Abstract
The linear “Ising” model, which has been around for nearly a century, treats the behavior of linear arrays of repetitive, interacting subunits. Linear “repeat-proteins” have only been described in the last decade or so, and their folding energies have only been characterized very recently. Owing to their repetitive structures, linear repeat-proteins are particularly well suited for analysis by the nearest-neighbor Ising formalism. After briefly describing the historical origins and applications of the Ising model to biopolymers, and introducing repeat protein structure, this chapter will focus on the application of the linear Ising model to repeat proteins. When applied to homopolymers, the model can be represented and applied in a fairly simplified form. When applied to heteropolymers, where differences in energies among individual subunits (i.e. repeats) must be included, some (but not all) of this simplicity is lost. Derivations of the linear Ising model for both homopolymer and heteropolymer repeat-proteins will be presented. With the increased complexity required for analysis of heteropolymeric repeat proteins, the ability to resolve different energy terms from experimental data can be compromised. Thus, a simple matrix approach will be developed to help inform on the degree to which different thermodynamic parameters can be extracted from a particular set of unfolding curves. Finally, we will describe the application of these models to analyze repeat-protein folding equilibria, focusing on simplified repeat proteins based on “consensus” sequence information.
Keywords: repeat protein, consensus sequence, Ising model, nearest-neighbor, folding
I. Historical overview of Ising models and motivation for the present review
A. Origins
The history of the “Ising” model, or perhaps more appropriately, the Ising-Lenz model, has been described extensively (Brush, 1967; Niss, 2005). Originally developed to study ferromagnetism, the model can be traced to the dissertation of Ernst Ising (Ising, 1925), and to an earlier proposal by Wilhelm Lenz (Lenz, 1920). At the time, Ising was directly connected to Lenz, as Ising carried out his dissertation work on the model under Lenz’s guidance at Hamburg University. Since that time, the model (with which Ising’s name is almost exclusively associated) has been applied to study a wide range of cooperative phenomena in one, two, and three-dimensions, including phase separation in mixtures, phase transitions in single-component systems (the lattice gas model), and cooperative phenomena in linear biopolymers. It seems unfortunate that Ising did not continue in this area, in part because he was discouraged that, in his view, the model could not capture ferromagnetic transitions (Brush, 1967).
B. Application to linear biopolymers
Although the Ising model has been used to describe order-disorder transitions in a wide variety of diverse systems, the one-dimensional Ising model has been particularly useful for conformational transitions in linear polymers. These transitions, which can be categorized as “helix-coil” transitions, include the equilibria between the α-helix- and coil in peptides (Schellman, 1958; Zimm and Bragg, 1959; Lifson and Roig, 1961), and various equilibria of DNA and RNA, including double-helix formation (Zimm, 1960; Crothers and Kallenbach, 1966), and stacking transitions of single strands (Applequist and Damle, 1965; Poland et al., 1966). This literature, along with a very clear development of analytical models, is presented in a beautiful monograph by Poland and Scheraga (Poland and Scheraga, 1970). More recent applications include binding of protein ligands to repetitive structures such as DNA and protein filaments (McGhee and von Hippel, 1974; De La Cruz, 2005).
In this review, we develop aspects of the nearest-neighbor or Ising model in the context of linear repeat proteins, emphasizing key features that are pertinent to recent experimental studies (including heterogeneous, homogeneous, and “capped” structures, see below). We focus both on the theory and on how it can be used to analyze experimental data. It is our aim to provide enough detail so that all steps of the derivation can be followed (from the basic model to the development of the partition function, and then to modeling equilibrium unfolding transitions), while avoiding specific features that apply exclusively to other types of linear biopolymers. In addition, we will include a discussion of some practical issues associated with determining the model-dependent parameters, emphasizing the relationship between these parameters and the data needed for their accurate determination.
II. Linear repeat proteins and their connection to linear Ising models
The structures and global stabilities of linear repeat proteins have been described in a number of reviews (Groves and Barford, 1999; Kobe and Kajava, 2000; Kajava, 2001; Mosavi et al., 2004; Main et al., 2005). The units of repeat proteins are constructed from tandem elements of secondary structure units (α-helix, β-strand, PII helix, turn), arranged in a large loop. The length of individual repeats is approximately 20-40 residues, depending on the type of repeat. Typically, individual repeats show primary sequence similarity, and in most cases repeats were identified by primary sequence before structural details were available. However, some repeats show little or no obvious repetition at the primary sequence level. Even when there is repetition, sequence identity from one repeat to the next is typically around twenty five percent. Thus, although consensus sequences can be identified, sequences of natural repeats differ significantly from the consensus.
Three types of repeat proteins that have been amenable to structural and thermodynamic analysis and simplification through consensus information are ankyrin- (ANK), leucine-rich- (LRR), and tetratricopeptide (TPR) repeat proteins (see (Kloss et al., 2008) for review; also (Courtemanche and Barrick, 2008; Kloss and Barrick, 2008)). TPR and ANK repeats are composed of α-helices and turns, with two short turns connecting the TPR helices, and one short turn and one extended loop connecting the ANK helices. In contrast, LRR proteins contain a β-strand that packs against strands of neighboring repeats to form a contiguous sheet. Depending on the subtype, LRRs contain either an α-helix, a 310 helix, or an extended PPII (Kajava, 2001).
In linear repeat proteins, adjacent repeat units pack against their neighbors in a roughly linear array (Figure 1). Depending on the shape and packing of repeats, different types of repeats typically show regular deviation from linearity (Kobe and Kajava, 2000), displaying twist from repeat to repeat (particularly pronounced for TPRs) and/or curvature along the entire stack (particularly pronounced for some LRR subtypes). For some repeat proteins, such as WD40 domains and TIM barrels, curvature is so extreme that a “closed” or circular structure is formed. Since such closed proteins have numerous sequence-distant interactions, they are not easily analyzed using nearest-neighbor thermodynamic models, and will not be discussed here.
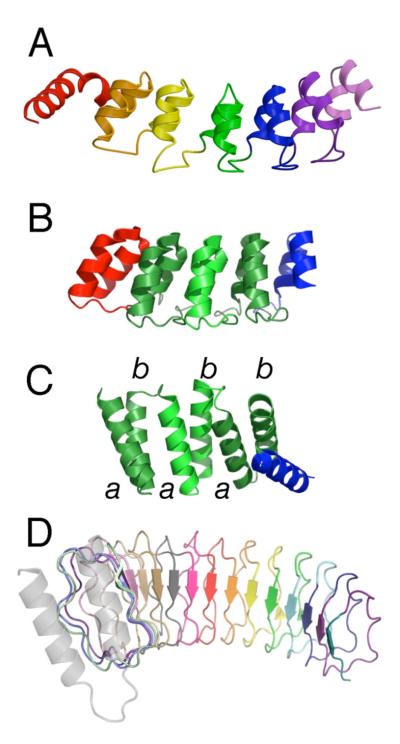
Figure 1. The modular architectures of repeat proteins.
(A) Crystal structure of the Notch ankyrin domain (1ot8.pdb, chain A) consisting of six structured ANK repeats (sequence repeats 2-7) and an N-terminal partly structured repeat. (B) Crystal structure of a consensus ankyrin repeat protein (2qyj.pdb) containing three consensus repeats (green) and N- and C-terminal caps (red and blue respectively). (C) Crystal structure of a consensus-based TPR protein (1na0.pdb, chain A) containing three consensus repeats and a C-terminal cap (blue). (D) Crystal structure of YopM, a leucine-rich repeat protein containing 15 full LRR repeats of the bacterial subtype (15jl.pdb). For naturally occurring (heterogeneous) proteins, individual repeats are shown in different colors; selection of boundaries between repeats (color changes) is somewhat arbitrary, and is based on considerations such as intron position, interresidue contact density, surface area, and visual impression. For the consensus ankyrin and TPR proteins, consensus repeats are shown with the same color but alternate in color saturation. This figure was prepared using PyMol (DeLano, 2003).
