Abstract
Long intergenic noncoding RNAs (lincRNAs) regulate chromatin states and epigenetic inheritance. Here we show that the lincRNA HOTAIR serves as a scaffold for at least two distinct histone modification complexes. A 5′ domain of HOTAIR binds Polycomb Repressive Complex 2 (PRC2) while a 3′ domain of HOTAIR binds the LSD1/CoREST/REST complex. The ability to tether two distinct complexes enables RNA-mediated assembly of PRC2 and LSD1, and coordinates targeting of PRC2 and LSD1 to chromatin for coupled histone H3 lysine 27 methylation and lysine 4 demethylation. Our results suggest that lincRNAs may serve as scaffolds by providing binding surfaces to assemble select histone modification enzymes, and thereby specify the pattern of histone modifications on target genes.
Long intergenic noncoding RNAs (lincRNAs) regulate dosage compensation, imprinting, and developmental gene expression by establishing chromatin domains in an allele- and cell-type specific manner [reviewed by (1, 2)]. LincRNAs are intimately associated with chromatin remodeling complexes (3–7), but molecular mechanisms of their functions are still lacking. Post-translational modifications of histones recruit DNA binding proteins and chromatin remodeling machinery, and are often coupled for combinatorial control [reviewed by (8)]. For instance in embryonic stem cells, many genes encoding developmental regulators, such as the HOX, are transcriptionally silent but possess bivalent histone H3 lysine 4 (H3K4) and lysine 27 (H3K27) methylation, which are resolved into univalent H3K4 or H3K27 methylation domains upon differentiation (9, 10). Here we show that a lincRNA can coordinate histone modifications by binding to multiple histone modification enzymes.
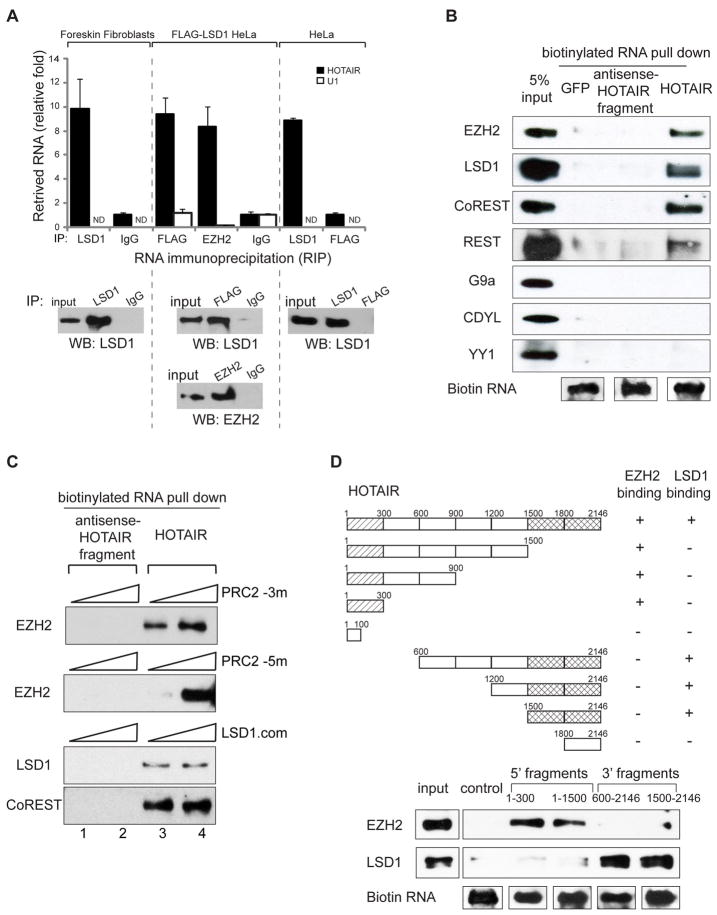
The lincRNA HOTAIR is transcribed from the HOXC locus and targets Polycomb Repressive Complex 2 (PRC2, comprised of H3K27 methylase EZH2, SUZ12, and EED) to silence HOXD and select genes on other chromosomes (7, 11). The genomic regions flanking HOXD are also bound by CoREST/REST repressor complexes (12), which contain LSD1 (KDM1/BHC110), a demethylase that mediates enzymatic demethylation of H3K4me2 (13) and that is required for proper repression of Hox genes in Drosophila (14). We therefore hypothesized that HOTAIR may coordinately interact with both PRC2 and LSD1. Immunoprecipitation (IP) of either endogenous LSD1 or FLAG-tagged LSD1 from primary foreskin fibroblasts or HeLa cells specifically retrieved endogenous HOTAIR RNA with comparable enrichment to EZH2 IP, the positive control (Fig. 1A and fig. S1A). IP