Abstract
Many mutations confer upon copper/zinc superoxide dismutase-1 (SOD1) one or more toxic function(s) that impair motor neuron viability and cause familial amyotrophic lateral sclerosis (FALS). Using a conformation-specific antibody that detects misfolded SOD1 (C4F6), we demonstrate that oxidized WT-SOD1 and mutant-SOD1 share a conformational epitope that is not present in normal WT-SOD1. In a subset of human sporadic ALS (SALS) cases, motor neurons in the lumbosacral spinal cord displayed striking C4F6 immunoreactivity, denoting the presence of aberrant WT-SOD1 species. Recombinant, oxidized WT-SOD1 and WT-SOD1 immunopurified from SALS tissues inhibited kinesin-based fast axonal transport in a manner similar to FALS-linked mutant SOD1. Studies here suggest that WT-SOD1 can be pathogenic in SALS and identifies an SOD1-dependent pathogenic mechanism common to FALS and SALS.
Amyotrophic lateral sclerosis (ALS) is an adult-onset, motor neuron disease that causes progressive degeneration of motor neurons and death within 3-5 years of diagnosis 1. The most prevalent factors associated with inherited forms of ALS (FALS) are mutations in the SOD1 gene that encodes cytosolic Cu/Zn superoxide dismutase 2. In FALS, cytotoxicity of motor neurons appears to result from a gain of toxic SOD1 function, rather than loss of dismutase activity 3. While the exact molecular mechanisms underlying mutant-SOD1-mediated motor neuron degeneration are unclear, prevailing hypotheses suggest a role for mutation-induced conformational changes that lead to SOD1 misfolding and subsequent aggregation 4-9.
The etiology of sporadic ALS (SALS), which accounts for ~90% of ALS, is largely unknown. In contrast, several genetic variants have been identified in association with FALS 2. That FALS and SALS are clinically and neuropathologically similar implies that the pathogenesis of these diseases must converge on a common pathogenic pathway and/or involve similar toxic factors, but such factors have remained elusive 1, 10. WT-SOD1 has been proposed as a potential link between SALS and FALS 11, 12, although the existence of a toxic WT-SOD1 species that is associated with SALS in vivo and that recapitulates the pathogenic features of mutant-SOD1 has not been demonstrated. One hypothesis states that defects in the normal post-translational modifications of WT-SOD1 or the introduction of aberrant covalent modifications to WT-SOD1 could induce conformational changes in WT-SOD1 that mimic structural features of FALS SOD1 mutants 13-15. Several lines of evidence support this view, including the reports that metal-depleted 16, 17 and oxidized 11, 18 WT-SOD1 exhibit enhanced propensities to misfold in vitro 19, and are toxic when exogenously administered to cells 11, 17. These observations suggest that aberrantly modified WT-SOD1 and FALS-linked SOD1 mutants share similar structural features; however, common pathogenic mechanisms triggered by FALS and SALS-related SOD1 species remain elusive.
Recently, a monoclonal antibody (mab C4F6) was generated against the FALS-linked SOD1 G93A mutant protein and shown to bind preferentially to several FALS-linked SOD1 mutant proteins, as compared to WT-SOD1 20. Thus, the reactivity of C4F6 appears to be specific for a particular conformation inherent in misfolded SOD1. If aberrant modifications to WT-SOD1 induce the protein to adopt a mutant-like conformation, we speculated that the C4F6 antibody could detect misfolded WT-SOD1 species associated with SALS. Moreover, if WT-SOD1 plays a pathogenic role in SALS, we expected these aberrant WT-SOD1 species to recapitulate one or more toxic effect(s) of FALS-linked SOD1 mutants. Here, we report investigations of SALS-associated WT-SOD1 species using the C4F6 antibody in vitro and in vivo. Our recent finding that FALS-linked, mutant SOD1 protein inhibits fast axonal transport (FAT) (Gerardo Morfini and Scott Brady, submitted and 10) led us to investigate potential toxic properties of WT-SOD1 that has undergone misfolding in association with SALS using the same experimental model 21. Our findings demonstrate that the SOD1 protein can become pathogenic via non-heritable modifications (e.g., oxidation), thus supporting the hypothesis that conformational abnormalities in WT-SOD1 can underlie SALS pathogenesis. In addition, functional experiments in isolated squid axoplasm showed that misfolded WT-SOD1 species inhibit conventional kinesin-based fast axonal transport (FAT) through a mechanism involving p38 kinase activation. These results provide evidence for a novel SOD1-dependent pathogenic mechanism that impairs axonal transport in both FALS and SALS.
RESULTS
Oxidized and mutant SOD1 are conformationally similar
The misfolded-SOD1 conformation-specific C4F6 monoclonal antibody, generated against the apo-SOD1 G93A antigen 20, 22, reacts with multiple FALS-linked mutant SOD1, but not WT-SOD1 proteins 20. When normal post-translational modifications of WT-SOD1 are altered or when new modifications (i.e., oxidation) are introduced, WT-SOD1 acquires some of the properties observed for FALS-linked SOD1 mutants, such as an enhanced propensity to misfold 11, 16-18. Based on these observations, we hypothesized that C4F6 may recognize aberrantly modified forms of WT-SOD1, and could thus be used as a tool to investigate the association of such species with SALS in vivo.
To test this hypothesis, WT-SOD1 was exposed to hydrogen peroxide (H2O2) to generate an oxidized form of WT-SOD1 (SOD1ox). Fourier-transform mass spectrometry (FT-MS) was employed to confirm the oxidation of WT-SOD1, as indicated by an increase of 48 Da for the predominant species in the spectrum of SOD1ox (monoisotopic mass of 15,892 Da; Fig. 1b) compared to unmodified WT-SOD1 (15,844 Da; Fig. 1a). SOD1ox was subjected to electron capture dissociation (ECD), which confirmed that the sulfhydryl group of Cys 111 encoded within exon 4 (Fig. 2) is fully and irreversibly oxidized to sulfonic acid 23 through the incorporation of 3 oxygens (Fig. 1c and Supplementary Table 1). No other oxidated forms of WT-SOD1 were observed.
Figure 1. Mass spectrometry confirms the oxidation of WT-SOD1 upon exposure to hydrogen peroxide (H2O2).
The Fourier-transform mass spectrometry (FT-MS) spectra for (a) untreated WT-SOD1 and (b) oxidized WT-SOD1 (SOD1ox). Data shown have been automatically deconvoluted and reconstructed into a mass domain. (a) The conditions under which the FT-MS analysis was performed reduced the integrity of the SOD1 dimer interface and the SOD1 metal binding capacity. Therefore, the apo form of WT-SOD1 (15,844 Da – average nominal mass) is the predominate species in the mass spectrum of unmodified WT-SOD. (a-b) Peaks representing SOD1 adducts containing sodium, potassium and phosphate ions from the buffers employed during the purification of SOD1 are indicated. (b) The predominant species in the SOD1ox spectrum has a mass increase of 48 Da (15,892 Da) relative to apo-SOD1, which corresponds to the incorporation of 3 oxygens (+3ox; +48 Da). (c) SOD1 proteins were subjected to gas-phase isolation followed by electron capture dissociation (ECD, shown for SOD1ox). MS/MS fragments were assigned using monoisotopic masses with a 5 ppm cutoff and superimposed upon the SOD1 primary sequence (top), where  indicates unmodified, c-type fragment ions that include the N-terminus,
indicates unmodified, c-type fragment ions that include the N-terminus,  indicates unmodified z-type fragment ions that include the C-terminus, and
indicates unmodified z-type fragment ions that include the C-terminus, and  indicates +48 Da modified z-type fragment ions corresponding to the conversion of the sulfhydryl group at Cys 111 into sulfonic acid (+3ox). Inset shows raw data for c729+ fragment. The SOD1ox peptides resulting from EDC that were used to deduce the Cys111 site of oxidation are shown in Supplementary Table 1.
indicates +48 Da modified z-type fragment ions corresponding to the conversion of the sulfhydryl group at Cys 111 into sulfonic acid (+3ox). Inset shows raw data for c729+ fragment. The SOD1ox peptides resulting from EDC that were used to deduce the Cys111 site of oxidation are shown in Supplementary Table 1.