Linear repeat proteins have two features that make them ideal subjects for simple nearest-neighbor models. First, as described above, they are constructed of a repeating unit at the level of secondary and tertiary structure; repetition can be extended to the level of primary sequence using consensus information (see below). This translational symmetry reduces the number and type of energy terms required to describe stability, allowing different regions of the molecule to be described in the same way. Second, as can be seen in inter-residue contact maps, direct contacts are limited to repeats that are immediately adjacent in sequence, which justifies using a nearest-neighbor approximation to describe folding.
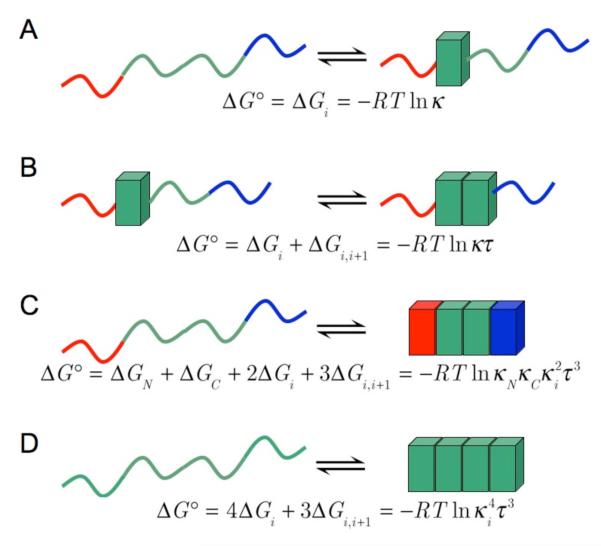
Given this structural simplicity, the free energy of repeat protein folding may be expected to have two dominant contributions: the intrinsic folding of individual units (which we will call ΔGi) and the interfacial interaction of neighboring repeats (ΔGi,i+1; Figure 2). Thermodynamically, the second term is similar to a cooperative term describing short-range interactions in the peptide helix-coil transition (although the statistics are often formulated differently to capture backbone hydrogen bonding between residues i andi+4), and to the stacking interactions in DNA duplex formation. As in these simpler systems, various levels of approximation can be used to analyze unfolding transitions with nearest-neighbor models. Because naturally occurring repeat proteins are quite heterogeneous at the primary sequence level, a homopolymer approach (treating all the repeats as identical) may not be appropriate. However, studies from a number of labs have shown that stable repeat proteins of various types (ANK, TPR, LRR) can be built of repeat arrays that are nearly identical in sequence, typically matching very closely to the consensus sequence for that particular repeat (Mosavi et al., 2002; Binz et al., 2003; Main et al., 2003). In principle, such consensus arrays can be well-modeled using a homopolymer approach (Figure 2B; (Main et al., 2003)), although in most cases polar substitutions at the terminal repeats are required to maintain solubility, introducing an intermediate level of heterogeneity (Wetzel et al., 2008).
Figure 2. A nearest-neighbor thermodynamic description of repeat-protein stability.
The first two lines (A, B) show single-repeat steps in folding (individual folded repeats are shown as blocks), whereas the last two lines (C, D) show overall folding reactions the fully denatured to the fully native state. Green repeats depict identical sequences, such as consensus repeats, the red and blue repeats represent N- and C-terminal caps.
III. Formulating a homopolymer partition function and the zipper approximation
The partition function, or sum over states, is central to analysis of the thermodynamic properties of repeat proteins, their populations, and their folding. Here the partition function will be developed for a homopolymeric linear system as a summation. As articulated by Zimm and Bragg in the late 1950s (Zimm and Bragg, 1959), this summation is particularly useful for short chains, thus keeping the number of terms in the sum manageable. The summation also simplifies to a useful approximate (closed) form in the high cooperativity limit.
One intuitive way to build a molecular partition function, q, for repeat protein folding, is to represent the statistical weight of each conformation (for a linear Ising model there will be 2n total) as the concentration of each conformation, compared (as a ratio) to an arbitrary reference conformation. By choosing the state in which all n repeats are unfolded (Un) as the reference state, such ratios are equivalent to equilibrium constants for folding, and are thus related exponentially to the intrinsic folding energy of each repeat (ΔGi) and the interfacial pairing energy between neighbors (ΔGi,i+1). With this reference, the molecular partition function can be written
| (1) |
The inside sum in equation 1 is taken over all microscopic configurations which have i folded repeats (Fi). Because of the dependence of overall folding energies on interfacial interactions, these microscopic configurations can differ in energy even though they have the same number of folded repeats. The number of interfaces is maximized when folded repeats are clustered together, whereas gaps separating folded repeats decrease the number of interfaces. Thus, converting equation 1 to a sum of equilibrium constants κ and τ for intrinsic folding and interfacial interaction (or exponentials in energies) requires the number of gaps between folded segments to be explicitly stated:
| (2) |
In this equation, Ωi,g is the number of ways that i out of n folded repeats can be arranged with g gaps.
Unfortunately, the degeneracy in equation 2 is rather complex even in open form, and is not particularly useful except for short arrays (low n), where each term in q can be given explicitly. However, in the limit of high interfacial stability, which eliminates gaps between folded repeats, the degeneracy (Ωi,g=0) and the partition function become particularly simple. When all i folded repeats are coalesced into one structured segment (g=0), there are n−i+1 ways to arrange the structured segment. This approximation is often referred to as the “zipper model” because structure (folded repeats in this case) zips up as a single block. The partition function for the zipper model can be written as
| (3) |
Both sums in the last line of equation (3) express partial geometric series in the variable κτ, which can be written in closed form as
Substituting this closed form expression into equation (3) gives
| (4) |
Differentiating the second term and rearranging gives a closed form of the partition function:
| (5) |
With this relatively simple expression for the partition function, populations and associated observable properties can be calculated. Of primary importance is the fraction of repeats that are folded, which is given as
| (6) |
where pi is the fractional population of the ith partly folded macrostate. Finding θ by differentiating q with respect to κ can be understood by recognizing that κ serves as a “counter” for folded repeats. For example, conformations with four folded repeats will have four powers of κ. The penultimate expression, which is general, and applies even when the zipper approximation does not hold, provides the simplest form for calculation of the fraction folded as a function of κ, τ, and n, given equation 5:
| (7) |
Equilibrium unfolding transitions can be derived from (or fitted using) equation (7) by introducing an explicit dependence on an external variable (temperature, pressure, or denaturant) to either κ, τ, or both parameters. In this review we will primarily focus on denaturant-induced unfolding. In Ising analysis of repeat protein unfolding, statistical weights have been have been assumed to vary exponentially with denaturant (linear in terms of free energy):
| (8A) |
| (8B) |
Here, [x] represents molar denaturant concentration, mi and mi,i+1 are denaturant sensitivities of the intrinsic and interfacial terms, and ΔGi,H2O and ΔGi,i+1,H2O are intrinsic folding and interfacial interaction energies in the absence of denaturant. This form of denaturant dependence has been used extensively for globular protein folding studies (Pace, 1986; Street et al., 2008).
Although in principle both the intrinsic and interfacial stability may be affected, most studies of repeat-protein denaturation have attributed the effect of denaturant solely to the intrinsic folding constant, κ (Mello and Barrick, 2004; Kajander et al., 2005; Wetzel et al., 2008). Assuming intrinsic folding involves formation of secondary structure elements (Figure 2), whereas the nearest-neighbor interaction corresponds to packing of neighboring repeats, this partitioning is consistent with a growing body of evidence suggesting that denaturants destabilize proteins largely by acting on the backbone, and thus should primarily destabilize units of secondary structure rather than packing interactions between such structures (Scholtz et al., 1995; Auton et al., 2007; Bolen and Rose, 2008). Moreover, this partitioning is consistent with recent global analysis from our laboratory on denaturant-induced unfolding of large numbers of consensus ankyrin repeat unfolding transitions (TA & DB, in preparation).