of three other chromatin proteins did not retrieve HOTAIR (fig. S1A), and LSD1, EZH2, or FLAG-LSD1 IP did not retrieve U1 RNA, a nuclear ncRNA that served as a negative control. Purified biotinylated HOTAIR RNA, but not GFP RNA or an antisense HOTAIR fragment, specifically retrieved EZH2, SUZ12, and LSD1 from HeLa cell nuclear extract (Fig. 1B and fig. S1B). LSD1 forms a complex with CoREST (15), which can bridge LSD1 to the neuronal gene silencer REST (16). REST is believed to mediate silencing through two distinct effector arms: one via LSD1-CoREST, and separately via the adaptor protein CDYL and the H3K9 KMT G9a (17). HOTAIR specifically bound to CoREST and REST, but not CDYL or G9a, nor to the putative PRC1 subunit YY1 (Fig. 1B). Further, biotinylated HOTAIR bound to purified PRC2 and LSD1 complexes in vitro (Fig. 1C and fig. S1C). These results suggest that HOTAIR directly interacts with PRC2 and LSD1 complexes.
Fig. 1.
5′ domain of HOTAIR binds PRC2 and 3′ domain of HOTAIR binds LSD1. (A) LSD1 IP specifically retrieves HOTAIR RNA. Data (mean ± SD, n = 3) is relative to mock-IP (IgG or FLAG). ND, not-detectable. (B) In vitro transcribed (IVT), biotinylated HOTAIR retrieves EZH2, LSD1, CoREST, and REST, but not G9a, CDYL, or YY1. (C) IVT biotinylated HOTAIR binds to purified PRC2 and LSD1 complexes. PRC2_3m: recombinant purified core PRC2 complex with 3 members (EZH2, SUZ12, EED). PRC2_5m: recombinant purified PRC2 complex with 5 members (+RbAP48, AEBP2). LSD1.com: Tandem affinity purified protein complex associated with FLAG-HA-LSD1 from HeLa cells. Composition of protein complexes are shown in fig. S1C. (D)The first 300 bp (lined boxes) of HOTAIR is necessary and sufficient to bind PRC2; the last 646 bp (meshed boxes) is necessary and sufficient to bind LSD1 complex. The profiles are established by RNA pull-down of HeLa extract; retrieved proteins are detected by immunoblotting.
Using a series of HOTAIR deletion mutants, the PRC2 binding activity mapped to nucleotides 1 to 300 of HOTAIR, while the LSD1 complex binding activity mapped to nucleotides 1500 to 2146 (Fig. 1D). Deletion mutants that retained nucleotides 1 to 300 bound EZH2 or SUZ12 with equal efficiency as full length HOTAIR, and deletion mutants that retained nucleotides 1500 to 2146 retained LSD1 binding activity. Thus, HOTAIR is a modular bifunctional RNA that has distinct binding domains for PRC2 and LSD1 complexes. Computational analysis and RNA footprinting showed that the PRC2 and LSD1 binding domains of HOTAIR are likely to possess extensive but distinct secondary structures (fig. S2).
