Summary
Prior to the availability of the genome sequence, the root of Arabidopsis had attracted a small but ardent group of researchers drawn to its accessibility and developmental simplicity. Roots are easily observed when grown on the surface of nutrient agar media, facilitating analysis of responses to stimuli such as gravity and touch. Developmental biologists were attracted to the simple radial organization of primary root tissues, which form a series of concentric cylinders around the central vascular tissue. Equally attractive was the mode of propagation, with stem cells at the tip giving rise to progeny that were confined to cell files. These properties of root development reduced the normal four-dimensional problem of development (three spatial dimensions and time) to a two-dimensional problem, with cell type on the radial axis and developmental time along the longitudinal axis. The availability of the complete Arabidopsis genome sequence has dramatically accelerated traditional genetic research on root biology, and has also enabled entirely new experimental strategies to be applied. Here we review examples of the ways in which availability of the Arabidopsis genome sequence has enhanced progress in understanding root biology.
Keywords: Arabidopsis, root biology, genomics, development, genetics, transcriptome
Genome-Assisted Genetic Approaches to Root Biology
Forward genetics
The Arabidopsis root is developmentally simple, with concentric rings of cell types continuously produced from a small set of stem cells near the root tip (Figures 1 and 2). In the pre-genome days of Arabidopsis research, this developmental simplicity and the ease of study of roots encouraged investigators to perform extensive genetic screens to define numerous loci that are important for various aspects of root development (reviewed by Benfey and Schiefelbein, 1994; Dolan and Roberts, 1995). However, a bottleneck quickly arose in understanding the functions of the genes, due to difficulties in cloning a gene based on a mutant phenotype alone (termed forward genetics). With the availability of the genome sequence and the associated molecular markers and DNA clones, genetic map-based cloning became routine. Starting from an interesting mutant phenotype, investigators have now identified dozens of root developmental genes, including ones affecting stem cell function, cell-type pattern formation, cell and organ morphogenesis, and the differentiation of specific cell types (reviewed by Guimil and Dunand, 2007; Ishida et al., 2008; Petricka and Benfey, 2008; Shaw and Dolan, 2008).
Figure 1.
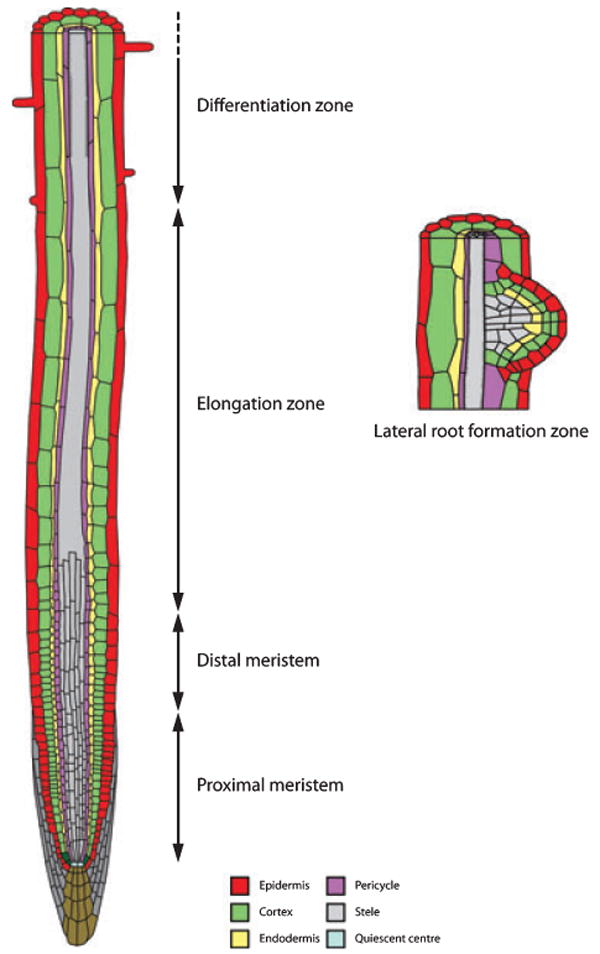
Schematic representation of Arabidopsis primary and lateral root tissues.
Single layers of epidermis (red), cortex (green), endodermis (yellow) and pericycle (purple) cells surround the central vascular tissue (grey). The outer layers are organized as concentric cylinders of tissues. Cells within these tissues exhibit a proximal–distal developmental gradient, originating from initial cells abutting quiescent centre cells (light blue) that transit through proximal and distal meristem zones during their division phase, then undergo rapid cell expansion in the elongation zone, prior to entering the differentiation zone. Lateral roots form from the pericyle. As lateral root primordia form (right), all of the tissues found in the primary root are generated de novo.
Figure 2.
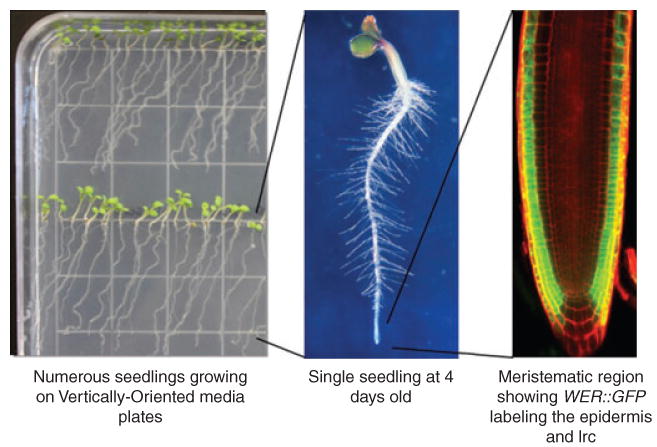
Basic features of arabidopsis root structure.
This series of progressively magnified images comprises an image of 4-day-old Arabidopsis seedlings grown on vertical plates (left), a magnified view of a single seedling illustrating the abundant root hairs (centre), and a fluorescent microscopy image of an Arabidopsis root tip with the epidermis and lateral root cap (lrc) cells expressing GFP (right).