The first application of the 1D-Ising model to repeat protein folding involved a series of constructs in which ankyrin repeats were deleted from one or both ends of the Notch ankyrin domain (Mello and Barrick, 2004). By analyzing the free energies of unfolding of these constructs using a set of linear equations, a free energy contribution originating from each repeat was obtained. Because of the way the deletion series was constructed, analysis yielded an estimate of the intrinsic stability (ΔGi) of one of the repeats of +6.6 kcal/mol and an average interfacial stability (ΔGi,i+1) of −9.1 kcal/mol. These parameters were used to evaluate the populations of folded, unfolded, and partly folded states as a function of denaturant concentration, using the zipper approximation, which confirmed the all-or-none nature of the unfolding transition observed experimentally (Mello and Barrick, 2004).
IV. Matrix approach: homopolymers
The zipper model assumes that the folding of each repeat is highly coupled to its neighbors. High coupling allows conformations in which stretches of folded repeats are separated by unfolded repeats to be ignored. However, if cooperativity between adjacent repeats is low, or repeat arrays are long, these intermediates will be significantly populated, and must be accounted for. In this section we will present a simple matrix-based derivation of the partition function for the folding reaction of “homopolymeric” repeat proteins (i.e. all repeats are the same) that accounts for all partly folded conformations in a very compact way. This “matrix-method” has been widely used to study one dimensional interacting biological systems (Zimm and Bragg, 1959; Poland and Scheraga, 1970). In addition to providing a full description of all partly folded states, this matrix-based form can be used to analyze experimental unfolding transitions to determine ΔGi and ΔGi,i+1.
Before we show how the matrix representation of the partition function can be manipulated to analyze unfolding curves, we will use a recursion-based approach that justifies the matrix form of the partition function. Although the matrix-based form of the partition function can easily be used without a detailed understanding of its origin, and its form is often justified simply by the fact that the rules of matrix multiplication combine statistical weights in the appropriate way, we feel that an understanding of the origins of the matrix method will result in a deeper understanding of its application.
In the homopolymer approximation, each repeat has the same intrinsic folding energy (ΔGi), and the same interaction energy with its neighbors (ΔGi,i+1 for all i repeats; we will retain the subscript i for use below, although the homopolymer approximation makes all n repeats identical). The free energy of any particular configuration, relative to the fully denatured state (Un above), can be written as
where δj=1 if repeat j is folded, 0 if it is unfolded. With this free energy relationship, the partition function of the homopolymer system with n identical repeats can written as:
| (9) |
Long repeat proteins (large n) leads to a very large number (2n) terms in the sum, and is impractical for calculations and analysis of data. Instead, a simpler, more compact form of q(n) in terms of ΔGi and ΔGi,i+1 and is needed. One approach to simplifying the sum is to derive an expression for q(n) in terms of the partition function of a construct that contains fewer repeats (q(n − 1), for example). Repeating this method recursively defines q(n) in terms of progressively smaller (and simpler) partition functions, and generates the matrix representation of the partition function in terms of ΔGi and ΔGi,i+1 in the process. Starting with q(n) in terms of q(n − 1), the nth repeat can be added to an n-1 array in one of the two states: folded (the partition function that counts all such states will be called qf(n)) or unfolded (qu(n)). Applying the same dichotomy to the n-1 state divides q(n − 1) into two halves, one in which the last (n-1) repeat is folded (qf(n − 1)), and one in which the last repeat is unfolded (qu(n − 1)).
When the nth repeat is added to the C-terminal end in a folded state, qf(n) can be written in terms of qf(n − 1) and qu(n − 1):
The equation above simply states that if repeat n-1 is folded (with partition function qf(n − 1)), adding a folded repeat (with intrinsic energy ΔGi) at position n creates a new interface (ΔGi,i+1). However, if repeat n-1 unfolded is (with partition function qu(n − 1)), adding an unfolded repeat at position n does not create a new interface. Likewise when the nth repeat is added to the C-terminal end in an unfolded state, qu(n) can be calculated using the same approach:
The expressions for qf(n) and qu(n) are linear equations in the variables qf(n − 1) and qu(n − 1):
and can be consolidated with a simple matrix relationship:
The second line comes from substituting statistical weights κ = e−ΔGi/RT and τ = e−ΔGi,i+1/RT for the free energy terms.
Continuing the recursion to the n-2 repeat gives
This recursion can continued all the way to the first (N-terminal) repeat to give
qf (1) and qf (1) are the statistical weights for a single N-terminal folded and unfolded repeats, and are simply
Thus
Multiplying the LHS by the row vector [1 1] sums qf(n) and qu(n) to give the full partition function, q(n), as
By expanding the column vector on the RHS in terms of the statistical weight matrix, q(n) can be expressed as the nth power of the matrix
One final rearrangement of q(n), which will be helpful for further calculations, is given by taking the transpose of the equation above (as q(n) is a scalar, it is unaffected by transposition):
where the weight matrix is represented using W. The above equation allows q(n) to be computed without having to enumerate all 2n terms explicitly. Moreover, it can be simplified by treating it as an eigenvalue problem, which greatly simplifies the product of the statistical weight matrices. In this treatment, W is substituted by a matrix product
where D is a diagonal matrix of the eigenvalues (λ1, λ2) of W, and T is an invertible matrix of its eigenvectors (Strang, 2005). This substitution leads to
| (10) |
The eigenvalues of W are obtained by solving the characteristic equation det(W − λI) = 0, yielding the two roots:
| (11A) |
| (11B) |
(the derivatives will be used below). Two corresponding eigenvectors of W are
and combine to give
Combining these eigenvalues and eigenvectors into equation 10 gives a relatively simple closed-form expression for q(n):
By differentiating q(n) with respect to κ as in equation (6) above, the fraction of folded repeats (θ) can be calculated as
| (12) |
Values of λ1 and λ2, along with derivatives with respect to κ, can be inserted into equation 12 from equations 11A and 11B above. The denaturant dependence of the fraction of folded repeats can be obtained by combining equations 8A (and if necessary, 8B) into equation 12. Finally, the fraction of folded repeats can be used to analyze experimental equilibrium denaturation curves to determine the underlying thermodynamic parameters through the equation
where Yobs represents an observed signal (often far-UV circular dichroism or tryptophan fluorescence). The A’s and B’s allow for a linear denaturant dependence of the signals from folded and unfolded repeats, and combine to give native and denatured baselines. In principle, when analyzing multiple repeat proteins of different length (n), all the baseline parameters should be describable using a single pair of values for each baseline. However, owing to modest uncertainties in concentration, fitting separate baseline parameters may be preferable to introducing such a constraint, which may degrade the quality of the fit in the equilibrium transition region and compromise fitted thermodynamic parameters (Johnson, 2008).
VI. Matrix approach: heteropolymers
A primary motivation for analyzing consensus repeat protein unfolding is that each repeat can be considered to have the same stability and the same interaction energy with its neighbors, greatly decreasing the number of unknown thermodynamic parameters. However, repeat protein arrays built of a single consensus sequence seem to have solubility problems, likely owing to large hydrophobic interfaces present at the ends of each array. In crystal structures of a fragment of the Notch ankyrin domain, a head-to head crystallographic dimer is seen (Lubman et al., 2005), suggesting that the end repeats can indeed mediate association by such an interface. Such associations are also seen crystallographically in superhelical consensus TPR arrays, and actually displace the C-terminal capping helix (Kajander et al., 2007). Capping one or both termini with repeats bearing polar or charge substitutions solves this problem, but introduces new thermodynamic parameters, and more importantly, requires more complex models for analysis.
In this section, we will describe how the partition function for a heterogeneous repeat protein can be manipulated to simulate populations and folding transitions, and more importantly, fitted to equilibrium folding transitions. As above, we will use a matrix representation of the partition function, which again can be simplified from an open sum that enumerates each conformation. For generality, our derivation will treat each repeat as different, having different intrinsic folding (ΔGi) and interaction energies (ΔGi,i+1). For many repeat protein folding studies (especially capped consensus arrays), an intermediate level of complexity, in which some terms are identical and some are unique, should be sufficient to model folding and determine underlying energetic parameters, and may provide a more convenient representation.