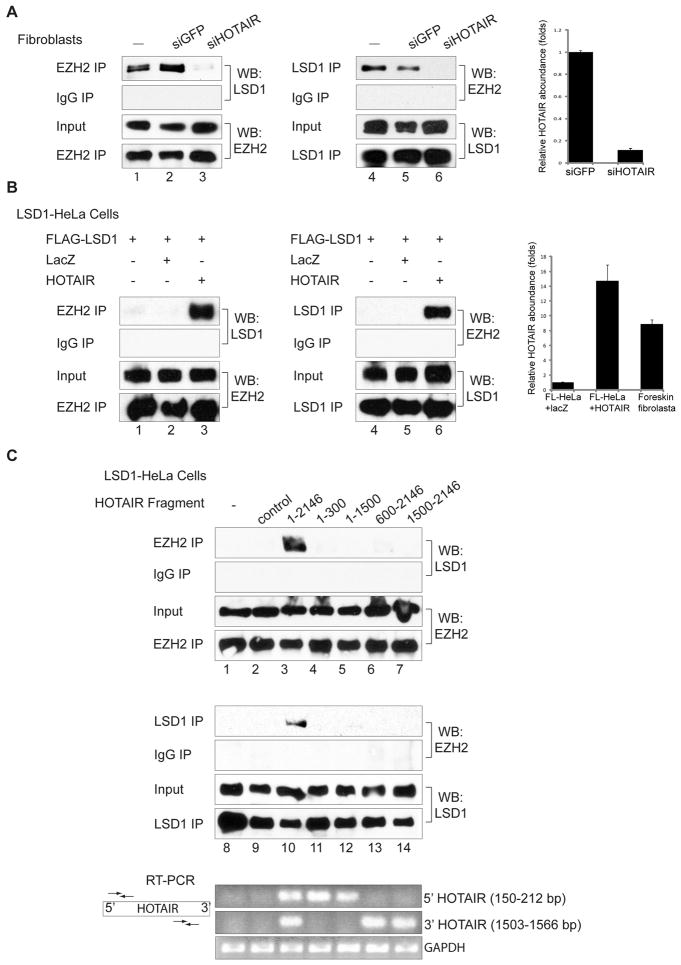
The presence of independent binding sites for PCR2 and LSD1 on HOTAIR suggests that HOTAIR may bridge PRC2 and LSD1 complexes. EZH2 IP retrieved LSD1, and conversely LSD1 IP retrieved EZH2 from foreskin fibroblasts (Fig. 2A). We estimate that less than 5% of the two complexes physically interact with each other, consistent with prior purification results that isolated PRC2 and CoREST-LSD1 as separate complexes (18, 19). RNAi of HOTAIR or RNase treatment of the IP abrogated the interaction between EZH2 and LSD1, suggesting that HOTAIR is required to bridge this interaction (Fig. 2A and fig. S3). Wild-type HeLa cells or HeLa cells stably expressing FLAG-LSD1 (FL-HeLa) expressed ~ten-fold less HOTAIR than foreskin fibroblasts and showed undetectable endogenous interaction between PRC2 and LSD1. Enforced expression of HOTAIR in FL-HeLa cells to a level comparable to foreskin fibroblasts allowed robust interaction between PRC2 and LSD1 (Fig. 2B). Gel filtration chromatography confirmed that HOTAIR expression shifts PRC2 subunits into a higher molecular weight complex coincident with the LSD1 complex, suggesting the formation of a higher ordered complex comprised of HOTAIR, PRC2, and LSD1 complexes in HOTAIR overexpressing cells (fig. S4). Moreover, expression of each HOTAIR mutant that lacked the ability to bind either PRC2 or LSD1 in vitro failed to induce PRC2-LSD1 interaction in cells (Fig. 2C and fig. S3C).
Fig. 2.
HOTAIR is necessary and sufficient for interaction between EZH2 and LSD1. (A) In foreskin fibroblasts, EZH2 interacts with LSD1 (lanes 1, 4). Knockdown of HOTAIR (lanes 3, 6), but not GFP (lanes 2, 5), abolishes this interaction. HOTAIR levels (mean ± SD) are shown on the right. (B) HOTAIR expression in FLAG-LSD1 HeLa cells induces EZH2 and LSD1 interaction (lanes 3 and 6). (C) Full length HOTAIR induces EZH2 and LSD1 interaction (lanes 3, 10) but not HOTAIR mutants lacking either 5′ or 3′ domain (lanes 4 to 7 and 11 to 14). Presence of indicated RNA domains is confirmed by RT-PCR (bottom panel).