The ease of molecular genetic analyses fostered by the availability of the Arabidopsis genome sequence prompted investigators to initiate a variety of new and imaginative mutant screens to define new root developmental genes. These include screens that use promoter trap constructs to define genes expressed in particular cell or tissue types (Aida et al., 2004), utilization of multiple cell-type marker lines to identify mutations altering a specific cell type (Willemsen et al., 2008), changes in the surface orientation of the medium to isolate mutants with alterations in the regulation of directional organ growth (Oliva and Dunand, 2007), and modifications in the composition of the growth medium to select mutants with abnormal responses to nutrients (Little et al., 2005). Root mutants have also been identified by profiling mutant populations for differences in shoot elemental composition (Lahner et al., 2003; Baxter et al., 2009). Genome information proved crucial in the latter case in order to map and clone the enhanced suberin 1 (esb1) mutant (Baxter et al., 2009). DNA prepared from mutant versus wild-type seedlings (segregating in an F2 mapping population) was first hybridized to an Arabidopsis ATH1 gene chip. Single feature polymorphisms enriched for the Col background of the esb1 mutation were identified, and centred around 11 Mb on chromosome 2. An Arabidopsis genome tiling array (ATTILE 1.0R) was then used to detect a deletion of approximately 1.1 kbp in At2g28670, whose gene product regulates suberin composition in the Casparian strip. This forward genetic screen provides direct evidence for the importance of suberin and the casparian strip in the root endodermis for maintaining proper shoot mineral composition.
Reverse genetics
Perhaps the most valuable knowledge provided by sequencing of the Arabidopsis genome is the complete list of predicted proteins produced by the plant. This information has enabled root biologists to select genes encoding proteins of potential interest and subject them to ‘reverse genetics’ analysis, by investigating the effect of inhibiting the desired gene's function using insertional mutations or RNAi. For example, a critical role for the Arabidopsis retinoblastoma-related (RBR) protein in regulating the root stem cell population, using a pathway seemingly shared by plants and animals, was established by subjecting the single Arabidopsis RBR gene to post-embryonic loss-of-function and gain-of-function alterations (Wildwater et al., 2005). In another study, a root-expressed relative of the CLAVATA3 signalling molecule, named CLV40, was shown to participate in the maintenance of root stem cells, with some similarities to the shoot meristem maintenance system (Stahl et al., 2009). In a third example of this approach, the Arabidopsis homologue of the human MMS21 SUMO E3 ligase was found to influence root cell proliferation and cause defects in the cytokinin response (Huang et al., 2009).
The availability of the entire gene/protein list of Arabidopsis has also enabled directed analysis of multiple members of gene families to assess their degree of redundant or unique function in root development. A particularly instructive example of this approach is provided by study of the PLETHORA (PLT) gene family, which showed that four related PLT transcription factors are together required for formation of the root, and that they act in an additive, dosage-dependent fashion to influence stem cell identity, cell division and cell-type differentiation (Galinha et al., 2007). Among many other examples, the relative roles of the five-member CAPRICE gene family in epidermal cell-type pattern formation have been defined by a complete analysis of all loss-of-function mutant combinations (Simon et al., 2007; Tominaga et al., 2008).
Reverse genetic screens have become much more focused as a result of the availability of high-resolution spatio-temporal transcript maps of individual cell types and developmental zones in the root (described in detail below) (Birnbaum et al., 2003; Brady et al., 2007). For example, the availability of the catalogues of genes expressed in the xylem pole pericycle cells (DeSmet et al., 2008) and in the outer tissues (Swarup et al., 2008) has greatly aided the identification of new genes regulating lateral root initiation and emergence processes, respectively. This approach led to identification of the receptor-like kinase ACR4, which promotes cell proliferation of newly initiated lateral root primordia while repressing the division of cells either adjacent to, or at the opposite pole to, the new lateral root primordia (DeSmet et al., 2008).
Quantitative and association genetics
Genome information has also greatly assisted recent efforts to exploit the natural variation in root traits exhibited by Arabidopsis accessions (Armengaud et al., 2008). The availability of the Columbia genome sequence initially enabled markers to be developed for other accessions by sequencing a large number of PCR-amplified genomic fragments (Schmid et al., 2003; Nordborg et al., 2005). In the latter case, 876 fragments were sequenced in 96 accessions. More recently, the genomes of 21 accessions have been completely sequenced (Clark et al., 2007), providing a rich source of information for marker development and gene identification. Current efforts to sequence the genomes of hundreds of accessions (http://www.1001genomes.org) promise to improve dramatically the ability to perform association genetic studies in Arabidopsis.
Genome sequence information has greatly assisted efforts to fine map and clone the genes encoding several root quantitative trait loci (QTL). For example, BREVIS RADIX (BRX) was identified as a strong-effect QTL accounting for approximately 80% of the variance in primary root length observed in the Umkirch-1 (Uk-1) accession. Map-based cloning led to identification of a region of approximately 45 kb containing approximately ten genes. At1g31880 was identified as the BRX gene by comparing the sequence of the gene in Uk-1 to that in Col, which revealed the existence of a single base pair difference, creating a premature stop codon. BRX encodes a novel gene product that plays an important integrative role in several plant hormone signalling pathways (Mouchel et al., 2006; Scacchi et al., 2009). Other examples of recently isolated root QTLs include a vacuolar invertase that regulates primary root elongation (Sergeeva et al., 2006) and a multi-copper oxidase that regulates the low phosphate growth response (Svistoonoff et al., 2007). Hence, this approach has successfully led to the identification of both novel genes and novel alleles in known genes.
The availability of genome sequence for hundreds of accessions (http://www.1001genomes.org) now makes it possible to perform association genetic studies in Arabidopsis. A recent example involved identification of the MOT1 QTL, which controls shoot molybdenum content (Baxter et al., 2008). Elemental analysis of 92 diverse Arabidopsis accessions identified 12 that exhibited low molybdenum content. Genome-based approaches led to fine mapping of MOT1 to a region containing approximately 80 genes. The promoter of the At2g25680 gene was observed to contain a 53 bp deletion that was common to seven of the 92 accessions tested, all of which contained low molybdenum levels. The MOT1 gene encodes a molybdenum transporter that was demonstrated to be active in the root, using grafting studies. This example clearly illustrates that natural accessions provide an excellent source of allelic variation suitable for the identification and functional characterization of novel root genes.
Assembling Root Biology Pathways Using Genetics and Genomics
Determination of the Arabidopsis genome sequence has enabled gene arrays to be manufactured and used to analyse the accumulation of gene transcripts across the entire genome. This approach has already had a major impact on our understanding of gene regulation during root development.
