∼37°, 5% CO2) (Applied Precision). Hepatocytes from 2-4 month old F1 hybrid mice (SJL × C57Bl), growing on collagen-coated ibi-Treat 8-well μ-slides (Ibidi), were imaged between days 2 and 4. Optimal conditions were utilized. Images were collected every 10 min over sessions lasting 16-24h. Cellular structures were visualized with DIC (∼0.05s exposure) and chromosomes visualized with Hoechst (0.6s exposure). Hoechst was originally used to identify defined ploidy populations by FACS; residual Hoechst remained intercalated during cell culture. The focal plane was optimally set at the beginning of each imaging session; however, due to movement and shape change, cells appeared to drift slightly out-of-focus. Time-lapse movies were formatted using SoftWoRx Explorer (Applied Precision) and exported in QuickTime (Apple). The amount of Hoechst signal (corresponding to nuclear content) was determined for a subset of dividing hepatocytes using Imaris image analysis software (Bitplane).
Statistical significance
Statistical significance was determined using 2-sided Student's t-test (Figs. 1e, 2b, 4a) or Fisher's exact test (Fig. 2c). P values less than 0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank Pamela Canaday (Flow Cytometry Resource at OHSU) for cell sorting; Aurelie Snyder and Stefanie Kaech Petrie (Advanced Light Microscopy Core at OHSU, Core grant S10-RR023432) for microscopy assistance; and the Morphology Core of the Texas Medical Center (DK56338) for histology support. Thanks also to Leslie Smith and Matt Thayer for helpful discussions. This work was supported by grants from the National Institute of Health to M.G. (R01DK067636) and A.W.D. (F32DK076232).
Footnotes
Author Contributions: A.W.D designed and performed most of the experiments, analyzed data and wrote the paper. M.H.T helped with imaging of dividing hepatocytes. R.D.H assisted with data analysis. A.E.H.N, M.L.L and S.B.O performed all of the cytogenetic analyses. Histological analyses were performed by M.J.F. M.G. supervised all aspects of this work. All authors discussed the results and edited the manuscript.
Author Information: Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Faktor VM, Uryvaeva IV. Progressive polyploidy in mouse liver following repeated hepatectomy. Tsitologiia. 1975;17:909–16. [PubMed] [Google Scholar]
- 2.Guidotti JE, et al. Liver cell polyploidization: a pivotal role for binuclear hepatocytes. J Biol Chem. 2003;278:19095–101. doi: 10.1074/jbc.M300982200. [DOI] [PubMed] [Google Scholar]
- 3.Kudryavtsev BN, Kudryavtseva MV, Sakuta GA, Stein GI. Human hepatocyte polyploidization kinetics in the course of life cycle. Virchows Arch B Cell Pathol Incl Mol Pathol. 1993;64:387–93. doi: 10.1007/BF02915139. [DOI] [PubMed] [Google Scholar]
- 4.Yim AP. Some flow-cytofluorimetric studies of the nuclear ploidy of mouse hepatocytes: iii. further observations on early changes in nuclear ploidy of mouse hepatocytes following various experimental procedures. Br J Exp Pathol. 1982;63:458–61. [PMC free article] [PubMed] [Google Scholar]
- 5.Barbason H, Van Cantfort J, Houbrechts N. Correlation between tissular and division functions in the liver of young rats. Cell Tissue Kinet. 1974;7:319–26. doi: 10.1111/j.1365-2184.1974.tb00414.x. [DOI] [PubMed] [Google Scholar]
- 6.Celton-Morizur S, Merlen G, Couton D, Margall-Ducos G, Desdouets C. The insulin/Akt pathway controls a specific cell division program that leads to generation of binucleated tetraploid liver cells in rodents. J Clin Invest. 2009;119:1880–7. doi: 10.1172/JCI38677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Margall-Ducos G, Celton-Morizur S, Couton D, Bregerie O, Desdouets C. Liver tetraploidization is controlled by a new process of incomplete cytokinesis. J Cell Sci. 2007;120:3633–9. doi: 10.1242/jcs.016907. [DOI] [PubMed] [Google Scholar]