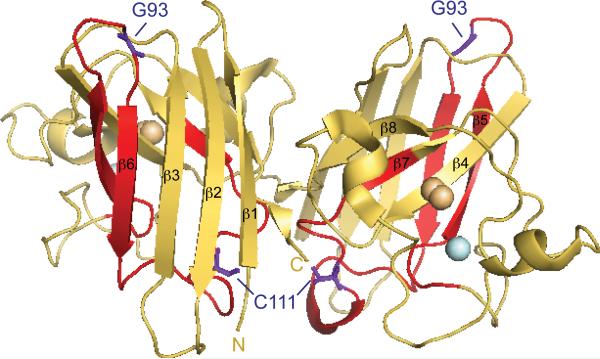
Figure 2. The structure of WT-SOD1.
The X-ray crystallographic structure of WT-SOD1 (pdb2C9V) 48 modeled in PyMOL. WT-SOD1 residues G93 and C111 within exon 2 are highlighted and labeled in purple. The zinc and copper atoms are shown in light cyan and orange, respectively. SOD1 conformation specific antibodies epitope map to the following regions: C4F6 to exon 4 (residues H80-V118 highlighted in red); A9G3 (Fig. 4c) to exons 1 and 2 (comprised of β-strands 1-4); SEDI 38 to β-strand 8; and USOD 33 to β-strand 4.
A native Western immunoblot analysis revealed that the reactivity of C4F6 is selective for recombinant SOD1ox, as compared to untreated WT-SOD1 protein. The commercial monoclonal SDG6 antibody, which is reactive for the native form of SOD1, recognized both SOD1ox and WT-SOD1 (Fig. 3a). SOD1ox migrated as a relatively diffuse band, indicative of a heterogeneous population of SOD1 molecules that migrate with both slower and faster motilities relative to untreated WT-SOD 24. The migration patterns of SOD1ox may be attributed to a loss of hydrodynamic volume due to misfolding (slower mobility), loss or exchange of metals (faster mobility), and/or a shift in the monomer-dimer equilibrium towards the monomer (faster mobility) 25. As expected, C4F6 was reactive with mutant SOD1 G93A but not WT-SOD1 by a native Western analysis of tissue lysates derived from the respective transgenic mice (Fig. 3b). C4F6 retained its reactivity towards SOD1 G93A under denaturing conditions; however, this monoclonal antibody was no longer reactive for SODox when this protein was denatured (Fig. 3c).
Figure 3. The C4F6 monoclonal antibody reacts with a conformational epitope shared by SOD1ox and mutant SOD1.
(a) Recombinant SOD1ox and WT-SOD1 (6 μg/lane) were subjected to a Western analysis using native (non-denaturing) gels with the C4F6 and SDG6 monoclonal antibodies. Native SOD1ox, but not WT-SOD1, is detected by C4F6, whereas SDG6 is reactive for both proteins. The samples were diluted (1 ng/lane) and subjected to an SDS (denaturing) Western analysis with a polyclonal anti-SOD1 antibody (Binding Site) to demonstrate equal gel loading. (b) The native Western shows that C4F6 is reactive only for native SOD1 G93A, whereas SDG6 is reactive only for native WT-SOD1 in lysates (30 μg total protein/lane) that were derived from the respective transgenic mouse. The SDS (denaturing) Western analysis, performed as in (a), demonstrates equal gel loading. (c) C4F6 is reactive for recombinant SOD1 G93A (55 ng/ lane), but not SODox (55 ng/ lane), whereas a polyclonal anti-SOD1 antibody (Calbiochem) detects both proteins. (d) Under denaturing conditions, C4F6 is reactive only for SOD1 G93A but not the other indicated SOD1 mutants. (e) C4F6 eptitope maps to exon 4. Lysates (30 μg total protein) from HEK 293 mammalian cells transfected with the indicated GST-tagged construct (Δ1-5 denote the respective exon deleted construct, FL = full length) were probed with C4F6 or a polyclonal anti-SOD1 antibody (Binding Site).
To address whether oxidation of Cys111 is necessary for detection by the C4F6 antibody under native conditions, we performed the native Western analysis described above for WT-SOD1 with the ‘AS-SOD1’ mutant containing the C6A/C111S point-mutations 26. In contrast to WT-SOD1, there was no evidence of AS-SOD1 oxidation as a result of H2O2 treatment, nor was C4F6 reactive for H2O2 treated AS-SOD1 (Supplementary Figure 1). Therefore, oxidation of Cys111 is required for the observed reactivity of C4F6 for SOD1ox.
The C4F6 reactive epitope, which is present in both SOD1 G93A and SOD1ox, was further investigated in an epitope mapping analysis. GST-tagged constructs encoding either the full-length SOD1 G93A gene or the SOD1 G93A gene lacking one of the five SOD1 exons were transfected into HEK-293 cells, and the respective cell lysates were subjected to a Western blot analysis using C4F6. The immunoblots in Figure 3e show that C4F6 reactivity requires the presence of exon 4 in GST-SOD1 G93A, which harbors the G93→A mutation (Fig. 2). As expected, C4F6 was not reactive towards HEK-293 endogenous WT-SOD1, whereas a commercial anti-SOD1 polyclonal antibody was reactive towards all SOD1 proteins (Fig. 3e).
That C4F6 only recognizes SOD1ox in the native conformation indicates that there is a conformational epitope within SODox, rather than the sulfonic moiety at Cys111, that is recognized by C4F6. Moreover, C4F6 is reactive for other FALS-linked SOD1 proteins in addition to SOD1 G93A under native conditions 20, and yet this antibody only detected SOD1 G93A but not SOD1 G93C, G93D, G93R, G93S G93V under denaturing conditions (Fig. 3d). Collectively, these data indicate that C4F6 recognizes an epitope within SOD1 G93A that contains both a conformational component and the G93A sequence component. The formation of this conformational epitope is induced by both the G93→A mutation and the Cys111 sulfonic acid moiety (Fig. 3), both of which are within exon 4 (Fig. 2). However, the conformational component of the epitope is lost when the SOD1 proteins are denatured, leaving only the G93A sequence element of the epitope to confer C4F6 reactivity (Figs. 3c and e).
SOD1ox inhibits kinesin-based fast axonal transport
Immunochemical analysis of WT-SOD1, SOD1ox, and FALS-related SOD1 mutants with the C4F6 antibody suggested a common conformational change shared by FALS-related SOD1 mutants and SOD1ox. These observations prompted us to compare the effects of SOD1ox with FALS-linked SOD1 mutant proteins in an ALS relevant biological assay. Results from vesicle motility assays in isolated squid axoplasm indicated that the FALS-linked SOD1 H46R mutant selectively inhibited conventional kinesin-based fast axonal transport (FAT) in the anterograde direction (Fig. 4a), whereas the WT-SOD1 protein did not affect FAT in either the anterograde or retrograde directions (Fig. 4b; Gerardo Morfini and Scott Brady, submitted and 10). This experimental system allows for quantitative analysis of membrane-bound organelles (MBOs) moving in both anterograde (conventional kinesin-dependent) and retrograde (cytoplasmic dynein-dependent) directions 27, 28. Further, the lack of plasma membrane in this preparation facilitates direct evaluation of the effects of neuropathogenic proteins on FAT 29, 30. Isolation of the squid axoplasm from the cell body and removal of plasma membrane allows one to investigate the effect of small molecules and proteins on FAT in a transcription-independent manner. Next, SOD1ox (5 μM) was perfused into isolated axoplasm and found to selectively inhibited anterograde FAT (Fig. 4c) to a similar extent as the SOD1 H46R mutant. Thus, SOD1ox mimics the toxic effect of the FALS-linked SOD1 mutant in these assays.
Figure 4. SOD1ox recapitulates the inhibitory effect of FALS-linked mutant SOD1 on anterograde FAT.