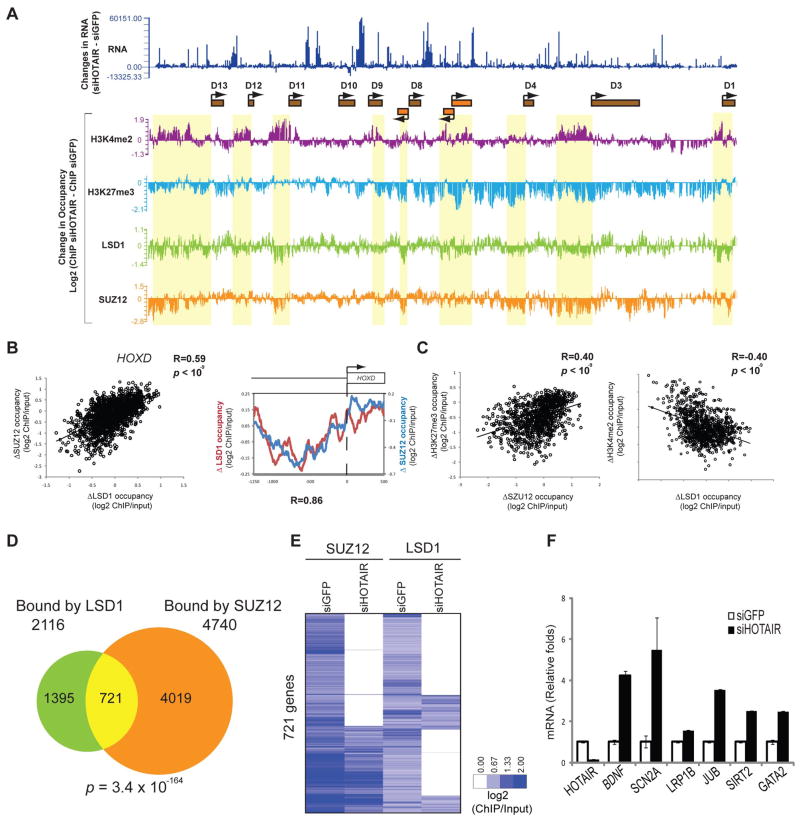
HOTAIR-mediated bridging of PRC2 and LSD1 complexes also enables their coordinate binding to target genes on chromatin. HOTAIR is required for H3K27 methylation and transcriptional silencing across the HOXD locus (7). Therefore, we mapped PRC2 (as indicated by SUZ12) and LSD1 occupancy across the HOX loci and on promoters genome-wide by chromatin IP followed by microarray analysis (ChIP-chip), in primary foreskin fibroblasts after control RNAi or HOTAIR knock down (Fig. 3 and figs. S5 to S7). HOTAIR knockdown decreased SUZ12 and LSD1 occupancy in a similar pattern across HOXD (Fig. 3, A and B, and fig. S6; R = 0.59, p < 10−9, t-test). Coordinate loss of SUZ12 and LSD1 occupancy caused by HOTAIR knockdown were concentrated in proximal promoters of HOXD genes (Fig. 3B). These regions correspondingly lost H3K27me3 and gained H3K4me2, the respective histone methylation products of PRC2 and LSD1 complexes (Fig. 3, A and C, and figs. S6 and S7; R = 0.40, p < 10−9, t-test). The loss of H3K27me3 occurred across broad domains encompassing multiple HOXD genes and intergenic regions, while the gain of H3K4me2 was concentrated near the transcriptional start sites of HOXD genes (8). Multiple independent siRNAs targeting HOTAIR gave the same results.
Fig. 3.
HOTAIR coordinates localization of PRC2 and LSD1 genome-wide. (A) Changes in mRNA and occupancy of H3K4me2, H3K27me3, LSD1, and SUZ12 across HOXD locus after RNAi of HOTAIR in foreskin fibroblasts. Yellow boxes indicate regions of notable correlation between gain of H3K4me2 and concordant loss of LSD1, H3K27me3, and SUZ12. (B) The patterns of change in LSD1 (x axis) and SUZ12 occupancy (y axis) upon HOTAIR knockdown across the HOXD locus are significantly correlated (Pearson correlation, R = 0.59, p < 10−9, t-test). This correlation is concentrated in proximal promoters of HOXD genes (R = 0.86). (C) Positive correlation of changes in SUZ12 (x axis) and H3K27me3 occupancy (y axis) and negative correlation of LSD1 (x axis) and H3K4me2 occupancy (y axis). (D) Venn diagram shows the genes occupied by SUZ12 (4740 genes), LSD1 (2116 genes), or both (721 genes). (E) Heatmap of SUZ12 and LSD1 co-occupied genes (721 genes). Each column is an experiment; each row is a gene. HOTAIR knockdown led to concordant loss of SUZ12 and LSD1 occupancy. Chromatin occupancy is indicated in blue per the scale bar. (F) HOTAIR knockdown leads to transcription de-repression of target genes. Mean ± SD of qRT-PCR data are shown.
Examining human promoters genome-wide, ChIP-chip analysis showed that PRC2 and LSD1 occupied 4740 and 2116 gene promoters, respectively (Fig. 3D). Nearly one third of LSD1 occupied promoters, comprised of 721 genes, were also occupied by SUZ12, revealing a significant overlap (257 overlap expected by chance alone. p = 3.4 × 10−164, hypergeometric distribution). Among these 721 genes co-occupied by SUZ12 and LSD1, the distances between the binding sites of SUZ12 and LSD1 were predominantly less than 500 base pairs, which is the fragmentation size of chromatin in our ChIP assay and the limit of resolution (fig. S8A).