A long-term goal is to integrate the large-scale gene expression data with the existing genetic information to assemble regulatory pathways that govern root development. The first steps in this direction have been taken by investigators using mutant lines or altered growth conditions as a way to perturb specific developmental pathways and thereby help to define new pathway genes and their relationships. For example, two related studies have defined a large set of new root hair pathway genes, by comparing transcript accumulation in hairless mutants to that in wild-type, and subsequently using reverse genetics to demonstrate the essential role of several of these genes in root hair differentiation (Jones et al., 2006; Won et al., 2009). As another example, components of the root's developmental response to phosphate (Pi) deficiency have been defined using comparative transcript analysis, and the critical role of the newly identified WRKY75 and ZAT6 transcription factors has been characterized by mutant analyses (Misson et al., 2005; Devaiah et al., 2007a,b). In a related study, the Pi-associated bHLH32 transcription factor was shown to interact with the root epidermal fate regulators TRANSPARENT TESTA GLABRA1 (TTG) and GLABRA3 (GL3). Moreover, the bhlh32 mutant exhibits a significant increase in transcription of Pi starvation-induced genes, suggesting that bHLH32 is a negative regulator of the plant's developmental response to nutrient availability (Chen et al., 2007).
Among the current challenges in the construction of transcriptional regulatory networks is to accurately define and functionally confirm the putative cis-acting regulatory elements that exist in gene promoters. Recent genome-aided studies of the root epidermis developmental pathway have begun to provide insights in this area. In one study, a previously defined root hair cell cis-element (RHE) was used in an in silico screen to assist in the identification of new genes that participate in root hair differentiation (Won et al., 2009). Although this element was only one of several filters, the success of this approach suggests that there is sufficient sequence conservation among some cis-elements to use them to define other members of the same transcriptional regulatory family. Several other studies of root epidermal transcription factors have defined additional cis-elements, including a putative GLABRA2-binding L1 box sequence (Tominaga-Wada et al., 2009) and a binding sequence for the WEREWOLF Myb protein (Koshino-Kimura et al., 2005; Ryu et al., 2005), which should prove useful for continuing network construction efforts.
A promising strategy for assembling transcriptional regulatory networks is to modulate or perturb a network using a variety of alterations (e.g. reduction of function, gain of function, mis-expression) of a gene(s) at a critical node(s), and then combine the multiple sets of results to infer network components and architecture. An example of this approach, known as meta-analysis, has recently been used to elucidate the SHORT ROOT (SHR) pathway in root stem cell and tissue type specification (Levesque et al., 2006). This led to the identification of several likely downstream target genes, a proposed architecture of the SHR-regulated network, and new connections to hormonal and signalling pathways (Levesque et al., 2006).
Cell Type-Specific Transcriptome Profiling
To obtain a deeper and systematic understanding of the nature of the networks regulating root biology, two types of data are needed: (i) mRNA expression data – the output of transcriptional networks, and (ii) transcription factor binding data to identify the causal connections in the network. If this information could be obtained for every cell type and every developmental stage of the root, it would provide an all-encompassing picture of the regulatory networks controlling root development.
Genomic sequence data were critical for developing genome-wide profiling technology. Until recently, the primary technology used was oligonucleotide-based microarrays, which require knowledge of genomic sequence to be constructed and interpreted. When this technology first became available in around 2000, it was used primarily to profile responses to various stimuli. Not surprisingly, these efforts used whole plants or plant parts such as leaves or roots. Useful insights were obtained, but there was a nagging doubt that responses specific to individual cell types or tissues would be missed, as they were diluted out in the mass of the whole plant or organ.
To address this concern, new approaches were needed to allow expression analysis of only one cell type or developmental stage. The three approaches that have been employed to generate these types of data use very different methods. Laser capture microdissection requires a microscopy platform designed specifically for this procedure. It has the advantage of being able to select cells based on morphology or position. Its primary disadvantage is that each cell or group of cells has to be individually excised. Current RNA isolation techniques generally require on the order of 500–1000 cells, making this a very tedious method for profiling cells of low abundance in the root. Nevertheless, the method has been successfully applied for profiling specific cell types in the root and shoot (Kerk et al., 2003). A second method involves expressing a tagged protein intrinsic to ribosomes behind a tissue-specific promoter. Immunoprecipitation from transgenic plants expressing the protein should yield mRNA that was in the process of being translated (Zanetti et al., 2005). This has the advantage of profiling only transcripts that are presumably being productively translated. A disadvantage is that its use is restricted to analysing polysomes, and it cannot be used to profile other cellular constituents such as metabolites and proteins. A third method relies on sorting marked cells using a fluorescence activated cell sorter (FACS) (Figure 3). This requires transgenic lines in which a fluorophore such as GFP is expressed in a tissue-specific manner. Examples are promoter fusions and enhancer trap lines. Prior to FACS sorting, cell walls are enzymatically digested, resulting in protoplasts. This is the main disadvantage of this approach, as the process of isolating cells disrupts intercellular signalling and thus could have an impact on the transcriptional state. The advantage is that it is easily scalable so that sufficient numbers of cells can be obtained from even very restricted cell populations, such as the quiescent centre (Nawy et al., 2005). Moreover, the same approach could be used to profile other cellular constituents such as metabolites, proteins or microRNAs. To date, only this third approach has been used extensively to profile expression in the Arabidopsis root. Proof-of-concept profiling was performed using laser capture microdissection on Arabidopsis (Kerk et al., 2003), but extensive expression analysis has only been performed on rice (Jiao et al., 2009). Proof-of-concept profiling has also been performed using a tagged ribosomal protein (Zanetti et al., 2005).
Figure 3.