Vesicle motility assays in isolated squid axoplasm. Individual fast axonal transport (FAT) velocity (μm/sec) measurements (arrowheads) are plotted as a function of time (minutes). Dark arrowheads and line represent anterograde, conventional kinesin-dependent FAT rates. Grey arrows and line represent retrograde, dynein-dependent FAT rates. (a) Perfusion of 5 μM of the FALS-linked H46R mutant into squid axoplasm caused a marked reduction in the rate of anterograde FAT (n=4). (b) In contrast to (a), perfusion of 5 μM WT-SOD1 in the squid axoplasm had no effect on anterograde or retrograde FAT rates (n=4). (c) Perfusion of 5 μM SOD1ox mimicked the inhibitory effect of SOD1 H46R on anterograde FAT (n=4).
Phosphorylation of the molecular motor conventional kinesin is known to regulate FAT in vivo 10. Further, FALS-linked mutant SOD1 inhibits FAT by a mechanism involving the activation of a kinase pathway (Gerardo Morfini and Scott Brady, submitted and 10). To evaluate the possibility that SOD1ox-mediated inhibition of anterograde FAT similarly involves the activation of axonal kinases, we screened for changes in the activity of various protein kinases in axoplasms perfused with either WT-SOD1 or SOD1ox by immunoblotting with activation-specific phosphoantibodies. No changes in the activities of GSK3 and ERK were observed between WT-SOD1 and SOD1ox-perfused axoplasms (Fig. 5a, b). In contrast, antibodies against phosphorylated, catalytically active p38 (pp38) revealed a dramatic increase in p38 activation in axoplasms perfused with SOD1ox, by comparison with those perfused with WT-SOD1 (Fig. 5a). Quantitative analysis of immunoblots indicated that SOD1ox induced an approximately 4-fold increase in p38 activation, compared to WT-SOD1 (n=6; P<0.05, Fig. 5b). Consistent with these data, co-perfusion of the highly specific p38 MAPK inhibitors SB203580 31 (5μM; Fig. 5c) or MW01-2-069SRM 32 (10μM; Fig. 5d) with SOD1ox prevented the inhibitory effect of SOD1ox on anterograde FAT. Thus, inhibition of anterograde FAT by SOD1ox requires activation of p38 kinase. Taken together, these data indicate that an aberrantly modified form of WT-SOD1, which share conformational motifs with FALS-linked SOD1 mutant proteins, inhibit conventional kinesin-based FAT through a mechanism involving p38 activation.
Figure 5. p38 mediates the inhibition of anterograde FAT induced by SOD1ox.
(a) Immunoblotting analysis using activation-specific phosphoantibodies reveals a marked activation of p38 (p-p38) in axoplasms perfused with recombinant oxidized SOD1 (SOD1ox), compared to those perfused with recombinant unmodified WT-SOD1 (WT). In contrast, no changes were found in the activities of ERK (pERK) and GSK3 (pGSK3) in association with a specific SOD1 species. A monoclonal antibody against SOD1 (D3H5) 22 confirmed similar levels of SOD1 perfusion, and antibodies against kinesin-1 (KHC) provided a loading control for total levels of axoplasmic protein. Results from three independent experiments are shown (Squid 1-3). (b) Quantitation of results in (a) reveals an approximately 4-fold increase in the phosphorylation of p38 kinase (indicative of p38 activation) in SOD1ox-perfused axoplasms, compared to unmodified WT-SOD1-perfused axoplasms (n=6, P<0.05 (*) by the pooled t-test of μ1-μ2). Error bars reflect the standard error of multiple measurements. Co-perfusion of the highly specific p38 inhibitors SB203580 (c) and MW01-2-069SRM (d) blocked the inhibitory effect of SOD1ox on anterograde FAT (compare to Fig. 4c). Similarly, FALS-linked mutant SOD1 polypeptides inhibit anterograde FAT through a mechanism involving activation of p38 kinase (Gerardo Morfini and Scott Brady, submitted and 10).
Misfolded SOD1 is present in SALS spinal cord tissues
To address the hypothesis that aberrantly modified forms of WT-SOD1 are associated with SALS in vivo, we performed immunohistochemistry on post-mortem human spinal cord (SpC) using the conformation-specific C4F6 antibody that reacts specifically with mutant SOD1 and SOD1ox but not unmodified WT-SOD1. The clinical and demographic information for the SALS and control cases analyzed in this study are shown in Supplementary Tables 2 and 3. In contrast to other studies that employ misfolded-SOD1 conformation-specific antibodies 33, 34, antigen retrieval approaches that can potentially alter the antigen conformation were avoided here (see Methods). Positive C4F6 staining was observed in 4/9 SALS cases (SALS1 to 4, Fig. 6a, d-f). Of the five remaining SALS cases, the SpC sections from two showed extensive degeneration in the motor regions to the extent that motor neurons could not be detected. Since motor neurons appear positively stained with C4F6 (Figs 6a, d-f), we cannot exclude the possibility that these two aforementioned cases lacking detectable motor neurons would have been C4F6-positive at an earlier stage of disease. Remarkably, no C4F6 staining was observed in 17 control cases (representative cases are shown in Fig. 6g and h) (P=0.008, two-tailed Fisher's exact test). Lack of C4F6 immunoreactivity in a FALS case without SOD1 mutations confirmed the specificity of staining (Fig. 6i). We note that the optimal IHC staining results were obtained with paraffin-fixed tissue, whereas IHC of frozen tissue specimens generally did not reveal strong, positive C4F6 staining (see Methods).
Figure 6. The C4F6 monoclonal antibody is reactive for WT-SOD1 in SALS tissues.
(a,d,e,f) C4F6 positive staining is shown for 4 SALS cases (SALS1-4). The positive staining observed for SALS1 (shown in panel a) is lost when C4F6 is excluded from the staining protocol (b) or when the alternative SOD1-mutant specific A9G3 antibody is employed (c). Representative control cases (g-h) and an SOD1-negative FALS case (i) illustrate the lack of positive C4F6 reactivity for such cases. In total, in 4/9 SALS cases exhibited positive C4F6 staining, whereas 0/17 control cases exhibited positive staining. For clinical and demographic information on these cases, see Supplementary Tables 2 and 3.
The SOD1 mutant-specific A9G3 monoclonal antibody (Jean-Pierre Julien, unpublished data), which was also generated against SOD1 G93A 22 but epitope maps to regions encoded by exons 1 and 2 (Bosco, unpublished data), was not reactive for WT-SOD1 in the SALS1 case (Fig. 6c). Thus, the WT-SOD1 present in our C4F6-positive SALS cases exposes a specific epitope that is not detectable by all misfolded-SOD1 conformation-specific antibodies, consistent with other reports of misfolded-SOD1 conformation-specific antibodies that epitope map outside the region encoded by exon 4 33, 34.
Aggregated WT-SOD1 is immunohistochemically detected within the Lewy body-like hyaline inclusions of some 35, 36, but not all 37, SALS cases. To assess the levels of aggregated WT-SOD1 in our C4F6-positive SALS cases (SALS1-4), we performed an extraction of insoluble SOD1 from our tissue lysates derived from frozen, post-mortem human SpC tissue. The protocol was first validated with tissue lysates derived from transgenic G93A and naïve mice. As expected, immunoblot analysis detected insoluble SOD1 in the tissue lysates of SOD1 G93A mice 4, but not naïve mice (Supplementary Fig. 2). The levels of insoluble SOD1 extracted from human tissues were not significantly different between human ALS cases and controls (Supplementary Fig. 2). The diffuse staining patterns of C4F6 and pan polyclonal anti-SOD1 antibody (Supplementary Fig. 3) observed in SALS cases, together with the lack of evidence for elevated levels of insoluble WT-SOD1, suggested that the misfolded WT-SOD1 species in SALS1-4 cases are relatively soluble. However we cannot exclude the possibility that smaller oligomeric aggregates may be present, or that SOD1 aggregates could be detected with other anti-SOD1 antibodies that reportedly detect such species 33, 38.