We will start with the same matrix formulation we presented for homopolymers, and define a unique weight matrix for repeat:
| (13) |
As demonstrated above for a homopolymer, the rules of matrix multiplication combine statistical weights in such a way as to produce the appropriate Boltzmann factor for each conformation. That derivation, which considered q(n) in terms q(n-1), q(n-2)…, can easily accommodate unique, position-specific coefficients, rather than a single value for κ and for τ, to generate q(n) as in equation 13. The index on the interaction parameter in equation 13 represents the interaction between repeat i and the previous repeat (i-1) because the rows of the statistical weight matrix represent the folding status of the previous repeat. In the partition function for the homopolymer, diagonalization provides a huge simplification, converting a product of n identical matrices to a product of only three (TDT−1). This is not possible for the heteropolymer partition function, because the n weight matrices are different (as are their eigenvalues and eigenvectors). Thus, we are stuck with a product of n matrices as the partition function for a heteropolymeric repeat protein. Although when multiplied out this product has no fewer terms than a general summation such as equation (9) above, owing to its compactness it is considerably easier to generate and manipulate using matrix manipulation programs such as Matlab (http://www.mathworks.com/) and Scilab (http://www.scilab.org/).
As described above, the quantity of greatest interest in terms of connecting with experiments is the fraction of the repeats folded, θ. For homopolymeric systems, an expression for θ could be generated by differentiating the partition function with respect to κ, and dividing by q (see equation 6). With the closed-form homopolymer partition function, this operation is mathematically quite simple. Here, not only is the partition function more complex, there is no single value of κ that can be used as a counter of folded repeats. Moreover, the option of calculating an open sum of populations for all possible conformations and multiplying by the number of folded repeats is cumbersome (2n terms) and for large arrays of repeats, fitting requires significant computer memory.
Instead, we favor a summation over the n positions of the folded repeat, calculating the probability that each of the n repeats is folded, instead of the probability of each of the 2n conformations. Clearly, the fraction of repeats that are folded is simply the average probability that each of the repeats is folded:
where θi is the probability of finding ith repeat in folded state. θi, can be connected to the q(n) through a sub-partition function qi, which sums over all the conformational states in which the ith repeat is folded. These quantities can be related by recognizing that the probability of finding the ith repeat folded is simply the sum of conformations where it is folded divided by all the conformations, or
giving
This summation emphasizes the fact that q(n) only needs to be calculated once. In contrast, qi needs to be calculated n times (once at each position), but it can also be calculated in matrix form:
In the statistical weight matrix, the second column corresponds to all of the conformations where the ith repeat is unfolded. Setting this column to zero in the Wi matrix of qi eliminates all of these conformations without affecting terms for conformations where the ith repeat is folded.
VI. Solvability criteria for Ising models applied to repeat protein folding
The above sections derive equations for nearest-neighbor partition functions for repeat protein folding. These partition functions can be used to evaluate populations of partly folded states, and generate folding curves, given a set of thermodynamic parameters (ΔGi, ΔGi,i+1, and denaturant dependences). Subsequent sections will show how these models can be applied to analyze experimental folding curves, and will analyze fitted thermodynamic parameters for different repeat types and sequences. However, in this section, we will describe a way to evaluate whether a set of thermodynamic parameters is likely to be determined with any meaningful accuracy, given a set of data (folding transitions for constructs of different length, and potentially different sequence). This analysis will also connect to a closely related issue of determining whether a chosen model is mechanistically correct.
Much has been written regarding criteria for testing different models and estimating uncertainties of parameter values, given a set of experimental data (see (Johnson, 2008) for a recent review). Models are typically rejected based on non-random residuals and/or physically unreasonable fitted parameter values. Confidence intervals on parameter values can be estimated by statistical methods such as bootstrapanalysis, jack-knife analysis, or simple repetition of the experiment (all resampling methods that differ in their severity), analysis of the parameter covariance matrix, systematic exploration of how the variance of the fit increases as parameters are varied, and Monte Carlo simulation (Johnson, 2008). It is an unfortunate fact that these critical tests usually come after data have been collected. Experimental analysis of repeat protein folding is a laborious undertaking (involving cloning of multiple genes, expression and purification of multiple proteins of different length, and quantitative analysis of each protein (preferably multiple times) by denaturant titrations), and it would be good to know in advance whether such efforts are likely to yield significant thermodynamic insight.
Although many aspects of the sequence in which data acquisition precedes parameter and model testing are largely unavoidable, it is often the case that experiments can be designed a priori so that parameters of interest can be determined with confidence, and alternative models can be compared and discriminated. This is particularly true for repeat proteins, given their simple linear architecture, and the simple form of the linear free energy relationships implicit in the linear Ising model. Here we will describe how equilibrium folding studies on repeat proteins can be designed to maximize the information content of the results, given the framework of a particular thermodynamic model. In addition to helping to design future experiments, these ideas help to interpret published studies on repeat-protein folding.
By considering the free energies of folding of a collection of repeat proteins of different length as a system of linear equations, simple ideas from linear algebra relating to solvability can be used to determine whether parameters are likely to be well-determined, and if not, what additional constructs would be required to improve the situation. For a set of repeat proteins of different length and composition, the free energy difference between the fully folded and fully unfolded states can be written as
The first sum takes into account the different intrinsic energy terms, and the second sum takes into account the different interaction terms. Table 1 provides some examples, both for a homopolymic repeat-protein and for a heteropolymeric repeat-protein with unique N- and C-terminal caps.
Table 1.
Free energies of folding of capped consensus repeat protein constructs.
| nrep | Construct | Folding free energy (D→N) | |
|---|---|---|---|
| A | 3 | R3 | ΔG° = 3ΔGR + 2ΔGi,i +1 |
| B | 4 | R4 | ΔG° = 4ΔGR + 3ΔGi,i +1 |
| C | 5 | R5 | ΔG° = 5ΔGR + 4ΔGi,i +1 |
| D | 3 | NRC | ΔG° = 1ΔGN + 1ΔGR + 1ΔGC + 2ΔGi,i +1 |
| E | 4 | NR2C | ΔG° = 1ΔGN + 2ΔGR + 1ΔGC + 3ΔGi,i +1 |
| F | 5 | NR3C | ΔG° = 1ΔGN + 3ΔGR + 1ΔGC + 4ΔGi,i +1 |
| G | 6 | NR4C | ΔG° = 1ΔGN + 4ΔGR + 1ΔGC + 5ΔGi,i +1 |
| H | 4 | NR3 | ΔG° = 1ΔGN + 3ΔGR + 3ΔGi,i +1 |
| I | 5 | NR4 | ΔG° = 1ΔGN + 4ΔGR + 4ΔGi,i +1 |
| J | 4 | R3C | ΔG° = 3ΔGR + 1ΔGC + 3ΔGi,i +1 |
| K | 5 | R4C | ΔG° = 4ΔGR + 1ΔGC + 4ΔGi,i +1 |
N- and C-terminal caps are assumed to differ in intrinsic folding energy from consensus repeats (R), but have the same interfacial energy (ΔGi,i+1). Relaxing this restriction would introduce two additional interfacial energy terms (N:R and R:C as well as R:R).
For a set of consensus repeats without caps (lines A-C, Table 1), the three free energy equations can be written as
where ΔG°A is the free energy difference between the native and denatured states for the reaction defined on line A, and other ΔG° values are analogously defined. Based on simple linear equation theory, this set of linear equations has a unique solution1, because the columns (and rows) of the matrix on the left-hand side are independent. As a result, the matrix has full column rank (r=2); that is, elimination produces a pivot in every column (Strang, 2005). This is fundamentally a result of the fact that linear repeat proteins have one more repeat than interface, and thus a length dependence can resolve these two parameters.