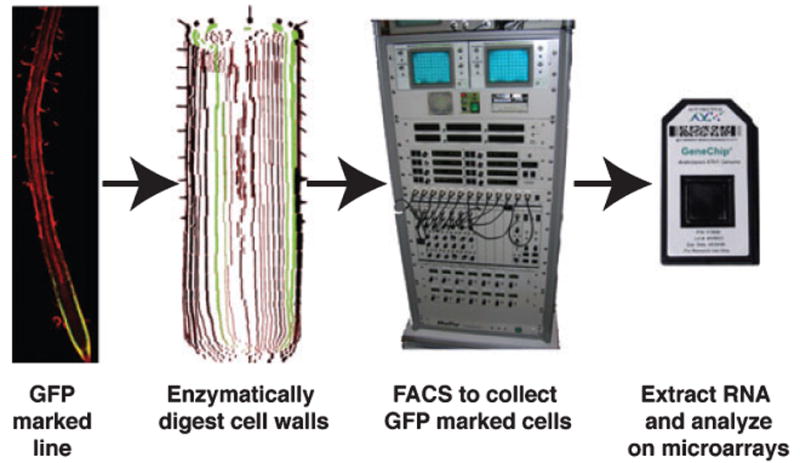
Pipeline for profiling mRNA expression in root cell types.
Roots of transgenic lines with cell type-specific GFP expression are subjected to enzymatic digestion of the cell walls. The resulting protoplasts are analysed in a fluorescent activated cell sorter (FACS), which separates the GFP-expressing cells from the non-expressing cells. RNA from the sorted cells is then used for labelling and hybridization to microarrays.
From Profiling to Networks
The simplicity of Arabidopsis root anatomy and development was the basis for the first effort to profile expression in specific cell populations. Five GFP-marked cell populations along the radial axis were FACS sorted, and three developmental zones were dissected (Birnbaum et al., 2003). Expression analysis was performed using Affymetrix microarrays. Comparison of expression in protoplasts with that in whole roots indicated that, within the time frame of the experiment, a relatively small number of genes were induced by the protoplasting procedure itself. The resulting data confirmed the prediction that whole-plant or organ profiling misses many genes that are expressed in a restricted manner. Many of the differentially expressed genes were not found in whole-plant or organ expression analyses. Clustering of the expression data revealed genes of similar biological function that were co-regulated in tissue- or developmental zone-specific patterns (Birnbaum et al., 2003). A subsequent effort focusing on cells of the quiescent centre provided insights into expression specific to the stem cell niche in the root (Nawy et al., 2005).
A substantially higher level of resolution was achieved through profiling of 15 cell types and 13 segments along the longitudinal axis of the root (Brady et al., 2007). Clustering of the genes in the latter dataset revealed the surprising finding that many had fluctuating expression patterns, such that they were turned on early in development, then were shut off, and then were turned back on at a later stage of development. This was unexpected because development is normally thought of as a progressive, uni-directional process.
As a result of the heightened resolution provided by the combined datasets, transcriptional network connections started to emerge. Within a specific cell type or expression cluster, genes were identified with known cis-elements over-represented in their regulatory regions. A search was then performed within the same cluster for a transcription factor that would potentially bind to that cis-element. In this way, several potential network connections were identified, one of which was already known, but to which additional potential downstream targets could be added (Brady et al., 2007).
Linking Developmental Responses to Environmental Stimuli Using Genomic Approaches
Profiling expression in different cell types and developmental zones revealed a wealth of information about roots grown under standard laboratory conditions, but little about what happens when they are subjected to the stress and stimuli they encounter in the wild. To begin to explore how plants respond to stimuli at the level of individual cell types or along a developmental timeline, plants were grown on either high salt or low iron, and GFP-marked cells were sorted or roots dissected, with subsequent microarray profiling (Dinneny et al., 2008). In the case of both stresses, the analysis revealed that there was a remarkable amount of cell type- and developmental zone-specific responses (Dinneny et al., 2008). Clustering suggested that certain biological functions were activated or repressed in specific cell types or developmental zones. Initial efforts suggested that these genes could be involved in responses in which specific cell types or zones carry out functions that allow the root to acclimatize or adapt to a particular stress. A similar approach was used to analyse the effects of nitrogen on root development at the level of individual cell types (Gifford et al., 2008). By combining information from cell sorting and metabolic pathways, a network was identified that regulates lateral root outgrowth upon stimulation by exogenous nitrogen (Gifford et al., 2008).
These results also pointed the way to a deeper understanding of network dynamics. Subjecting the plant to external stimuli is equivalent to perturbing the networks. Observing how networks respond to perturbation can reveal how robust they are, which is critical to our understanding of network function and evolution. We are only at the early stages of this effort, and new technologies are required to allow real-time recording of responses at the resolution of individual cell types and developmental zones.
Computational and mathematical models are set to play an increasingly important role in probing network dynamics. Researchers at the Centre for Plant Integrative Biology (http://www.cpib.ac.uk) have recently developed the first mathematical model of the auxin response network employing ordinary differential equations (Middleton et al., 2009). The model of the auxin response network captures all of the key components identified to date, including Aux/IAA, auxin response factor (ARF) and transport inhibitor resistant (TIR) proteins, together with their known regulatory relationships. Crucially, the model is able to accurately predict the dynamic behavior of GFP-tagged auxin-responsive reporters expressed in root tissues (T. Vernoux, ENS-Lyon, France; A. Middleton, A. Larrieu and M. Bennett, Centre for Plant Integrative Biology, unpublished data). Such experimental validation is essential for any model as it provides experimental evidence that the network being studied contains all of the key components necessary to account for its dynamic behaviour.
A Beginning of Network Analysis
Researchers are beginning to develop quantitative, rather than purely qualitative, models in the Arabidopsis root. Models simulating auxin fluxes within and between root tissues are helping to realize this important goal (Swarup et al., 2005; Grieneisen et al., 2007; Jones et al., 2008; Laskowski et al., 2008). These simulations of auxin fluxes employ realistic transport parameters, carrier distributions and cellular geometries (Swarup et al., 2005; Grieneisen et al., 2007; Laskowski et al., 2008). Such models provide an invaluable tool for researchers by generating new predictions that can be tested experimentally. For example, simulations of a gravity-induced auxin gradient in the root elongation zone predicted that the hormone accumulated preferentially in expanding epidermal cells (as opposed to cortical and endodermal cells) on the lower side of the root, prompting experiments that later revealed the primary importance of the epidermis for gravitropic curvature (Swarup et al., 2005). Similarly, simulations of auxin fluxes in root proximal tissues following a gravitropic stimulus provided an elegant mechanism to explain the non-intuitive observation that new lateral root primordia initiate on the upper side of a bending root (Laskowski et al., 2008). In the near future, such models will provide an invaluable tool for researchers to relate the impact of allelic variation in network components at the genome level to root growth and developmental processes occurring at higher physical scales.