SALS-derived, misfolded WT-SOD1 mimics mutant SOD1
The IHC analysis described above suggested that misfolded, C4F6-positive WT-SOD1 is significantly associated with many SALS cases. We evaluated the possibility that endogenous misfolded WT-SOD1 from SpC tissue inhibits FAT as observed with SOD1ox (Fig. 4c), To this end, WT-SOD1 was immunopurified from both SALS and control SpC tissues under detergent-free conditions (Methods) and perfused in isolated squid axoplasm. Mass spectrometry confirmed that the purified SOD1 preparations were 99% free of contaminating proteins. Perfusion of WT-SOD1 (1 μM) immunopurified from SALS tissues selectively inhibited anterograde, but not retrograde FAT, a pattern of FAT inhibition consistent with that induced by both FALS-linked mutant SOD1 (Fig. 4a; Gerardo Morfini and Scott Brady, submitted and 10) and SOD1ox (Fig. 4c). By contrast, WT-SOD1 immunopurified from control tissues had no effect on FAT (Fig. 7b). Co-perfusion of the conformation specific C4F6 monoclonal antibody (22.5 ng) blocked the inhibitory effect of SALS-derived SOD1 (Fig. 7c), demonstrating that C4F6-reactive SOD1 species mediate the inhibitory effect on FAT. Results shown in Figure 7 are representative of 3-5 independent squid axoplasm assays, each performed with human-derived WT-SOD1 proteins from two separate immunopurification preparations (SALS1, 2 and 4, and control 6). The fact that mild, detergent-free conditions (Methods) were employed to immunopurify WT-SOD1 proteins from human SpC lysates further suggested that the toxic, SALS-derived WT-SOD1 species are relatively soluble.
Figure 7. WT-SOD1 purified from SALS tissues inhibits anterograde FAT.
hSOD1 immunopurified from spinal cords of SALS (SALS hSOD1) and control (Ctrl hSOD1) were perfused into isolated squid axoplasm, and the effects on FAT evaluated as in Fig. 4. (a) Perfusion of SALS-derived hSOD1 (1 μM) selectively inhibits anterograde FAT (dark lines, right arrowheads) while retrograde FAT (gray lines, left arrowheads) remains unchanged (n=5 motility plots, from 2 independent immunopurifications of hSOD1). The inhibitory effect of SALS-derived hSOD1 on FAT mimics that of FALS-SOD1 H46R and SOD1ox (Fig. 4). (b) Perfusion of control-derived hSOD1 has no effect on FAT (n=3 motility plots, from 2 independent immunopurifications). (c) Co-perfusion of the C4F6 monoclonal antibody (22.5 ng) with SALS-derived hSOD1 blocked the inhibitory effect of SOD1 on anterograde FAT (n=3 axoplasms), demonstrating that the C4F6-reactive SOD1 species mediate the inhibitory effect on FAT.
DISCUSSION
Approximately 150 mutations dispersed throughout the SOD1 sequence have been linked to FALS (http://alsod.iop.kcl.ac.uk/). In many cases, these represent subtle, conservative (e.g. Gly→Ala) amino acid substitutions 3; nonetheless, all these mutations lead to an ALS phenotype. While pathogenic mechanisms underlying FALS-linked SOD1-mediated toxicity have not been definitively elucidated, a prevailing hypothesis is that FALS-linked mutations induce an altered or misfolded conformation in SOD1 4, 5, 7-9 that modifies its interactions with other proteins and perturbs its cellular localization 39-41.
Given the common pathological effects of diverse FALS-linked SOD1 mutations, it seems plausible that alterations to the non-Mendelian, post-translational modifications of SOD1 may similarly lead to an ALS phenotype 13-15. For example, disruptions of the normal SOD1 post-translational modifications (Fig. 2), such as subunit dimerization, the intrasubunit disulfide bond between residues Cys57 and Cys146, and the coordination of copper and zinc, have all been shown to cause WT-SOD1 to aggregate 16, 17, 25, 38, 42. Moreover, aberrant post-translational modifications of SOD1, such as oxidation, have adverse effects on WT-SOD1 protein conformation 11, 15, 18. By employing the C4F6 monoclonal antibody 20, we found that both oxidation of Cys111 (Fig.1) and mutagenesis of G93→A induce the formation of a conformational epitope that includes elements of exon 4 (Fig. 2) and that is not normally exposed by WT-SOD1 (Fig. 3). In addition to this conformational component of the C4F6 epitope that is shared by SOD1ox and SOD1 G93A, there is an aminoacid sequence component that includes G93A, which could be expected based on the fact that the SOD1 G93A antigen was used to raise the C4F6 antibody 20, and which likely explains the stronger reactivity of C4F6 for G93A relative to SOD1ox under native conditions (Supplementary Fig. 1). Under denaturing conditions, the conformational epitope is lost and only the G93A sequence element remains to confer reactivity with C4F6, thus explaining a lack of C4F6 reactivity for SODox and other G93 variants under denaturing conditions (Figs. 3c and e).
Studies of human spinal cord tissues with C4F6 revealed the presence of aberrantly folded WT-SOD1 species in approximately half of the available SALS cases, but not in control cases (Fig. 6). These results indicate that at least a subset of SALS cases contain WT-SOD1 proteins that are structurally similar to FALS-linked SOD1 mutant proteins. The lack of C4F6 reactivity in the remaining SALS cases may indicate that misfolded SOD1 is not associated with ALS pathogenesis for these cases, suggesting that modified WT-SOD1 plays a role in a subset of SALS in an analogous manner to the role that mutant-SOD1 plays in a subset of FALS. However, we cannot confirm that the C4F6 antibody is reactive for all possible misfolded forms of WT-SOD1.
Our finding that misfolded SOD1 is associated with SALS is consistent with the report that an aberrant 32-kDa, SOD1-containing species is indirectly detected through a biotinylation cross-linking reaction with homogenized tissue lysates from both SALS and FALS 12. Here, we probe for a misfolded SOD1 conformation in SALS using an IHC approach on fixed tissues with C4F6, a conformation-specific monoclonal antibody. Moreover, we demonstrate that these misfolded WT-SOD1 species derived from C4F6-positive SALS cases recapitulate the toxic effect of FALS-linked mutant SOD1 on FAT (Fig. 7). Further, we show that activation of p38 MAPK is a common feature of both SALS- and FALS-linked SOD1 inhibition of FAT (Fig. 5 and 10).
While we have shown that C4F6 is reactive for an oxidized form of WT-SOD1 (Fig. 3a), it is possible that other modifications to WT-SOD1, including alterations in normal post-translational modifications 16, 17, 25, 38, might also induce an altered conformation that confers C4F6 reactivity. That different modifications to SOD1 can induce similar structural consequences is supported by observations that C4F6 reacts with different FALS-linked SOD1 mutants 20, and by a recent hydrogen/deuterium exchange study that revealed enhanced flexibility within the same SOD1 electrostatic loop region (residues 133-144, located between β-strands 7 and 8; Fig. 2) for a panel of 13 different FALS-linked SOD1 mutants 7. Additional studies with alternate misfolded forms of WT-SOD1 and different SOD1 conformation specific antibodies will provide greater insight into the conformational similarities of these proteins and their prevalence in SALS.
Although the C4F6 antibody recognizes a conformation-dependent epitope common to both mutant and WT-SOD1 proteins associated with ALS pathology, the critical epitope is not detected by all antibodies that recognize other misfolded SOD1 species. For example, our A9G3 antibody detects a subset of FALS-linked mutant SOD1 species, but does not react with spinal cord sections from SALS patients that are immunoreactive for C4F6 (Fig. 6c). Similarly, both the SEDI (SOD1 Exposed Dimer Interface) and USOD (Unfolded SOD1) antibodies failed to detect WT-SOD1 in SALS cases 33, 34. There are several significant distinctions between the epitopes recognized by C4F6 and the SEDI and USOD antibodies. C4F6 is reactive for a conformational epitope that includes G93 encoded within exon 4 (Fig. 3), which is distal to those epitopes recognized by SEDI and USOD (Fig. 2). Moreover, both SEDI and USOD are reactive for linear sequences that can become exposed in aggregated SOD1 inclusions in FALS cases, whereas the misfolded WT-SOD1 species in our SALS cases is relatively soluble as evidenced by the diffuse C4F6 staining pattern (Fig. 6), the low levels of insoluble WT-SOD1 detected in our SALS cases (Supplementary Fig. 2), and the ability to purify these species under detergent-free conditions while maintaining their inhibitory effect on anterograde FAT (Fig. 7). Importantly, our IHC methods do not require the harsh conditions used for antigen-retrieval. Such treatments may disrupt the C4F6-like conformational-epitope, but enhance the detection of epitopes like those for the SEDI and USOD antibodies that are otherwise buried 33, 34. Thus, the differing results obtained with the C4F6, SEDI and USOD antibodies are likely due to the different epitopes recognized by these antibodies.