HOTAIR knockdown led to concordant loss of SUZ12 and LSD1 occupancy in 289 of the 721 genes normally co-occupied by SUZ12 and LSD1 (almost 40%) (Fig. 3E and table S1). Additional genes showed more exclusive loss of LSD1 occupancy (33%) or SUZ12 occupancy (16%), suggesting that HOTAIR may be involved in other LSD1- or SUZ12-dependent pathways. ChIP followed by qPCR confirmed the requirement of HOTAIR for PRC2 and LSD1 localization for all six genes tested (fig. S8C). HOTAIR knockdown did not change the chromatin occupancy by PRC2 and LSD1 at hundreds of other genes, nor did it affect the protein or mRNA level of the subunits of PRC2 or LSD1 complexes (Fig. 2A and fig. S9, A to C). The functional consequence of coordinate targeting of PRC2 and LSD1 by HOTAIR is gene repression: genes co-occupied by SUZ12-LSD1 in a HOTAIR-dependent manner are also significantly induced upon HOTAIR knockdown as measured by microarray or qRT-PCR [p < 0.05, Gene Set Enrichment Analysis (20); Fig. 3F and fig. S8D]. These results suggest that a single lincRNA—HOTAIR—may be required to target both PRC2 and LSD1 to hundreds of genes across the genome in order to coordinate histone modifications for gene silencing.
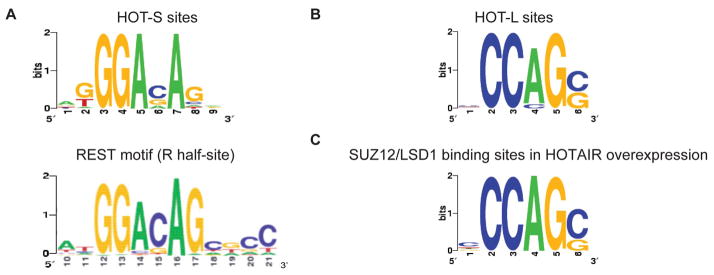
Both PRC2 and LSD1 can bind multiple proteins that are thought to provide DNA target specificity (16, 21). A possible consequence of the HOTAIR-mediated bridging is that PRC2 may be recruited to LSD1-CoREST-REST binding sites, and conversely, LSD1 may be recruited to PRC2 binding sites. Prior genome-scale mapping studies of PRC2 already identified the REST motif as one of the most enriched DNA sequence motifs within PRC2 binding sites but with no mechanistic explanation (22). We searched for enriched sequence motifs in SUZ12 binding sites lost upon HOTAIR knock down (“HOT-S sites” for short) and identified several enriched motifs (23), including a motif that corresponds to the right half of the canonical REST motif (p = 1.05 × 10−12; Fig. 4A and fig. S10). REST is able to bind only one half site of the canonical REST motif (24), and genes containing HOT-S sites are enriched for experimentally measured REST occupancy (p < 1.27 × 10−16, hypergeometric distribution; fig. S9D and table S2) (24). The most significantly enriched motif in LSD1 binding sites that are lost upon HOTAIR knockdown (termed “HOT-L sites”) is a CG-rich motif (p = 3.66 × 10−10; Fig. 4B and fig. S10), which is important for PRC2 binding (22, 25, 26). Thus, the enrichment of the CG-rich motif may reflect the HOTAIR-dependent recruitment of LSD1 complexes to PRC2 bound sites, which are often in CpG islands. We examined the gain of SUZ12 and LSD1 occupancy on chromatin when HOTAIR is overexpressed in primary lung fibroblasts, which do not express endogenous HOTAIR. HOTAIR overexpression caused ectopic occupancy of LSD1 and SUZ12 that significantly overlapped (p < 7.31 × 10−95). Further, motif analysis of the ectopically gained binding sites recovered an almost identical CG-rich motif (p = 7.9 × 10−37; Fig. 4C), suggesting that this motif is involved in HOTAIR target selection. Nonetheless, the REST half site and the CG-rich motif are currently not sufficient for de novo prediction of all HOTAIR-dependent genes, suggesting that additional motifs, binding partners, and/or motif arrangements may be important.