Attempts to model many signals and their transduction pathways need to go beyond the network level and capture the cellular, tissue and organ levels, thereby better reflecting the various levels of organization of the root system. Indeed, regulatory networks frequently function across multiple root cells and tissues. For example, several transcription factors have been shown to move between root cells and tissues, where they alter cell fate by altering the expression of target genes. For example, specification of Arabidopsis root endodermal/cortical cell identity is dependent on the mobile transcription factor SHORT ROOT (SHR), which originates from the stele (Nakajima et al., 2001). Similarly, root hair or non-root hair cell fate in the Arabidopsis root epidermis is dependent on the mobile transcription factors CAPRICE (CPC) and GLABRA3 (GL3) (Wada et al., 2002; Bernhardt et al., 2005; Kurata et al., 2005). Savage et al. (2008) recently employed a mathematical model to simulate the dynamic behaviour of the network components regulating root hair patterning. Modelling revealed that the currently accepted model for root hair patterning involving local activation of the MYB transcription factor WEREWOLF (WER) through its auto-regulation (summarized in Fig. 3A of Savage et al., 2008) cannot account for all of the experimental evidence available. Instead, epidermal cell fate appears to be dependent on the active exchange of GL3 and CPC proteins between cell files, underpinning a mechanism based on mutual support rather than on local activation and lateral inhibition (summarized in Fig. 3B of Savage et al., 2008). Such modelling approaches could be applied to study other trans-cellular signalling components and networks such as the SHR-regulated radial patterning of root tissues to probe their mechanistic basis.
Beyond Arabidopsis: Understanding Root Biology in Other Plants
The Arabidopsis genome sequence provides a foundation to compare mechanistic aspects of root biology in this reference plant to that of other plants. Like many dicotyledonous plants, Arabidopsis has a primary root that repeatedly branches to generate several orders of lateral roots, whereas the root systems of cereal crops such as rice and maize have substantially greater complexity (Hochholdinger et al., 2004; Osmont et al., 2007). Comparative studies are likely to define core mechanisms shared by all plant roots as well as developmental pathways unique to particular plant lineages. At present, given the rudimentary understanding of root development and genomic information in other plants, there are only a handful of examples of this approach. Recent genetic studies in rice (Oryza sativa) and maize (Zea mays) have identified several genes that control root branching that are shared with Arabidopsis (reviewed by Hochholdinger and Zimmermann, 2008). For example, LATERAL ORGAN BOUNDARIES-DOMAIN16/ASYMMETRIC LEAVES2-LIKE18 (LBD16/ASL18) and LBD29/ASL16 (Okushima et al., 2007) and their rice homologue CROWN-ROOTLESS1/ADVENTITIOUS ROOTLESS1 (CRL1/ARL1) (Inukai et al., 2005; Liu et al., 2005) regulate lateral and crown root formation, respectively. Both crl1/arl1 and lbd16 lbd29 mutants exhibit auxin-related phenotypes in roots, including reduced lateral root number and root gravitropism, and their gene expression is induced by auxin (Inukai et al., 2005; Liu et al., 2005; Okushima et al., 2007). Hence, the function and regulation of LBD16- and LBD29-related genes appear to be conserved between monocot and dicot root development.
A rich source of information for comparative root developmental analysis is likely to come from comparison of cell-specific transcripts in various plants. A recently published ‘atlas’ of cell type-specific transcriptomes in rice, including several root cell types, offers one of the first opportunities for identifying common and distinct cell type-specific expressed genes in Arabidopsis versus rice (Jiao et al., 2009).
At a more fundamental level, it should be possible to use comparative genomics to explore the evolutionary origin of roots. A recent investigation suggests that microRNA-based regulation of HD-ZIP transcription factor gene expression is crucial for the root–shoot decision during Arabidopsis embryogenesis (Grigg et al., 2009), and an analysis of this pathway in various plant lineages may indicate whether it is principally responsible for the molecular origin of the root.
In addition to uncovering the mechanisms of root development, the genes and associated processes elucidated in these genome-assisted studies have, in some instances, provided novel insights into general aspects of biology of broad interest. For example, analysis of the role of the Arabidopsis RHD3 (Root Hair Defective 3) GTPase in polarized cell growth (Wang et al., 1997; Zheng et al., 2004) has provided clues to the role of a related GTPase involved in a common form of hereditary spastic paraplegia (SPG3A) in humans. The putative RHD3 orthologue in humans (known as Atlastin-1) is required for proper ER dynamics during directional cell expansion in neurons, and this finding suggests a fascinating conservation in cellular growth regulation in plant root hairs and mammalian neurons (Hu et al., 2009).
Conclusions and Future Directions
Our understanding of root biology has advanced dramatically in the last 10 years, thanks in large part to completion of the Arabidopsis genome and the experimental tools and resources that have resulted from this key event.
Mutant studies continue to play a key role in dissecting the molecular mechanisms regulating Arabidopsis root development. Nevertheless, reverse genetic screens are becoming increasingly important as a result of the availability of transcriptome datasets for individual root cell types and developmental zones (Laplaze et al., 2005; Birnbaum, 2003; Brady, 2007). Such transcriptomic resources have proved particularly useful for studies of lateral root development. For example, new genes regulating lateral root initiation and emergence processes have been identified in expression datasets for xylem pole pericycle cells (DeSmet et al., 2008) and the outer tissues (Laskowski et al., 2006; Swarup et al., 2008), respectively. Nevertheless, there is an increasing need for conditional approaches to aid the discovery of novel genes in order to overcome issues relating to embryo lethality or genetic redundancy, such as inducible RNAi and chemical genetic approaches (Lehar et al., 2008). Quantitative and association genetic approaches are set to play an increasingly important role in the study of root biology as genome sequences for hundreds of Arabidopsis accessions become available in the coming months and years. These genome resources will enable researchers to tap this rich seam of allelic variation much more efficiently. A comprehensive protein–protein interaction map would be another valuable resource that would allow the identification of network connections.
During the last several years, the field has moved beyond studying the functions of individual gene products, focusing instead on determining the functional relationships between the multiple components that constitute macromolecular complexes and/or regulatory pathways that control root biology. Computer and mathematical modelling approaches are set to become much more important as our understanding of networks becomes increasingly complex and their behaviour and outputs less intuitive. Nevertheless, the cutting-edge technologies employed to address these new and challenging questions continue to rely heavily on the original Arabidopsis genome information. For example, the output of chromatin immunoprecipitation sequencing (ChIP-Seq), a new technique that is revolutionizing identification of the nuclear targets for transcription factors (Yant et al., 2009), remains reliant on the original Arabidopsis genome sequence.