The foregoing analyses of immunoreactivity patterns for the C4F6 antibody reveal that genetic variants transmitted as Mendelian traits and non-inherited modifications to SOD1 can both induce similar structural perturbations within the protein, and that non-inherited modifications of SOD1 can be associated with SALS. A critical question that follows directly from these observations is whether these SALS-linked modifications confer upon WT-SOD1 the same toxic properties that are elicited by FALS-linked SOD1 mutations, including activation of p38 and inhibition of FAT. Both recombinant SOD1ox (Fig. 4c) and WT-SOD1 derived from C4F6-postive SALS spinal cord tissues (Fig. 7a) recapitulate this pattern of mutant-SOD1 mediated FAT inhibition, whereas untreated recombinant WT-SOD1 and WT-SOD1 derived from control spinal cord tissues had no effect (Figs. 4b and 7b, respectively). The effect of SOD1 from C4F6-postive SALS spinal cord on anterograde FAT was abolished by incubation with the C4F6 antibody prior to perfusion (Fig. 7c). C4F6 also abolished the ability of FALS mutant SOD1 to inhibit FAT (data not shown). Biochemical and pharmacological experiments further indicated that the inhibition of anterograde FAT induced by SOD1ox involved activation of p38 kinase (Fig. 5). FALS-linked mutant SOD1 mediated defects in axonal transport have been reported previously 10, 43, 44, and were thought to represent an early pathogenic event in mutant-SOD1 transgenic mice that contributes to a “dying back” mode of motor neuron degeneration 45, 46, 47. That the inhibition of FAT is selectively in the anterograde direction demonstrates that this effect of mutant SOD1 and SOD1ox shows specificity, as this effect is not universally observed for all toxic, neurodegenerative disease associated proteins 10. The pattern of FAT inhibition is largely determined by the nature of the regulatory kinases that become activated by the toxic protein 10. Both modified WT-SOD1 (Fig. 5) and several mutant-SOD1 proteins (Gerardo Morfini and Scott Brady, submitted and 10) inhibit FAT through a mechanism involving specific activation of p38 MAPK.
Concurrent studies showed that p38 directly phosphorylates kinesin-1 subunits of conventional kinesin and dramatically inhibited the translocation of this motor protein along axonal microtubules, thereby providing a common molecular basis for the effects of activated p38 kinase activity on anterograde FAT 10 (Morfini and Brady, submitted). Thus, SALS-associated WT-SOD1 species induce the same defects on conventional kinesin-based FAT as FALS-linked SOD1 mutants, and by the same molecular mechanism.
The concept that proteins can become pathogenic via both inheritable and non-heritable modifications has precedence in the context of other neurodenegerative diseases, as exemplified by α-synuclein in Parkinson's disease, Aβ or tau in Alzheimer's disease and frontotemporal dementia (FTDP). Although our data do not rule out potential toxic effects of aggregated WT-SOD1 species in ALS pathogenesis, they reveal toxic effects associated with relatively soluble, misfolded WT-SOD1 species in SALS. While aberrantly modified WT-SOD1 is aggregation-prone in vitro 16, 17, 25, 38, the toxic species in vivo may in fact be a pre-aggregated, conformationally mis-folded form of the protein.
In conclusion, our investigations indicate that misfolded, SALS-linked WT-SOD1 proteins activate the same neurotoxic mechanism that is invoked by FALS-linked SOD1 mutants, strongly suggesting that conformational abnormalities and post-translational modifications in WT-SOD1 can contribute to SALS pathogenesis. These studies identify a novel pathogenic mechanism for ALS common to both mutant-SOD1-mediated FALS and many cases of SALS.
Supplementary Material
Acknowledgments
We are grateful to John Landers and Peter Sapp for DNA sequencing analysis of the SALS cases employed in this study; Lawrence Hayward, Ashutosh Tiwari and Ru-Ju Chain for help with expression of recombinant WT-SOD1; Sarah Berth, Alena Leitman and Maria Saparauskaite for help with axoplasm vesicle transport assays; Agnieszka Kaminska and Linda Molla for help with biochemical experiments in squid axoplasm; Mercedes Prudencio and David Borchelt for cell lysates containing SOD1 G93 mutants; Charles Vanderburg, Eric Tamrazian, Anna Bialik and the DERC core laboratory (UMMS) for assistance with immunohistochemistry; Karlotta Fitch and the Massachusetts Alzheimer Disease Research Center (P50AG005134) for assistance with human tissue samples; Jill Zitzewitz for C6A/C111S-SOD1 protein; Alexandra Weiss for assistance with mice; Karin Green at the UMMS Proteomics & Mass Spectrometry Facility for analysis of C6A/C111S-SOD1; and Greg Petsko for insightful dialogue and support. We acknowledge financial support from the ALS Therapy Alliance-CVS Pharmacy (DAB, GM), 2007/2008 MBL research fellowships (GM), the ALS Association (DAB, RHB, GM and STB), NIH (RHB, STB, JNA), the Angel Fund (RHB) and Project ALS (RHB).
METHODS
Antibodies and reagents
In our experiments, we used antibodies to SOD1 (PC077, the Binding Site; Calbiochem, 574597; SDG6 clone, Sigma), mutant-SOD1 (C4F6 and A9G3 20), KHC (H2) 30, phospho-p38 MAPK (Cell Signaling #9215), phospho-ERK (Santa Cruz #7383), and phospho-GSK3 (Santa Cruz #11757). Secondary antibodies included horseradish peroxidase (HRP)-conjugated rabbit anti-sheep IgG (Upstate, 12-342) and HRP-conjugated rabbit anti-Mouse IgG (Sigma, A9044). SB203580 was obtained from Calbiochem and handled as described 30, and MW01-2-069SRM was a generous gift from (M. Watterson, Northwestern University. Total protein concentration was determined using bicinchoninic acid (BCA) assay (Pierce #23225).
Human samples
Human control and SALS paraffin-embedded spinal cord tissues for IHC analysis, and frozen tissues for immunopurification of WT-SOD1, were obtained through the Massachusetts Alzheimer's Disease Research Center (ADRC) at Massachusetts General Hospital, with the appropriate IRB approval. All experiments were performed with either lumbar or thoracic SpC tissue sections. All available demographics and clinical information for the SALS and control cases analyzed throughout this study are in Supplementary Tables 1 and 2.
Immunoblots
Samples for native Western analysis were prepared with 2X loading buffer (Invitrogen, LC2673), separated by PAGE with 12% Tris-Glycine Gels (Invitrogen, EC60052BOX) in Tris-Glycine buffer (Invitrogen, LC2672) for 3 hours at 4°C, and transferred to PVDF (Pierce #88518) in transfer buffer, pH 9.2 (Invitrogen, LC3675) for 90 min at 4°C. Membranes were fixed for 5 minutes with Ponceau S stain (Sigma # P7170) and blocked for 1 hour at 25°C in 5% nonfat dry-milk (BioRad #170-6404). Blots were probed with either C4F6 or with SDG6 overnight at 4°C followed by HRP-conjugated rabbit anti-Mouse IgG. Blots were visualized by chemiluminescence (Pierce #34095). Samples for denaturing Western analyses were prepared in 4X loading buffer (Invitrogen #NP0007), separated by SDS-PAGE with 4-12% Bis-Tri gels (Invitrogen) in MES running buffer (Invitrogen #NP0002), and transferred to PVDF with transfer buffer (Invitrogen #NP0006-1). Unless otherwise noted, blots were processed as described above for native Westerns.