For a set of consensus repeats with caps (lines D-G, Table 1), the free energy equations can be written as
Although there are enough equations to solve four unknowns (the column vector on the left-hand side), the columns are not independent. The first and third columns are equal; moreover, the sum of the first and second columns is equal to the fourth. Thus, the matrix lacks full column rank (again, r=2). As a result, this set of linear equations has an infinite number of solutions. Thus, the parameters cannot be uniquely determined by elimination. This problem will not be rectified by including additional equations (constructs) that retain both N- and C-terminal capping repeats.
Instead, if a set of four (or more) constructs is considered in which the caps vary along with the length, unique intrinsic folding energies can be determined for both the N- and C-terminal caps. For example, lines B, F, H, and J of Table 1 define the system of equations
The columns of this matrix are now independent, showing full column (and row) rank (r=4). Thus, the four thermodynamic parameters can be uniquely determined (although adding equations by including additional constructs will likely improve the robustness of the solution, given uncertainties in free energy measurements).
In principle, this type of analysis could be applied directly to experimental unfolding free energies determined by linear extrapolation from denaturant-induced unfolding transitions (Pace, 1986; Street et al., 2008) assuming a two-state (high cooperativity) model. However, if partly folded states are populated in the transition, either because of moderate values of ΔGi,i+1 or because stability is unevenly distributed along the repeat array, such free energy estimates will be incorrect. In such cases, globally fitting the denaturation transitions directly using an Ising model, which takes partly folded states into account, may improve estimates of free energy terms, in favorable cases providing access to parameters that could not be determined based on considerations of matrix rank above (see discussion of consensus ankyrin arrays below). Nonetheless, this simple analysis is extremely useful both for thinking about what constructs need to be studied to analyze a particular model, and for thinking about why certain parameters don’t appear to be well-determined, given a set of data. This type of rank analysis can also be applied to models that can accommodate differences between interfaces, models that include non-nearest-neighbor interactions, and by differentiation with respect to denaturant concentration, partitioning of m-values into intrinsic and interfacial components.
V. Matrix homopolymer analysis of consensus TPR folding
The first study in which a homopolymeric Ising model was used to analyze repeat protein folding involved a collection of consensus TPR arrays of different lengths (Kajander et al., 2005). As described above, TPR units are composed of two anti parallel α-helices (termed A and B) and are arranged in a linear array in which adjacent repeats twist along the long axis of the domain, like the steps in a spiral staircase (Figure 1C). Using TPR units of identical consensus sequence (termed CTPRan by the authors, where n represents the number of full 34 residue TPR units in a given construct), Regan and coworkers created a series of constructs of different lengths that were amenable to analysis using a homopolymeric Ising model (section III above). However, as with other consensus repeat arrays, to make their CTPR proteins soluble, the authors added an additional polar C-terminal capping helix (a variant of helix A with four polar substitutions).
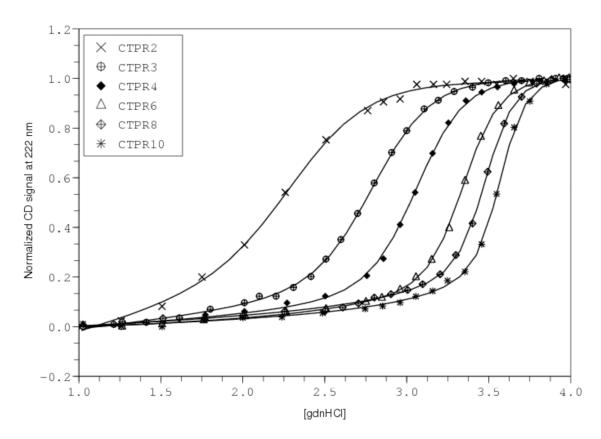
By monitoring helical structure using CD spectroscopy as a function of guanidine hydrochloride concentration, Kajandar et al. were able to generate and analyze unfolding transitions for constructs containing from two to ten full TPR repeats, as well as the C-terminal cap (CTPRa2 to CTPRa10; data reproduced from Fig. 2 of (Kajander et al., 2005)). The authors developed a homopolymer partition function in which each helix, rather than each repeat, is treated as the single repeating unit. Applying the homopolymer approximation at the single-helix level treats the A and B helices (and the C-terminal capping helix) as energetically equivalent, both in terms of intrinsic stability and in terms of nearest-neighbor interaction. Using this model, Kajandar et al were able to globally fit all of these transitions (and in a subsequent paper included even longer constructs (Kajander et al., 2007)) to a single intrinsic folding and interfacial interaction term (Kajander et al., 2005), clearly demonstrating the applicability of the linear Ising model to repeat protein folding.
Several aspects of this seminal study warrant further discussion. First, Kajandar et al. phrased the interaction energies in a way that is closer to the original magnetic spin-spin interactions than that described above (Kajander et al., 2005). Although at first glance the two representations look different, they can be shown to be identical, and the CTPR unfolding data can be fitted equally well with the two formulations of the homopolymer Ising model. The curves in Figure 3 were generated by fitting the model derived above to data from (Kajander et al., 2005); nearly identical fits and χ2 values are obtained with their representation of the model. Moreover, parameters from the two different formulations are nearly identical, when converted using relationships given previously (Kloss et al., 2008).
Figure 3. Unfolding and 1D-Ising analysis of consensus TPR proteins.
Data are from Kanandar et al. (Kajander et al., 2005), and were obtained using the the program DigitizeIt 1.5 for Mac OSX (http://www.digitizeit.de). Solid lines result from fitting a homopolymer partition function, with single helices as individual lattice sites, to the guanidine unfolding transitions. Fitted parameters are very close to those determined by Kajandar et al. (Table 2; fitted parameters are re-cast to ΔGi and ΔGi,i+1), and are well-determined by the data.
Second, fitted parameter values (ΔGi, ΔGi,i+1, and the denaturant dependence, which the authors assigned entirely to intrinsic folding) appear to be very well-determined. Kajandar et al. reported errors of 1% (Kajander et al., 2005), although no description was given for how these error margins were determined. To help compare the confidence levels of these parameters with those from other studies and from other models, we have evaluated parameter confidence intervals by Bootstrap analysis (Johnson, 2008). Briefly, the best-fitted parameters for the model used by Kajandar were used to generate “error-free” data at each experimental denaturant concentration for each construct. Residuals (observed minus fitted) were then used as a source of random error, by randomly sampling (with replacement) from the experimental residual set. The new data set (error free plus randomized residuals) was then re-fitted using the same model to generate a new set of fitted parameters. By repeating this procedure many times (1000-10000, depending on the distribution in parameter space) for different random data sets, a distribution of fitted values was generated, from which confidence intervals were approximated at the 95% level. Using the bootstrap method, we find fitted values of ΔGi, ΔGi,i+1, and mi to be determined to within 2-3% at the 95% confidence level (Table 2), quite similar to the bounds provided by Kajandar et al (Kajander et al., 2005). These narrowly bounded parameters provide significant insight into the origins of TPR folding and cooperativity. The parameters indicate that each helix has an unfavorable free energy of folding (+2.2 kcal·mol−1; Table 2), which is more than offset by a favorable helix-helix pairing energy (−4.5 kcal·mol−1). As was found for the Notch ankyrin domain, and also for consensus ankyrin constructs (see below), this leads to cooperative folding.
Table 2.
Paramaters from 1D-Ising analysis of consensus repeat proteins.
| Consensus TPR (Kajander et al., 2005)a |
Consensus Ankyrin (Wetzel et al., 2008)b |
Consensus Ankyrind | |
|---|---|---|---|
| ΔGN | n.d. | 10.6±0.6c 9.2±1.1 |
5.2±0.1 5.2±0.1 |
| ΔGR | 2.30±0.04 2.26±0.07 |
3.3±0.2 1.9±1.2 |
4.4±0.1 4.4±0.1 |
| ΔGC | n.d. | 10.6±0.6c 9.3±1.9 |
6.8±0.1 6.8±0.1 |
| ΔGi,i+1 | −4.52±0.04 −4.43±0.1 |
−14.2±0.7 −11.8±1.5 |
−11.2±0.2 −11.2±0.2 |
| mi | 0.57±0.01 0.57±0.01 |
1.1±0.1 1.1±0.2 |
0.75±0.01 0.75±0.02 |
| mcap | n.d. | 0.83±0.04 0.65±0.09 |
n.d. |
Energies are in kcal·mol−1; m-values are in kcal·mol−1·M−1. n.d., not determined in the model used.