In summary, during the last decade, the Arabidopsis genome sequence has, is and will continue to underpin many of the key scientific breakthroughs and technical innovations in root biology. We wish the Arabidopsis genome sequence a very happy tenth birthday!
Acknowledgments
We apologize to our colleagues for having to omit many outstanding research findings in root biology due to lack of space. We thank Benjamin Peret (CPIB, University of Nottingham) for preparing Figure 1. Work on root biology in the Benfey lab is funded by grants from the US National Science Foundation, the National Institutes of Health, and the Defense Advanced Research Projects Agency. Root research in the Schiefelbein laboratory is funded by the US National Science Foundation. Root research in the Bennett laboratory is funded by grants from the Biotechnology and Biological Sciences Research Council/Engineering and Physical Sciences Research Council Centres for Integrative Systems Biology initiative and a Biotechnology and Biological Sciences Research Council Professorial Research Fellowship.
References
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell. 2004;119:109–120. doi: 10.1016/j.cell.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Armengaud P, Zambaux K, Hills A, Sulpice R, Pattison RJ, Blatt MR, Amtmann A. Ez-Rhizo: integrated software for the fast and accurate measurement of root system architecture. Plant J. 2008;57:945–956. doi: 10.1111/j.1365-313X.2008.03739.x. [DOI] [PubMed] [Google Scholar]
- Baxter I, Muthukumar B, Park HC, et al. Variation in molybdenum content across broadly distributed populations of Arabidopsis thaliana is controlled by a mitochondrial molybdenum transporter (MOT1) PLoS Genet. 2008;4(2):e1000004. doi: 10.1371/journal.pgen.1000004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter I, Hosmani PS, Rus A, Lahner B, Borevitz JO, Muthukumar B, Micklebart MV, Schreiber L, Franke RB, Salt DE. Root suberin forms an extracellular barrier that affects water relations and mineral nutrition in Arabidopsis. PLoS Genet. 2009;5(5):e1000492. doi: 10.1371/journal.pgen.1000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey PN, Schiefelbein JW. Getting to the root of plant development: the genetics of Arabidopsis root formation. Trends Genet. 1994;10:84–88. doi: 10.1016/0168-9525(94)90230-5. [DOI] [PubMed] [Google Scholar]
- Bernhardt C, Zhao M, Gonzalez A, Lloyd A, Schiefelbein J. The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development. 2005;132:291–298. doi: 10.1242/dev.01565. [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN. A gene expression map of the Arabidopsis root. Science. 2003;302:1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. A high-resolution root spatio-temporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- Chen ZH, Nimmo GA, Jenkins GI, Nimmo HG. BHLH32 modulates several biochemical and morphological processes that respond to Pi starvation in Arabidopsis. Biochem J. 2007;405:191–198. doi: 10.1042/BJ20070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RM, Schweikert G, Toomajian C, et al. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science. 2007;317:338–342. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- DeSmet I, Vassileva V, DeRybel B, et al. Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science. 2008;322:594–597. doi: 10.1126/science.1160158. [DOI] [PubMed] [Google Scholar]
- Devaiah BN, Karthikeyan AS, Raghothama KG. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 2007a;143:1789–1801. doi: 10.1104/pp.106.093971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Nagarajan VK, Raghothama KG. Phosphate homeostasis and root development in Arabidopsis are synchronized by the zinc finger transcription factor ZAT6. Plant Physiol. 2007b;145:147–159. doi: 10.1104/pp.107.101691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, Barron C, Brady SM, Schiefelbein J, Benfey PN. Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science. 2008;320:942–945. doi: 10.1126/science.1153795. [DOI] [PubMed] [Google Scholar]
- Dolan L, Roberts K. Plant development: pulled up by the roots. Curr Opin Genet Dev. 1995;5:432–438. doi: 10.1016/0959-437x(95)90045-i. [DOI] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature. 2007;449:1053–1057. doi: 10.1038/nature06206. [DOI] [PubMed] [Google Scholar]
- Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD. Cell-specific nitrogen responses mediate developmental plasticity. Proc Natl Acad Sci USA. 2008;105:803–808. doi: 10.1073/pnas.0709559105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieneisen VA, Xu J, Maree AFM, Hogeweg P, Scheres B. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature. 2007;449:1008–1013. doi: 10.1038/nature06215. [DOI] [PubMed] [Google Scholar]
- Grigg SP, Galinha C, Kornet N, Canales C, Scheres B, Tsiantis M. Repression of apical homeobox genes is required for embryonic root development in Arabidopsis. Curr Biol. 2009;19:1485–1490. doi: 10.1016/j.cub.2009.06.070. [DOI] [PubMed] [Google Scholar]
- Guimil S, Dunand C. Cell growth and differentiation in Arabidopsis epidermal cells. J Exp Bot. 2007;58:3829–3840. doi: 10.1093/jxb/erm253. [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Zimmermann R. Conserved and diverse mechanisms in root development. Curr Opin Plant Biol. 2008;11:70–74. doi: 10.1016/j.pbi.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Park WJ, Sauer M, Woll K. From weeds to crops: genetic analysis of root development in cereals. Trends Plant Sci. 2004;9:42–48. doi: 10.1016/j.tplants.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Hu J, Shibata Y, Zhu PP, Voss C, Rismanchi N, Prinz WA, Rapoport TA, Blackstone C. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell. 2009;138:549–561. doi: 10.1016/j.cell.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Yang S, Zhang S, et al. The Arabidopsis SUMO E3 ligase AtMMS21, a homologue of NSE2/MMS21, regulates cell proliferation in the root. Plant J. 2009;60:666–678. doi: 10.1111/j.1365-313X.2009.03992.x. [DOI] [PubMed] [Google Scholar]