Production of recombinant SOD1 proteins
To produce human WT-SOD1 recombinant protein, the detailed protocol for insect cell expression and subsequent purification described by Hayward et al was employed 49. The human WT-SOD1 baculovirus stock was a generous gift from Drs. Lawrence Hayward and Ashutosh Tiwari (University of Massachusetts Medical School). The established protocol was modified to use TN5B1-4 (High 5) cells from Trichoplusia ni, which were a generous gift from Drs. Jonathan Francis and Ru-Ju Chain (Massachusetts General Hospital).
Recombinant SOD1ox was prepared by exposing human WT-SOD1 to 10 mM hydrogen peroxide 11 (Sigma) at room temperature for 16-24 hours, after which the reaction was quenched by buffer exchanging SOD1ox into PBS using a PD-10 column (GE Healthcare). SOD1 G93A was prepared as described above for SOD1 WT. SOD1 H46R was a generous gift from Drs. Lawrence Hayward and Ashutosh Tiwari (University of Massachusetts Medical School) 49. AS-SOD1 was a generous gift from Dr. Jill Zitzewitz (University of Massachusetts Medical School) 26. AS-SOD was oxidized as described above for WT-SOD1. Working aliquots of SOD1 were stored at -80°C.
Epitope mapping analysis
GST-tagged SOD G93A full-length, and GST-tagged SOD G93A lacking one of the five exons were cloned into the pcDNA3.1 vector, and transfected into HEK-293T cells (American Type Culture Collection) with Lipofectamine 2000 (Invitrogen) according to manufacture's instructions. Cells were then lysed with radio immunoprecipitation assay (RIPA; Boston BioProducts) buffer supplemented with protease inhibitors (Roche, 11873580001). Extracts from cells expressing SOD1 G93C, G93D, G93R, G93S G93V mutants 8 were a generous gift from Drs. David Borchelt and Mercedes Prudencio (University of Florida).
Immunohistochemistry
Tissues were de-paraffinized through histology grade xylenes (3 times for 5 minutes each) and a graded alcohol series (100%, 95% and 70% ethanol; 2 times for 2 minutes each). A method for antigen retrieval (ie acid, heat or proteolysis induced) was avoided when C4F6, the conformation-specific monoclonal antibody was used. Sections were incubated in 3% hydrogen peroxide (Sigma), blocked with 10% normal goat serum (Jackson ImmunoResearch Laboratories, Inc), and incubated with C4F6 (1 μg/mL) prepared in antibody diluent (Dako, 3022) overnight at 4°C. Sections were incubated with biotinylated anti-mouse secondary antibody followed by VECTASTAIN RTU ABC reagent according to the manufacture's instructions (Vector Laboratories, PK-2200). Sections were colorimetrically developed using the 3, 3'-diaminobenzidine (DAB) substrate kit (SK-4100), and cover-slipped with paramount.
For IHC with the pan anti-SOD1 antibody, the following procedure was used. Antigen retrieval was carried out with 0.01 mol/L citrate buffer at pH 6.0, in an 800-W microwave oven for 10 minutes, two times before immunostaining. The slides were stained on the DAKO Autostainer (DAKO Corporation, Carpinteria, CA). The sections were first blocked for endogenous protein binding and peroxidase activity with an application of Dual Endogenous Block (DAKO) for 10 minutes, and then with goat serum for 60 minutes. The sections were then incubated with a sheep polyclonal antibody (Calbiochem, 574597) at 1:100 overnight at 4°C. Sections were then incubated with HRP conjugated with anti-sheep-IgG (Upstate, 12-342) at 1:500 for 60 minutes, and treated with diaminobenzidine (DAB) for 5 minutes. The sections were counterstained with hematoxylin before sealing with permanent media.
Immunopurification and extraction WT-SOD1 from human tissues
Individual immunopurification columns were prepared by coupling 3.5 mg anti-SOD1 antibody (Binding Site, PC077) to 100 mg cyanogen bromide-activated-Sepharose 4B resin (Sigma, C9142) according to the manufacture's instructions, and the antibody cross-linked resin was then transferred into Bio-Spin Columns (BioRad, 732-6008). Frozen SALS and control, lumbar or thoracic SpC tissue sections (~100 mg) were homogenized (Wheaton, 903475) in lysis buffer (25 mM Tris, pH 7.8 supplemented with protease inhibitors; Roche, 11873580001) at 4°C and cleared by centrifugation at 13k rpm and 4°C. The pellet was stored at -80°C, and the cleared lysate was applied to an individual immunoaffinity column (1 unused column per sample), and the flow-through saved on ice. The column was washed 4 times with 600 μL (~20 column volumes total) wash buffer (25 mM Tris, 100 mM NaCl, pH 7.8), followed by elution of WT-SOD1 proteins with 2 × 500 μL gentle antibody elution buffer, pH 6.6 (Pierce, 21027). The columns were re-equilibrated in lysis buffer, the flow-through re-applied, and the purification repeated for a total of 3 times in order to deplete the lysate of WT-SOD1. Elutions were buffer exchanged into 25 mM HEPES, pH 7.4 using 10K MWCO Vivaspin concentrators (Sartorius, VS15RH01), concentrated to ~100 μL with microcon concentrators (Millipore, 42406). The concentrations of WT-SOD1 proteins were determined by Western and densitometry (Image J) analyses with recombinant WT-SOD1 standards.
WT-SOD1 was extracted from the insoluble pellet using a modified protocol of Wang et al 50. Briefly, pellets were re-suspended in 1mL washing buffer (50mM Tris HCL 100mM NaCl pH 7.4, 10% glycerol, 1% Triton, 0.5% NP-40, protease inhibitor) and centrifuged (10 minutes) at 13000 rpm at 4°C for a total of 4 times. The resulting pellet was re-suspended in re-solublization buffer (50mM Tris HCL 100mM NaCl pH 7.4, 10% glycerol, 1% Triton, 0.25mM DTT, 1mM EDTA, 2.5% SDS, protease inhibitor), heated to 100°C for 20 minutes, sonicated (Transsonic 310) for 30 minutes, and heated again before centrifuging at 13krpm (10 minutes) at room temperature. Five μg total re-solubilized protein was subjected to a Western analysis with an anti-SOD1 antibody (Calbiochem, #574597) as described above, except using the secondary anti-sheep IRDye800cw antibody (Rockland, 613-431-002), the Odyssey Infrared Imager (LiCor, Model 9120) and the Odyssey Software (LiCor, V3.0) for densitometry analysis. One μg total re-solublized protein and 0.43 μg of the fourth wash that preceded the re-solubilization was visualized by silver stain analysis (Bio-Rad Laboratories’ Silver Stain Plus kit, 161-0449) to check the total protein loaded to each well qualitatively.
The protocol above was used to extract SOD1 from fresh spinal cord tissue isolated from transgenic SOD1 G93A mice (B6/SJL, the Jackson Laboratory, Bar Harbor, ME) and naïve mice (C57/BL6, the Jackson Laboratory, Bar Harbor, ME).
Vesicle motility assays in isolated squid axoplasm
Vesicle motility assays and immunoblot analysis of axoplasmic kinases were performed as described 29, 30.
Mass spectrometry analysis of SOD1 proteins
Reversed phase liquid chromatography (LC) was performed for WT-SOD1 and SOD1ox (Fig. 1) using a two-dimensional nanoflowrate LC (Eksigent); 5mm, 300 μm ID guard column (LC Packings, Part Number 160454); and a self-packed 14cm, 100μm ID column, with 5 μm beads (taken from a larger Targa column). Buffer A was 0.1% formic acid in HPLC grade water, Buffer B was 100% HPLC grade acetonitrile. Samples were diluted to a final formic concentration of 0.1% and injected. Following injection, samples were washed on the guard column with 160 column volumes of Buffer A (8 μL/min), and eluted at 650 nL/min using a 0-40% gradient over 30 minutes. For intact protein MS/MS, SOD1 was disulfide reduced in 50mM tris(2-carboxyethyl)phosphine and 0.1mM EDTA, desalted using C18 ZipTips (Millipore), and infused directly. Samples were introduced via a nanospray ion source with dual ion funnel (Apollo II) connected to a hybrid quadrupole Fourier transform ion cyclotron resonance (FT-ICR; FT-MS) mass spectrometer (apex Qe-94, Bruker Daltonics). The instrument was equipped with a hollow cathode for electron capture dissociation. External calibration of m/z scale was performed using ES tuning mix (Agilent, G2431A) using peaks at m/z 622, 922, 1522, and 2122.