Parameters for CTPR folding are for single helices. Parameters in the top line for CTPR arrays have been converted from the original formulation to ΔGi and ΔGi,i+1 to allow comparison with the other studies. Errors were propagated assuming the published H and J values to be uncorrelated.
The top line for each parameter gives estimated parameter values and uncertainties given by the authors. To facilitate comparison, the bottom line (italics) gives parameter values based on our fits, with uncertainties determined by bootstrap analysis as described in the text.
For the consensus ankyrin repeats of Wetzel et al., ΔGN and ΔGC are assumed identical, and are fitted as a single parameter.
For the consensus ankyrin repeats from our laboratory, parameters and errors in the top line come from resampling of guanidine titrations as described; errors in the bottom line (italics) come from bootstrap analysis as described.
Third, although treatment of the A and B helices as identical is clearly consistent with the published data, it would be surprising if the two helices were thermodynamically identical. The A and B helices have virtually no sequence similarity in the consensus design (Main et al., 2003). Moreover, structural analysis shows that the packing interactions of helices A and B differ substantially. Whereas the B-helices interact mostly with A-helices, lacking contacts with one another, the A-helices contact neighboring A-helices from adjacent TPRs, as well as their flanking B-helices, as can be seen from the zig-zag patterns in CTPR contact maps (Kajander et al., 2007). Adjacent A-helices have a two unit separation in a single-helix Ising model; thus, close contacts between adjacent A-helices would suggest a more complex model that has non-nearest neighbor terms (ΔGi,i+2). In addition, the C-terminal polar cap may be expected to introduce further complexity, as its folding energy may differ significantly even from the A-helix from which it is derived.
Given all of these sequence complexities, why not use a more complicated model to describe CTPR folding? One answer to this question is that a simple model works just fine. But does that mean the simple model is right? Given the differences between the two types of helices, a more complex model in which the A and B helices are treated differently makes more physical sense. Unfortunately, all of the CTPR constructs in Kajandar et al. have the same number of A and B helices, and thus it is not possible to separate the relative contributions of the two. Consideration of the free energy equations describing these constructs in terms of separate A and B helices makes this clear:
The matrix on the right hand side only has a rank of 2, and thus there are an infinite number of solutions to the set of equations. Treating each helix as identical simply adds column 1 and 2, making the unknown corresponding to this column the sum of ΔGA and ΔGB. Such a treatment gives full column rank, allowing a unique solution to be obtained, although it is a solution that is blind to the differences between helices. If instead a single A helix were deleted from one of the constructs (for example from the longest construct, making the last row [10 10 19]), the three-column matrix above would gain full column rank (r=3), and ΔGA and ΔGB would be resolved. This illustrates that in order to determine a particular parameter, the structural element corresponding to that parameter must be varied relative to elements defining the other fitted parameters.
VII. Matrix heteropolymer analysis of consensus ankyrin repeat folding
Consensus ankyrin repeats have been available for some time (Mosavi et al., 2002; Binz et al., 2003), and have been used successfully as a platform for protein design (Steiner et al., 2008). However, the application of Ising analyis to the folding of consensus ankyrin repeats has been relatively recent (Wetzel et al., 2008). To maintain solubility, Pluckthun and coworkers added capping repeats on both termini (called N and C respectively). This modification is similar to the C-terminal TPR-capping helix of Regan and coworkers, although the capping N and C ankyrin repeats designed by Pluckthun and coworkers are significantly different from their consensus sequences, with only 15/33 and 8/24 identities, respectively.
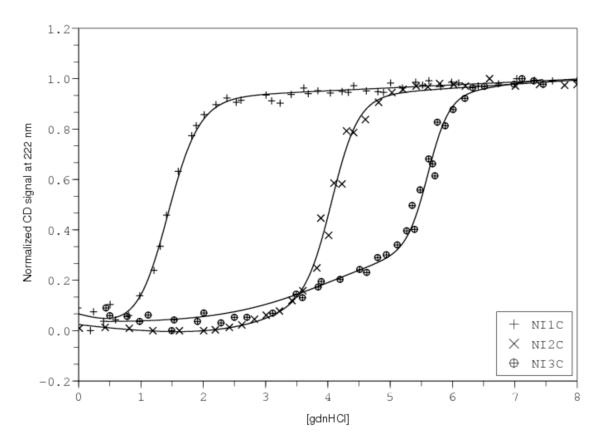
Using guanidine hydrochloride-induced unfolding, Pluckthun and coworkers obtained complete reversible unfolding transitions that could be used for Ising analysis for three constructs, NI1C, NI2C and NI3C (where I denotes internal consensus ankyrin repeats; (Wetzel et al., 2008), Figure 4). These three transitions were analyzed using a linear Ising model in which the N- and C-terminal capping repeats have intrinsic free energies (ΔGcap) that differ from the internal consensus repeats, but are identical to one another. In contrast, a single interfacial interaction energy was used (given the large number of sequence changes in the capping repeats, this may or may not be a valid assumption). As with the CTPR analysis, the denaturant dependence was attributed entirely to intrinsic parameters, although different denaturant sensitivities were assumed for the cap and internal repeats (mcap and mi, respectively).
Figure 4. Unfolding and 1D-Ising analysis of capped consensus ankyrin repeat proteins.
Data are from Pluckthun et al. (Wetzel et al., 2008), and were obtained using the the program DigitizeIt 1.5 for Mac OSX (http://www.digitizeit.de). Solid lines result from fitting a heteropolymer partition function, assuming the N- and C-caps have identical intrinsic folding energies that differ from the value for the internal repeats. Likewise, the effect of guanidine is partitioned into intrinsic folding energies and is allowed to vary between the capping and internal repeats. The pre-transition for NI3C appears to partly resolve the parameters from the capping and internal repeats.
As can be seen from the solid lines in Figure 4, this model describes the three fitted unfolding transitions reasonably well. Fitted parameters from Wetzel et al. (Wetzel et al., 2008) are listed in Table 2, along with confidence intervals provided by the authors. Again, there is no description of how these confidence intervals were determined. Using the heteropolymer partition function described above, and the same bootstrap method for error analysis described to analyze the CTPR array, we obtain intrinsic and interfacial energies that agree within 1-2.5 kcal/mol, although we find significantly larger margins of uncertainty on the fitted parameters than the authors; these values are also higher than those obtained by the same error analysis of the CTPR data. One reason for the high level of parameter uncertainty may be none of the three analyzed constructs have their caps removed, making it difficult to separate their contribution to free energy from the other parameters. Representing the constructs as a system of linear equations with a single cap free energy gives
In the coefficient matrix, half the first column plus the second column is equal to the third column, giving a rank of only 2, and again, an infinite number of solutions. Although at face value, this would severely compromise the accuracy of the fitted parameters, one feature of the unfolding transitions of Wetzel et al. may significantly narrow parameter confidence intervals: the appearance of a partial unfolding transition in the long native baseline of NI3C. Interpreted as a separate unfolding event involving one or both caps, this pre-transition provides additional information about the stability of the caps relative to the internal repeats. It is as if, from this region of the unfolding transition, the authors have prepared the construct I3 for analysis, which would give the coefficient matrix above full column rank. As described in section VI above, this study illustrates the value of analyzing complete denaturant unfolding transitions using the full partition function. Nonetheless, it appears that even with this information, the fitted parameter values are not as well determined as for the analysis of CTPR folding.