- Inukai Y, Sakamoto T, Ueguchi-Tanaka M, Shibata Y, Gomi K, Umemura I, Hasegawa Y, Ashikari M, Kitano H, Matsuoka M. Crown rootless1, which is essential for crown root formation in rice, is a target of an auxin response factor in auxin signaling. Plant Cell. 2005;17:1387–1396. doi: 10.1105/tpc.105.030981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, Kurata T, Okada K, Wada T. A genetic regulatory network in the development of trichomes and root hairs. Annu Rev Plant Biol. 2008;59:365–386. doi: 10.1146/annurev.arplant.59.032607.092949. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Tausta SL, Gandotra N, et al. A transcriptome atlas of rice cell types uncovers cellular, functional and developmental hierarchies. Nat Genet. 2009;41:258–263. doi: 10.1038/ng.282. [DOI] [PubMed] [Google Scholar]
- Jones MA, Raymond MJ, Smirnoff N. Analysis of the root-hair morphogenesis transcriptome reveals the molecular identity of six genes with roles in root-hair development in Arabidopsis. Plant J. 2006;45:83–100. doi: 10.1111/j.1365-313X.2005.02609.x. [DOI] [PubMed] [Google Scholar]
- Jones AR, Kramer EM, Knox K, Swarup R, Bennett MJ, Lazarus CM, Leyser HMO, Grierson CS. Auxin transport through non-hair cells sustains root-hair development. Nat Cell Biol. 2008;11:78–84. doi: 10.1038/ncb1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerk NM, Ceserani T, Tausta SL, Sussex IM, Nelson TM. Laser capture microdissection of cells from plant tissues. Plant Physiol. 2003;132:27–35. doi: 10.1104/pp.102.018127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshino-Kimura Y, Wada T, Tachibana T, Tsugeki R, Ishiguro S, Okada K. Regulation of CAPRICE transcription by MYB proteins for root epidermis differentiation in Arabidopsis. Plant Cell Physiol. 2005;46:817–826. doi: 10.1093/pcp/pci096. [DOI] [PubMed] [Google Scholar]
- Kurata T, Ishida T, Kawabata-Awai C, et al. Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development. 2005;132:5387–5398. doi: 10.1242/dev.02139. [DOI] [PubMed] [Google Scholar]
- Lahner B, Gong J, Mahmoudian M, et al. Genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana. Nat Biotechnol. 2003;21:1215–1221. doi: 10.1038/nbt865. [DOI] [PubMed] [Google Scholar]
- Laplaze L, Parizot B, Baker A, Ricaud L, Martiniere A, Auguy F, Franche C, Nussaume L, Bogusz D, Haseloff J. GAL4–GFP enhancer trap lines for genetic manipulation of lateral root development in Arabidopsis thaliana. J Exp Bot. 2005;56:2433–2442. doi: 10.1093/jxb/eri236. [DOI] [PubMed] [Google Scholar]
- Laskowski M, Biller S, Stanley K, Kajstura T, Prusty R. Expression profiling of auxin treated Arabidopsis roots: toward a molecular analysis of lateral root emergence. Plant Cell Physiol. 2006;47:778–792. doi: 10.1093/pcp/pcj043. [DOI] [PubMed] [Google Scholar]
- Laskowski M, Grieneisen VA, Hofhuis H, TenHove CA, Hogeweg P, Marer AFM, Scheres B. Root system architecture from coupling cell shape to auxin transport. PLoS Biol. 2008;6:e307. doi: 10.1371/journal.pbio.0060307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehar J, Stockwell B, Giaever G, Nislow C. Combination chemical genetics. Nat Chem Biol. 2008;4:674–681. doi: 10.1038/nchembio.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque MP, Vernoux T, Busch W, et al. Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol. 2006;4:e143. doi: 10.1371/journal.pbio.0040143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little DY, Rao H, Oliva S, Daniel-Vedele F, Krapp A, Malamy JE. The putative high-affinity nitrate transporter NRT2.1 represses lateral root initiation in response to nutritional cues. Proc Natl Acad Sci USA. 2005;102:13693–13698. doi: 10.1073/pnas.0504219102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang S, Yu X, Yu J, He X, Zhang S, Shou H, Wu P. ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J. 2005;43:47–56. doi: 10.1111/j.1365-313X.2005.02434.x. [DOI] [PubMed] [Google Scholar]
- Middleton AM, King JR, Bennett MJ, Owen MR. Mathematical modelling of the Aux/IAA negative feedback loop. Bull Math Biol. 2009 doi: 10.1007/s11538-009-9497-4. [DOI] [PubMed] [Google Scholar]
- Misson J, Raghothama KG, Jain A, et al. A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA. 2005;102:11934–11939. doi: 10.1073/pnas.0505266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchel CF, Osmont KS, Hardtke CS. BRX mediates feedback between brassinosteroid levels and auxin signalling in root growth. Nature. 2006;443:458–461. doi: 10.1038/nature05130. [DOI] [PubMed] [Google Scholar]
- Nakajima K, Sena G, Nawy T, Benfey PN. Intercellular movement of the putative transcription factor SHR in root patterning. Nature. 2001;413:307–311. doi: 10.1038/35095061. [DOI] [PubMed] [Google Scholar]
- Nawy T, Lee JY, Colinas J, Wang JY, Thongrod SC, Malamy JE, Birnbaum K, Benfey PN. Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell. 2005;17:1908–1925. doi: 10.1105/tpc.105.031724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M, Hu TT, Ishino Y, et al. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 2005;3:e196. doi: 10.1371/journal.pbio.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell. 2007;19:118–130. doi: 10.1105/tpc.106.047761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva M, Dunand C. Waving and skewing: how gravity and the surface of growth media affect root development in Arabidopsis. New Phytol. 2007;176:37–43. doi: 10.1111/j.1469-8137.2007.02184.x. [DOI] [PubMed] [Google Scholar]
- Osmont KS, Sibout R, Hardtke CS. Hidden branches: developments in root system architecture. Annu Rev Plant Biol. 2007;58:93–113. doi: 10.1146/annurev.arplant.58.032806.104006. [DOI] [PubMed] [Google Scholar]