After desolvation, the ions were transferred from a source hexapole to the quadrupole mass filter where isolation could occur in a second hexapole (collision cell). Ions accumulated in the second hexapole were then transferred through ion optics region of the instrument to the ICR cell. Frequency sweep excitation was followed by image charge detection. For ECD experiment, activation of in the ICR cell was performed using a 105 Vpp, 1000 Hz off-resonant, 4 ms sustained off-resonance irradiation (SORI) during ECD pulse. Important instrument operation parameters include source declustering potential 40V, hexapole 1 accumulation time = 0.1 ms, collision cell accumulation time = 1 s, time of flight = 1.8 milliseconds (D2), sidekick extraction voltages = -1.0 V (EV1, EV2, DEV2), RF excitation voltage = 130 V, ICR trapping potential 1.2 V, for MS/MS: Q1 m/z = 887.0 (for SOD1ox), isolation window (m/z) = 5, ECD bias = 6 V, ECD lens = 20 V, ECD duration = 4 ms.
Intact protein masses were reconstructed using the “deconvolution” function from DataAnalysis (Bruker Daltonics, version 3.4), and monoisotopic masses were determined using the Snap II algorithm (Bruker Daltonics). MS/MS data was automatically searched in human protein sequence database (MSDB) using Mascot TD and was matched to human SOD1 with trioxidation at Cys111.
For the analysis of AS-SOD1 proteins (Supplementary Fig. 1), samples were diluted in 0.1% trifluoracetic acid (TFA) to 1μg/μL and 5μL was injected using a 10μl loop injector onto a μ-precolumn C4 (Vydac) 15mm × 1mm, 3μm, 300Ǻ (LC-Packings) using a flow rate of 50μL/min with 10% acetonitrile (ACN) and 0.1% formic acid (FA). The mass spectra were acquired on a Q-Tof using W-mode (Waters, Inc.) and the electro spray voltage set to 3500V. During the gradient, from 10% ACN, 0.1% FA to 95% ACN, 0.1% FA over 5 minutes, 3 second scans were collected from m/z 500 – 2000. The spectra under the eluted peak were summed, and deconvoluted using MaxEnt1 (Waters, Inc).
Statistical analysis
A two-tailed Fisher exact test was used to determine the significance between SALS and control cases positively stained with the C4F6 antibody (Fig. 6). The activation of kinases in squid axoplasm and effects of FAT by SOD1ox (Fig. 5b) and insoluble levels of SOD1 in control and human tissues were tested using a pooled t-test of μ1-μ2 (Datadesk). A P <0.05 was considered statistically significant for all analyses.
References
- 1.Tandan R, Bradley WG. Amyotrophic lateral sclerosis: Part 1. Clinical features, pathology, and ethical issues in management. Ann Neurol. 1985;18:271–280. doi: 10.1002/ana.410180302. [DOI] [PubMed] [Google Scholar]
- 2.Valdmanis PN, Daoud H, Dion PA, Rouleau GA. Recent advances in the genetics of amyotrophic lateral sclerosis. Curr Neurol Neurosci Rep. 2009;9:198–205. doi: 10.1007/s11910-009-0030-9. [DOI] [PubMed] [Google Scholar]
- 3.Selverstone Valentine J, Doucette PA, Zittin Potter S. Copper-zinc superoxide dismutase and amyotrophic lateral sclerosis. Annu Rev Biochem. 2005;74:563–593. doi: 10.1146/annurev.biochem.72.121801.161647. [DOI] [PubMed] [Google Scholar]
- 4.Bruijn LI, et al. Aggregation and motor neuron toxicity of an ALS-linked SOD1 mutant independent from wild-type SOD1. Science. 1998;281:1851–1854. doi: 10.1126/science.281.5384.1851. [DOI] [PubMed] [Google Scholar]
- 5.Chattopadhyay M, Valentine JS. Aggregation of Copper-Zinc Superoxide Dismutase in Familial and Sporadic ALS. Antioxid Redox Signal. 2009 doi: 10.1089/ars.2009.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furukawa Y, Fu R, Deng HX, Siddique T, O'Halloran TV. Disulfide cross-linked protein represents a significant fraction of ALS-associated Cu, Zn-superoxide dismutase aggregates in spinal cords of model mice. Proc Natl Acad Sci U S A. 2006;103:7148–7153. doi: 10.1073/pnas.0602048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Molnar KS, et al. A common property of amyotrophic lateral sclerosis-associated variants: Destabilization of the Cu/Zn superoxide dismutase electrostatic loop. J Biol Chem. 2009 doi: 10.1074/jbc.M109.023945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prudencio M, Hart PJ, Borchelt DR, Andersen PM. Variation in aggregation propensities among ALS-associated variants of SOD1: correlation to human disease. Hum Mol Genet. 2009;18:3217–3226. doi: 10.1093/hmg/ddp260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Q, Johnson JL, Agar NY, Agar JN. Protein aggregation and protein instability govern familial amyotrophic lateral sclerosis patient survival. PLoS Biol. 2008;6:e170. doi: 10.1371/journal.pbio.0060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morfini GA, et al. Axonal transport defects in neurodegenerative diseases. J Neurosci. 2009;29:12776–12786. doi: 10.1523/JNEUROSCI.3463-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ezzi SA, Urushitani M, Julien JP. Wild-type superoxide dismutase acquires binding and toxic properties of ALS-linked mutant forms through oxidation. J Neurochem. 2007;102:170–178. doi: 10.1111/j.1471-4159.2007.04531.x. [DOI] [PubMed] [Google Scholar]
- 12.Gruzman A, et al. Common molecular signature in SOD1 for both sporadic and familial amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2007;104:12524–12529. doi: 10.1073/pnas.0705044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beckman JS, Estevez AG, Crow JP, Barbeito L. Superoxide dismutase and the death of motoneurons in ALS. Trends Neurosci. 2001;24:S15–20. doi: 10.1016/s0166-2236(00)01981-0. [DOI] [PubMed] [Google Scholar]
- 14.Bredesen DE, Ellerby LM, Hart PJ, Wiedau-Pazos M, Valentine JS. Do posttranslational modifications of CuZnSOD lead to sporadic amyotrophic lateral sclerosis? Ann Neurol. 1997;42:135–137. doi: 10.1002/ana.410420202. [DOI] [PubMed] [Google Scholar]
- 15.Kabashi E, Valdmanis PN, Dion P, Rouleau GA. Oxidized/misfolded superoxide dismutase-1: the cause of all amyotrophic lateral sclerosis? Ann Neurol. 2007;62:553–559. doi: 10.1002/ana.21319. [DOI] [PubMed] [Google Scholar]
- 16.Durazo A, et al. Metal-free superoxide dismutase-1 and three different ALS variants share a similar partially unfolded {beta}-barrel at physiological temperature. J Biol Chem. 2009 doi: 10.1074/jbc.M109.052076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Estevez AG, et al. Induction of nitric oxide-dependent apoptosis in motor neurons by zinc-deficient superoxide dismutase. Science. 1999;286:2498–2500. doi: 10.1126/science.286.5449.2498. [DOI] [PubMed] [Google Scholar]
- 18.Rakhit R, et al. Oxidation-induced misfolding and aggregation of superoxide dismutase and its implications for amyotrophic lateral sclerosis. J Biol Chem. 2002;277:47551–47556. doi: 10.1074/jbc.M207356200. [DOI] [PubMed] [Google Scholar]
- 19.Banci L, et al. Metal-free superoxide dismutase forms soluble oligomers under physiological conditions: a possible general mechanism for familial ALS. Proc Natl Acad Sci U S A. 2007;104:11263–11267. doi: 10.1073/pnas.0704307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urushitani M, Ezzi SA, Julien JP. Therapeutic effects of immunization with mutant superoxide dismutase in mice models of amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2007;104:2495–2500. doi: 10.1073/pnas.0606201104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brady ST, Lasek RJ, Allen RD. Fast axonal transport in extruded axoplasm from squid giant axon. Science. 1982;218:1129–1131. doi: 10.1126/science.6183745. [DOI] [PubMed] [Google Scholar]