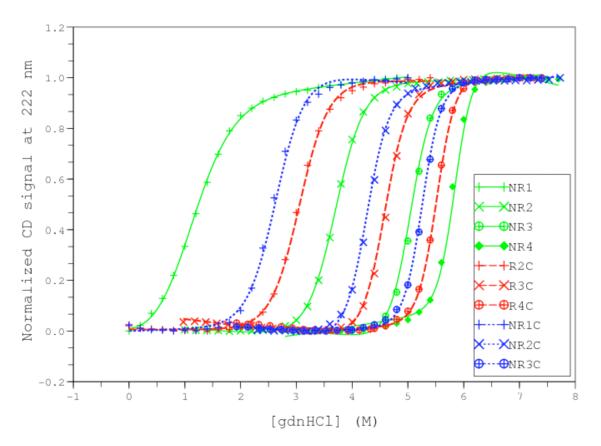
A more direct way to obtain information on the contribution of the caps would be to prepare constructs that lack the caps. Although ankyrin consensus arrays lacking both caps show poor solubility, we have been able to prepare arrays that lack either one cap or the other (we will refer to these as NRn and RnC, since in this partly exposed context these proteins lose their internal (I) nature; TA & DB, manuscript in preparation). Unlike the cap sequences of Wetzel et al., these caps differ by only four nonpolar→polar/charged substitutions on the “outside” face of the array, which should result in NR and RC interfaces that are much closer to full consensus (RR) interfaces. By combining these constructs with NRnC constructs, we have been able to obtain guanidine-induced folding transitions for ten constructs that are fully resolved and fully reversible (Figure 5). The difference in the contributions of the three types of repeats (N, R, C) to stability is clearly illustrated in the unfolding transitions. For constructs that have the same number of repeats (same symbols, Figure 5), the least stable construct has both caps, indicating that the caps are less stable than the consensus repeats. Between any pair of single-cap constructs, the one with the N-terminal cap is more stable than the one with the C-terminal cap.
Figure 5. Unfolding and 1D-Ising analysis of consensus ankyrin repeat proteins with and without terminal caps.
Constructs are described in the text (TA and DB, in preparation). Lines result from fitting a heteropolymer partition function, assuming N- and C-caps and the internal consensus repeats (R) all have different intrinsic folding energies. The effect of guanidine is partitioned into intrinsic folding energies and is assumed to be the same for all types of repeats. Different contributions of the N-, C- and R repeats can be seen by noting the shifts between constructs containing the same number of repeats, but different identities.
By independently removing the capping repeats, we have been able to test a number of different parameterizations of the Ising model to determine the relative intrinsic stabilities and contributions of the caps to denaturant-induced unfolding. The fits shown in Figure 5 are from an Ising model with separate intrinsic free energies for each cap and consensus sequence (ΔGN, ΔGR, ΔGC), a single interfacial energy (ΔGi,i+1), and a single m-value that affects only intrinsic folding energies. Fitted parameters are included in Table 2. In matrix form, the linear free energy equations for this data set show full column rank, allowing each parameter to be determined with minimal parameter correlation. To permit comparison to the other analyses described above, we have calculated errors using the bootstrap method2. Using this method, uncertainties (at the 95% confidence level) on fitted energy values are approximately 2% of the fitted parameters, about the same as for the CTPR studies, but significantly better than for analysis of NIC-NI3C.
Overall, the two consensus ankyrin repeat studies show a similar view of cooperativity in which the individual repeats are unstable, and the interfacial interaction is highly stabilizing (Table 2). Again, this is consistent with the high degree of cooperativity seen in solution, because single folded repeats should be rarified, and conformations with a large number of interfaces (blocks of consecutive folded repeats) should be maximized. Although this is qualitatively similar to what was seen in the CTPR study, cooperativity is much higher for the consensus ankyrin arrays. This is especially clear when the fitted Ising parameters from the CTPR studies are converted to whole-repeat (rather than single-helix) parameters. The intrinsic folding energy of an entire CTPR (ΔGi,helix+ΔGi,i+1,helix) is ~0.1 kcal/mol (nearly half-folded), whereas the interfacial energy is −4.5 kcal/mol. Thus, individual CTPR repeats are moderately less stable than consensus ankyrin repeats (the latter at ~3-4 kcal/mol), whereas the interfaces for CTPRs are significantly less stable than those of consensus ankyrin repeats (−12 to −14 kcal/mol).
The fitted Ising parameters for the two ankyrin consensus arrays are in reasonable agreement (Table 2). Fitted ΔGi values for consensus repeats and ΔGi, i+1 values are within 1-2 kcal/mol. Given the differences between consensus sequences from the two studies (~67% identity), this modest difference is not surprising. These two parameters both favor folding more than the parameters extracted from the deletion analysis (by about 3 kcal/mol; (Mello and Barrick, 2004)), which may also be a reflection of the substantial deviation from consensus seen for naturally occurring ankyrin repeat proteins. Fitted ΔGi values for the capping repeats for the two ankyrin studies show larger differences: the capping repeats of Wetzel et al are considerably less stable. This difference may result from the greater number of sequence differences, compared to the consensus, in that study.
VIII. Summary and future directions
The studies featured in this article show quite clearly that a simple nearest-neighbor model that has been highly successful in describing a wide variety of cooperative phenomena can be used to study repeat-protein folding, and extract quantitative interaction energies from real data. Although Ising-like models have been applied to model globular protein folding (for example, see (Munoz, 2001)), the heterogeneity of globular proteins and their intrachain contacts makes such models overparameterized, requiring assumptions about energy terms that come from informatics or from native state structures, rather than from first principles or measurements. A recent retrospective from Harold Scheraga, one of the major contributors to the application of Ising analysis to biopolymers, states of his epic research trajectory “it was soon realized that the helix–coil transition is not a good model for conformational changes in globular proteins, because the one-dimensional Ising model does not capture the cooperative features, embodied in the interplay between short- and long-range interactions, of the folding/unfolding transition of globular proteins” (Scheraga, 2008). Although repeat proteins differ from globular proteins in that they have structural simplicity and are somewhat elongated, they are the same in many other key respects. They have large, continuous hydrophobic cores, they have significant medium and long-range electrostatic interactions (Kloss and Barrick, 2008; Merz et al., 2008), and they are highly cooperative (Kloss et al., 2008). Thus, repeat-proteins provide a unique experimental system to dissect protein folding using this elegant model.
One of the most exciting aspects of the work featured here is that it provides an opportunity to understand protein folding cooperativity in quantitative and structural detail. Determination of ΔGi,i+1 provides a direct measure of long-range coupling within a folded protein. Further analysis of repeat proteins using the 1D Ising model should reveal not only the structural origins of this cooperativity, but how such cooperativity influences the kinetics of folding.
ACKNOWLEDGMENTS
This work was supported by NIH grant RO1GM068462 to DB.
Footnotes
If there are experimental errors associated with the column on the right hand side, the solution will be inexact, but can be found using least-squares.
To obtain a more rigorous measure of parameter uncertainties, we have measured each unfolding transition at least three times, allowing us to use resampling methods to fit separate transitions and compare results. This resampling approach, which employs more data, cannot be directly compared with the other studies analyzed here, but it gives similar confidence intervals to those from the bootstrap method.