- Petricka JJ, Benfey PN. Root layers: complex regulation of developmental patterning. Curr Opin Genet Dev. 2008;18:354–361. doi: 10.1016/j.gde.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu KH, Kang YH, Park YH, Hwang I, Schiefelbein J, Lee MM. The WEREWOLF MYB protein directly regulates CAPRICE transcription during cell fate specification in the Arabidopsis root epidermis. Development. 2005;132:4765–4775. doi: 10.1242/dev.02055. [DOI] [PubMed] [Google Scholar]
- Savage N, Walker T, Wieckowski T, Schiefelbein J, Dolan L, Monk NAM. A mutual support mechanism through intercellular movement of CAPRICE and GLABRA3 can pattern the Arabidopsis root epidermis. PLoS Biol. 2008;6:e235. doi: 10.1371/journal.pbio.0060235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scacchi E, Osmont KS, Beuchat J, Salinas P, Navarrete-Gómez M, Trigueros M, Ferrándiz C, Hardtke CS. Dynamic, auxin-responsive plasma membrane-to-nucleus movement of Arabidopsis BRX. Development. 2009;136:2059–2067. doi: 10.1242/dev.035444. [DOI] [PubMed] [Google Scholar]
- Schmid KJ, Sorenson TR, Stracke R, Torjek O, Altmann T, Mitchell-Olds T, Weisshaar B. Large scale identification of genome wide single nucleotide polymorphisms for mapping in Arabidopsis thaliana. Genome Res. 2003;13:1250–1257. doi: 10.1101/gr.728603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeeva LI, Keurentjes JJB, Bentsink L, Vonk J, Van der Plas LW, Koornneef M, Vreughenhil D. Vacuolar invertase regulates elongation of Arabidopsis thaliana roots as revealed by QTL and mutant analysis. Proc Natl Acad Sci USA. 2006;103:2994–2999. doi: 10.1073/pnas.0511015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P, Dolan L. Chromatin and Arabidopsis root development. Semin Cell Dev Biol. 2008;19:580–585. doi: 10.1016/j.semcdb.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Simon M, Lee MM, Lin Y, Gish L, Schiefelbein J. Distinct and overlapping roles of single-repeat MYB genes in root epidermal patterning. Dev Biol. 2007;311:566–578. doi: 10.1016/j.ydbio.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Stahl Y, Wink RH, Ingram GC, Simon R. A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr Biol. 2009;19:909–914. doi: 10.1016/j.cub.2009.03.060. [DOI] [PubMed] [Google Scholar]
- Svistoonoff S, Creff A, Reymond M, Sigoillot-Claude C, Ricaud L, Blanchet A, Nussaume L, Desnos T. Root tip contact with low-phosphate media reprograms plant root architecture. Nat Genet. 2007;39:792–796. doi: 10.1038/ng2041. [DOI] [PubMed] [Google Scholar]
- Swarup R, Kramer EM, Perry P, Knox K, Leyser HMO, Haseloff J, Beemster GTS, Bhalerao R, Bennett MJ. Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat Cell Biol. 2005;7:1057–1065. doi: 10.1038/ncb1316. [DOI] [PubMed] [Google Scholar]
- Swarup K, Benkova E, Swarup R, et al. The auxin influx carrier LAX3 promotes lateral root emergence. Nat Cell Biol. 2008;10:946–954. doi: 10.1038/ncb1754. [DOI] [PubMed] [Google Scholar]
- Tominaga R, Iwata M, Sano R, Inoue K, Okada K, Wada T. Arabidopsis CAPRICE-LIKE MYB 3 (CPL3) controls endoreduplication and flowering development in addition to trichome and root hair formation. Development. 2008;135:1335–1345. doi: 10.1242/dev.017947. [DOI] [PubMed] [Google Scholar]
- Tominaga-Wada R, Iwata M, Sugiyama J, Kotake T, Ishida T, Yokoyama R, Nishitani K, Okada K, Wada T. The GLABRA2 homeodomain protein directly regulates CESA5 and XTH17 gene expression in Arabidopsis roots. Plant J. 2009;60:564–574. doi: 10.1111/j.1365-313X.2009.03976.x. [DOI] [PubMed] [Google Scholar]
- Wada T, Kurata T, Tominaga R, Koshino-Kimura Y, Tachibana T, Goto K, Marks MD, Shimura Y, Okada K. Role of a positive regulator of root hair development, CAPRICE, in Arabidopsis root epidermal cell differentiation. Development. 2002;129:5409–5419. doi: 10.1242/dev.00111. [DOI] [PubMed] [Google Scholar]
- Wang H, Lockwood SK, Hoeltzel MF, Schiefelbein JW. The ROOT HAIR DEFECTIVE3 gene encodes an evolutionarily conserved protein with GTP-binding motifs and is required for regulated cell enlargement in Arabidopsis. Genes Dev. 1997;11:799–811. doi: 10.1101/gad.11.6.799. [DOI] [PubMed] [Google Scholar]
- Wildwater M, Campilho A, Perez-Perez JM, Heidstra R, Blilou I, Korthout H, Chatterjee J, Mariconti L, Gruissem W, Scheres B. The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell. 2005;123:1337–1349. doi: 10.1016/j.cell.2005.09.042. [DOI] [PubMed] [Google Scholar]
- Willemsen V, Bauch M, Bennett T, Campilho A, Wolkenfelt H, Xu J, Haseloff J, Scheres B. The NAC domain transcription factors FEZ and SOMBRERO control the orientation of cell division plane in Arabidopsis root stem cells. Dev Cell. 2008;15:913–922. doi: 10.1016/j.devcel.2008.09.019. [DOI] [PubMed] [Google Scholar]
- Won SK, Lee YJ, Lee HY, Heo YK, Cho M, Cho HT. Cis-element- and transcriptome-based screening of root hair-specific genes and their functional characterization in Arabidopsis. Plant Physiol. 2009;150:1459–1473. doi: 10.1104/pp.109.140905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant L, Mathieu J, Schmid M. Just say no: floral repressors help Arabidopsis bide the time. Curr Opin Plant Biol. 2009;12:580–586. doi: 10.1016/j.pbi.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Zanetti ME, Chang IF, Gong F, Galbraith DW, Bailey-Serres J. Immunopurification of polyribosomal complexes of Arabidopsis for global analysis of gene expression. Plant Physiol. 2005;138:624–635. doi: 10.1104/pp.105.059477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Kunst L, Hawes C, Moore I. A GFP-based assay reveals a role for RHD3 in transport between the endoplasmic reticulum and Golgi apparatus. Plant J. 2004;37:398–414. doi: 10.1046/j.1365-313x.2003.01969.x. [DOI] [PubMed] [Google Scholar]