Fig. 4.
HOTAIR-dependent SUZ12 and LSD1 binding motifs. (A) SUZ12 occupancy sites lost upon HOTAIR knockdown (HOT-S sites) are enriched for a DNA motif very similar to the right half of canonical REST motif. (B) LSD1 occupancy sites lost upon HOTAIR knockdown (HOT-L sites) are enriched for a CG-rich motif. (C) A nearly identical CG-rich motif is enriched in LSD1/SUZ12 binding sites gained upon HOTAIR overexpression, suggesting that this motif is involved in HOTAIR target selection.
In this report, we demonstrate that the lincRNA HOTAIR can link a histone methylase and a demethylase by acting as a modular scaffold (fig. S11). Other lincRNAs may also contain multiple binding sites for distinct protein complexes that direct specific combinations of histone modifications on target gene chromatin. Some lincRNAs may be “tethers” that recruit several chromatin modifications to their sites of synthesis (2), while other lincRNAs can act on distantly located genes as “guides” to affect their chromatin states (2). Based on their dynamic patterns of expression (27), specific lincRNAs can potentially direct complex patterns of chromatin states at specific genes in a spatially and temporally organized manner during development and disease states.
Acknowledgments
Microarray data are deposited in Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/) under accession number GSE22345. We thank members of the D. Herschlag lab for assistance with RNA footprinting; X. Tan, P. Khavari, and J. Wysocka for critical reading of the manuscript. Supported by California Institute for Regenerative Medicine (RN1-00529-1 to H.Y.C.), NIH (R01-HG004361 to H.Y.C. and E.S, R01-CA118487 to Y.S), Susan G. Komen Foundation (M.-C.T.), Azrieli Foundation (O.M.), National Science Foundation (J.K.W.), and A-STAR (Y.W.). E.S. is the incumbent of the Soretta and Henry Shapiro career development chair. Y.S. is co-founder and on the scientific advisory board of Constellation Pharmaceuticals. H.Y.C. is an Early Career Scientist of the Howard Hughes Medical Institute.
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/science.1192002/DC1
Materials and Methods
Figs. S1 to S11
Tables S1 and S2
References and Notes
References and Notes
- 1.Ponting CP, Oliver PL, Reik W. Cell. 2009;136:629. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Lee JT. Genes Dev. 2009;23:1831. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khalil AM, et al. Proc Natl Acad Sci USA. 2009;106:11667. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pandey RR, et al. Mol Cell. 2008;32:232. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 5.Nagano T, et al. Science. 2008;322:1717. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 6.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Science. 2008;322:750. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rinn JL, et al. Cell. 2007;129:1311. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rando OJ, Chang HY. Annu Rev Biochem. 2009;78:245. doi: 10.1146/annurev.biochem.78.071107.134639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernstein BE, et al. Cell. 2006;125:315. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 10.Mikkelsen TS, et al. Nature. 2007;448:553. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta RA, et al. Nature. 2010;464:1071. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lunyak VV, et al. Science. 2002;298:1747. doi: 10.1126/science.1076469. [DOI] [PubMed] [Google Scholar]
- 13.Shi Y, et al. Cell. 2004;119:941. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 14.Di Stefano L, Ji JY, Moon NS, Herr A, Dyson N. Curr Biol. 2007;17:808. doi: 10.1016/j.cub.2007.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee MG, Wynder C, Cooch N, Shiekhattar R. Nature. 2005;437:432. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- 16.Ooi L, Wood IC. Nat Rev Genet. 2007;8:544. doi: 10.1038/nrg2100. [DOI] [PubMed] [Google Scholar]
- 17.Mulligan P, et al. Mol Cell. 2008;32:718. doi: 10.1016/j.molcel.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi YJ, et al. Mol Cell. 2005;19:857. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 19.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Genes Dev. 2002;16:2893. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramanian A, et al. Proc Natl Acad Sci USA. 2005;102:15545. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerppola TK. Trends Cell Biol. 2009;19:692. doi: 10.1016/j.tcb.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ku M, et al. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharon E, Lubliner S, Segal E. PLoS Comput Biol. 2008;4:e1000154. doi: 10.1371/journal.pcbi.1000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson DS, Mortazavi A, Myers RM, Wold B. Science. 2007;316:1497. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 25.Peng JC, et al. Cell. 2009;139:1290. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li G, et al. Genes Dev. 2010;24:368. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guttman M, et al. Nature. 2009;458:223. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]