- 8.Duncan AW, et al. Ploidy reductions in murine fusion-derived hepatocytes. PLoS Genet. 2009;5:e1000385. doi: 10.1371/journal.pgen.1000385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, et al. Cell fusion is the principal source of bone-marrow-derived hepatocytes. Nature. 2003;422:897–901. doi: 10.1038/nature01531. [DOI] [PubMed] [Google Scholar]
- 10.Willenbring H, et al. Myelomonocytic cells are sufficient for therapeutic cell fusion in liver. Nat Med. 2004;10:744–8. doi: 10.1038/nm1062. [DOI] [PubMed] [Google Scholar]
- 11.Overturf K, et al. Hepatocytes corrected by gene therapy are selected in vivo in a murine model of hereditary tyrosinaemia type I. Nat Genet. 1996;12:266–73. doi: 10.1038/ng0396-266. [DOI] [PubMed] [Google Scholar]
- 12.Jorquera R, Tanguay RM. The mutagenicity of the tyrosine metabolite, fumarylacetoacetate, is enhanced by glutathione depletion. Biochem Biophys Res Commun. 1997;232:42–8. doi: 10.1006/bbrc.1997.6220. [DOI] [PubMed] [Google Scholar]
- 13.Yannoutsos N, et al. A membrane cofactor protein transgenic mouse model for the study of discordant xenograft rejection. Genes Cells. 1996;1:409–19. doi: 10.1046/j.1365-2443.1996.d01-244.x. [DOI] [PubMed] [Google Scholar]
- 14.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–82. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gimelbrant A, Hutchinson JN, Thompson BR, Chess A. Widespread monoallelic expression on human autosomes. Science. 2007;318:1136–40. doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- 16.Rancati G, et al. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell. 2008;135:879–93. doi: 10.1016/j.cell.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manning K, Al-Dhalimy M, Finegold M, Grompe M. In vivo suppressor mutations correct a murine model of hereditary tyrosinemia type I. Proc Natl Acad Sci U S A. 1999;96:11928–33. doi: 10.1073/pnas.96.21.11928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitchell C, Willenbring H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat Protoc. 2008;3:1167–70. doi: 10.1038/nprot.2008.80. [DOI] [PubMed] [Google Scholar]
- 19.Ko MA, et al. Plk4 haploinsufficiency causes mitotic infidelity and carcinogenesis. Nat Genet. 2005;37:883–8. doi: 10.1038/ng1605. [DOI] [PubMed] [Google Scholar]
- 20.Bayani J, Squire JA. Fluorescence in situ Hybridization (FISH) Curr Protoc Cell Biol. 2004;Chapter 22:4. doi: 10.1002/0471143030.cb2204s23. Unit 22. [DOI] [PubMed] [Google Scholar]
- 21.Overturf K, et al. Adenovirus-mediated gene therapy in a mouse model of hereditary tyrosinemia type I. Hum Gene Ther. 1997;8:513–21. doi: 10.1089/hum.1997.8.5-513. [DOI] [PubMed] [Google Scholar]
- 22.Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G. Cyclin A is required at two points in the human cell cycle. EMBO J. 1992;11:961–71. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–23. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 24.Grompe M, et al. Loss of fumarylacetoacetate hydrolase is responsible for the neonatal hepatic dysfunction phenotype of lethal albino mice. Genes Dev. 1993;7:2298–307. doi: 10.1101/gad.7.12a.2298. [DOI] [PubMed] [Google Scholar]
- 25.Lagasse E, et al. Purified hematopoietic stem cells can differentiate into hepatocytes in vivo. Nat Med. 2000;6:1229–34. doi: 10.1038/81326. [DOI] [PubMed] [Google Scholar]
- 26.Darlington GJ, Kelley JH, Buffone GJ. Growth and hepatospecific gene expression of human hepatoma cells in a defined medium. In Vitro Cell Dev Biol. 1987;23:349–354. doi: 10.1007/BF02620991. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