- 22.Gros-Louis F, Soucy G, Lariviere R, Julien JP. Intracerebroventricular infusion of monoclonal antibody or its derived Fab fragment against misfolded forms of SOD1 mutant delays mortality in a mouse model of ALS. J Neurochem. 2010 doi: 10.1111/j.1471-4159.2010.06683.x. [DOI] [PubMed] [Google Scholar]
- 23.Fujiwara N, et al. Oxidative modification to cysteine sulfonic acid of Cys111 in human copper-zinc superoxide dismutase. J Biol Chem. 2007;282:35933–35944. doi: 10.1074/jbc.M702941200. [DOI] [PubMed] [Google Scholar]
- 24.Tiwari A, et al. Metal deficiency increases aberrant hydrophobicity of mutant superoxide dismutases that cause amyotrophic lateral sclerosis. J Biol Chem. 2009 doi: 10.1074/jbc.M109.043729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rakhit R, et al. Monomeric Cu,Zn-superoxide dismutase is a common misfolding intermediate in the oxidation models of sporadic and familial amyotrophic lateral sclerosis. J Biol Chem. 2004;279:15499–15504. doi: 10.1074/jbc.M313295200. [DOI] [PubMed] [Google Scholar]
- 26.Svensson AK, Bilsel O, Kondrashkina E, Zitzewitz JA, Matthews CR. Mapping the folding free energy surface for metal-free human Cu,Zn superoxide dismutase. J Mol Biol. 2006;364:1084–1102. doi: 10.1016/j.jmb.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 27.Brady ST, Lasek RJ, Allen RD. Video microscopy of fast axonal transport in extrudedaxoplasm: a new model for study of molecular mechanisms. Cell Motil. 1985;5:81–101. doi: 10.1002/cm.970050203. [DOI] [PubMed] [Google Scholar]
- 28.Morfini G, Szebenyi G, Elluru R, Ratner N, Brady ST. Glycogen synthase kinase 3 phosphorylates kinesin light chains and negatively regulates kinesin-based motility. EMBO J. 2002;21:281–293. doi: 10.1093/emboj/21.3.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morfini G, et al. JNK mediates pathogenic effects of polyglutamine-expanded androgen receptor on fast axonal transport. Nat Neurosci. 2006;9:907–916. doi: 10.1038/nn1717. [DOI] [PubMed] [Google Scholar]
- 30.Morfini GA, et al. Pathogenic huntingtin inhibits fast axonal transport by activating JNK3 and phosphorylating kinesin. Nat Neurosci. 2009;12:864–871. doi: 10.1038/nn.2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fabian MA, et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 32.Munoz L, et al. A novel p38 alpha MAPK inhibitor suppresses brain proinflammatory cytokine up-regulation and attenuates synaptic dysfunction and behavioral deficits in an Alzheimer's disease mouse model. J Neuroinflammation. 2007;4:21. doi: 10.1186/1742-2094-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kerman A, et al. Amyotrophic lateral sclerosis is a non-amyloid disease in which extensive misfolding of SOD1 is unique to the familial form. Acta Neuropathol. 2010 doi: 10.1007/s00401-010-0646-5. [DOI] [PubMed] [Google Scholar]
- 34.Liu HN, et al. Lack of evidence of monomer/misfolded superoxide dismutase-1 in sporadic amyotrophic lateral sclerosis. Ann Neurol. 2009;66:75–80. doi: 10.1002/ana.21704. [DOI] [PubMed] [Google Scholar]
- 35.Shibata N, Asayama K, Hirano A, Kobayashi M. Immunohistochemical study on superoxide dismutases in spinal cords from autopsied patients with amyotrophic lateral sclerosis. Dev Neurosci. 1996;18:492–498. doi: 10.1159/000111445. [DOI] [PubMed] [Google Scholar]
- 36.Shibata N, et al. Cu/Zn superoxide dismutase-like immunoreactivity in Lewy body-like inclusions of sporadic amyotrophic lateral sclerosis. Neurosci Lett. 1994;179:149–152. doi: 10.1016/0304-3940(94)90956-3. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe M, et al. Histological evidence of protein aggregation in mutant SOD1 transgenic mice and in amyotrophic lateral sclerosis neural tissues. Neurobiol Dis. 2001;8:933–941. doi: 10.1006/nbdi.2001.0443. [DOI] [PubMed] [Google Scholar]
- 38.Rakhit R, et al. An immunological epitope selective for pathological monomer-misfolded SOD1 in ALS. Nat Med. 2007;13:754–759. doi: 10.1038/nm1559. [DOI] [PubMed] [Google Scholar]
- 39.Pasinelli P, et al. Amyotrophic lateral sclerosis-associated SOD1 mutant proteins bind and aggregate with Bcl-2 in spinal cord mitochondria. Neuron. 2004;43:19–30. doi: 10.1016/j.neuron.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 40.Urushitani M, et al. Chromogranin-mediated secretion of mutant superoxide dismutase proteins linked to amyotrophic lateral sclerosis. Nat Neurosci. 2006;9:108–118. doi: 10.1038/nn1603. [DOI] [PubMed] [Google Scholar]
- 41.Vande Velde C, Miller TM, Cashman NR, Cleveland DW. Selective association of misfolded ALS-linked mutant SOD1 with the cytoplasmic face of mitochondria. Proc Natl Acad Sci U S A. 2008;105:4022–4027. doi: 10.1073/pnas.0712209105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lindberg MJ, Normark J, Holmgren A, Oliveberg M. Folding of human superoxide dismutase: disulfide reduction prevents dimerization and produces marginally stable monomers. Proc Natl Acad Sci U S A. 2004;101:15893–15898. doi: 10.1073/pnas.0403979101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Vos KJ, Grierson AJ, Ackerley S, Miller CC. Role of axonal transport in neurodegenerative diseases. Annu Rev Neurosci. 2008;31:151–173. doi: 10.1146/annurev.neuro.31.061307.090711. [DOI] [PubMed] [Google Scholar]
- 44.Strom AL, et al. Retrograde axonal transport and motor neuron disease. J Neurochem. 2008;106:495–505. doi: 10.1111/j.1471-4159.2008.05393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Collard JF, Cote F, Julien JP. Defective axonal transport in a transgenic mouse model of amyotrophic lateral sclerosis. Nature. 1995;375:61–64. doi: 10.1038/375061a0. [DOI] [PubMed] [Google Scholar]
- 46.Williamson TL, Cleveland DW. Slowing of axonal transport is a very early event in the toxicity of ALS-linked SOD1 mutants to motor neurons. Nat Neurosci. 1999;2:50–56. doi: 10.1038/4553. [DOI] [PubMed] [Google Scholar]
- 47.Fischer LR, Glass JD. Axonal degeneration in motor neuron disease. Neurodegener Dis. 2007;4:431–442. doi: 10.1159/000107704. [DOI] [PubMed] [Google Scholar]
- 48.Strange RW, et al. Variable metallation of human superoxide dismutase: atomic resolution crystal structures of Cu-Zn, Zn-Zn and as-isolated wild-type enzymes. J Mol Biol. 2006;356:1152–1162. doi: 10.1016/j.jmb.2005.11.081. [DOI] [PubMed] [Google Scholar]
- 49.Hayward LJ, et al. Decreased metallation and activity in subsets of mutant superoxide dismutases associated with familial amyotrophic lateral sclerosis. J Biol Chem. 2002;277:15923–15931. doi: 10.1074/jbc.M112087200. [DOI] [PubMed] [Google Scholar]
- 50.Wang L, et al. Wild-type SOD1 overexpression accelerates disease onset of a G85R SOD1 mouse. Hum Mol Genet. 2009;18:1642–1651. doi: 10.1093/hmg/ddp085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.