LITERATURE CITED
- Applequist J, Damle V. Thermodynamics of the Helix-Coil Equilibrium in Ologoadenylic Acid from Hypochromicity Studies. J. Am. Chem. Soc. 1965;87(7):1450–1458. [Google Scholar]
- Auton M, Holthauzen LM, Bolen DW. Anatomy of energetic changes accompanying urea-induced protein denaturation. Proc Natl Acad Sci U S A. 2007;104(39):15317–15322. doi: 10.1073/pnas.0706251104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binz HK, Stumpp MT, Forrer P, Amstutz P, Pluckthun A. Designing repeat proteins: well-expressed, soluble and stable proteins from combinatorial libraries of consensus ankyrin repeat proteins. J Mol Biol. 2003;332(2):489–503. doi: 10.1016/s0022-2836(03)00896-9. [DOI] [PubMed] [Google Scholar]
- Bolen DW, Rose GD. Structure and energetics of the hydrogen-bonded backbone in protein folding. Annu Rev Biochem. 2008;77:339–362. doi: 10.1146/annurev.biochem.77.061306.131357. [DOI] [PubMed] [Google Scholar]
- Brush SG. History of the Lenz-Ising Model. Rev. Mod. Phys. 1967;39(4):883–893. [Google Scholar]
- Courtemanche N, Barrick D. Folding thermodynamics and kinetics of the leucine-rich repeat domain of the virulence factor Internalin B. Protein Sci. 2008;17(1):43–53. doi: 10.1110/ps.073166608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crothers DM, Kallenbach NR. On the Helix-Coil Transition in Heterogeneous Polymers. J Chem. Phys. 1966;45(3):917–927. [Google Scholar]
- De La Cruz EM. Cofilin Binding to Muscle and Non-muscle Actin Filaments: Isoform-dependent Cooperative Interactions. J. Mol. Biol. 2005;346:557–564. doi: 10.1016/j.jmb.2004.11.065. [DOI] [PubMed] [Google Scholar]
- DeLano WL. MacPyMOL: PyMOL Enhanced for Mac OS X. DeLano Scientific; Palo Alto: 2003. [Google Scholar]
- Groves MR, Barford D. Topological characteristics of helical repeat proteins. Curr Opin Struct Biol. 1999;9(3):383–389. doi: 10.1016/s0959-440x(99)80052-9. [DOI] [PubMed] [Google Scholar]
- Ising E. Z.Physik. 1925;31:253. Title Unavailable. [Google Scholar]
- Johnson ML. Nonlinear least-squares fitting methods. Methods Cell Biol. 2008;84:781–805. doi: 10.1016/S0091-679X(07)84024-6. [DOI] [PubMed] [Google Scholar]
- Kajander T, Cortajarena AL, Main ER, Mochrie SG, Regan L. A new folding paradigm for repeat proteins. J Am Chem Soc. 2005;127(29):10188–10190. doi: 10.1021/ja0524494. [DOI] [PubMed] [Google Scholar]
- Kajander T, Cortajarena AL, Mochrie S, Regan L. Structure and stability of designed TPR protein superhelices: unusual crystal packing and implications for natural TPR proteins. Acta Crystallogr D Biol Crystallogr. 2007;63(Pt 7):800–811. doi: 10.1107/S0907444907024353. [DOI] [PubMed] [Google Scholar]
- Kajava AV. Review: proteins with repeated sequence--structural prediction and modeling. J Struct Biol. 2001;134(2-3):132–144. doi: 10.1006/jsbi.2000.4328. [DOI] [PubMed] [Google Scholar]
- Kloss E, Barrick D. Thermodynamics, kinetics, and salt dependence of folding of YopM, a large leucine-rich repeat protein. J Mol Biol. 2008;383(5):1195–1209. doi: 10.1016/j.jmb.2008.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss E, Courtemanche N, Barrick D. Repeat-protein folding: new insights into origins of cooperativity, stability, and topology. Arch Biochem Biophys. 2008;469(1):83–99. doi: 10.1016/j.abb.2007.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe B, Kajava AV. When protein folding is simplified to protein coiling: the continuum of solenoid protein structures. Trends Biochem Sci. 2000;25(10):509–515. doi: 10.1016/s0968-0004(00)01667-4. [DOI] [PubMed] [Google Scholar]
- Lenz W. Physik. Z. 1920;21:613. Title Unavailable. [Google Scholar]
- Lifson S, Roig A. On the Theory of Helix-Coil Transition in Polypeptides. J. Chem. Phys. 1961;34(6):1963–1974. [Google Scholar]
- Lubman OY, Kopan R, Waksman G, Korolev S. The crystal structure of a partial mouse Notch-1 ankyrin domain: repeats 4 through 7 preserve an ankyrin fold. Protein Sci. 2005;14(5):1274–1281. doi: 10.1110/ps.041184105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Main ER, Lowe AR, Mochrie SG, Jackson SE, Regan L. A recurring theme in protein engineering: the design, stability and folding of repeat proteins. Curr Opin Struct Biol. 2005;15(4):464–471. doi: 10.1016/j.sbi.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Main ER, Xiong Y, Cocco MJ, D’Andrea L, Regan L. Design of stable alpha-helical arrays from an idealized TPR motif. Structure. 2003;11(5):497–508. doi: 10.1016/s0969-2126(03)00076-5. [DOI] [PubMed] [Google Scholar]
- McGhee JD, von Hippel PH. Theoretical Aspects of DNA-protein interadtions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 1974;86(469-489) doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- Mello CC, Barrick D. An experimentally determined protein folding energy landscape. Proc Natl Acad Sci U S A. 2004;101(39):14102–14107. doi: 10.1073/pnas.0403386101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz T, Wetzel SK, Firbank S, Pluckthun A, Grutter MG, Mittl PR. Stabilizing ionic interactions in a full-consensus ankyrin repeat protein. J Mol Biol. 2008;376(1):232–240. doi: 10.1016/j.jmb.2007.11.047. [DOI] [PubMed] [Google Scholar]
- Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004;13(6):1435–1448. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosavi LK, Minor DL, Jr., Peng ZY. Consensus-derived structural determinants of the ankyrin repeat motif. Proc Natl Acad Sci U S A. 2002;99(25):16029–16034. doi: 10.1073/pnas.252537899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz V. What can we learn about protein folding from Ising-like models? Curr Opin Struct Biol. 2001;11(2):212–216. doi: 10.1016/s0959-440x(00)00192-5. [DOI] [PubMed] [Google Scholar]
- Niss M. History of the Lenz-Ising Model 1920--1950: From Ferromagnetic to Cooperative Phenomena. Arch. Hist. Exact. Sci. 2005;59(3):267–318. [Google Scholar]
- Pace CN. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- Poland D, Scheraga HA. Theory of Helix-Coil Transitions in Biopolymers. Academic Press; New York: 1970. [Google Scholar]
- Poland D, Vournakis JN, Scheraga HA. Cooperative Interactions in Single-Strand Oligomers of Adenylic Acid. Biopolymers. 1966;4:223–235. doi: 10.1002/bip.1966.360040209. [DOI] [PubMed] [Google Scholar]
- Schellman JA. The Factors Affecting the Stability of Hydrogen-Bonded Polypeptide Structures in Solution. J. Phys. Chem. 1958;62(12):1485–1494. [Google Scholar]
- Scheraga HA. From helix-coil transitions to protein folding. Biopolymers. 2008;89(5):479–485. doi: 10.1002/bip.20890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholtz JM, Barrick D, York EJ, Stewart JM, Baldwin RL. Urea unfolding of peptide helices as a model for interpreting protein unfolding. Proc Natl Acad Sci U S A. 1995;92(1):185–189. doi: 10.1073/pnas.92.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner D, Forrer P, Pluckthun A. Efficient selection of DARPins with subnanomolar affinities using SRP phage display. J Mol Biol. 2008;382(5):1211–1227. doi: 10.1016/j.jmb.2008.07.085. [DOI] [PubMed] [Google Scholar]
- Strang G. Introduction to Linear Algebra. Wellesly-Cambridge Press; Wellesly, MA: 2005. [Google Scholar]
- Street TO, Courtemanche N, Barrick D. Protein folding and stability using denaturants. Methods Cell Biol. 2008;84:295–325. doi: 10.1016/S0091-679X(07)84011-8. [DOI] [PubMed] [Google Scholar]
- Wetzel SK, Settanni G, Kenig M, Binz HK, Pluckthun A. Folding and unfolding mechanism of highly stable full-consensus ankyrin repeat proteins. J Mol Biol. 2008;376(1):241–257. doi: 10.1016/j.jmb.2007.11.046. [DOI] [PubMed] [Google Scholar]
- Zimm B, Bragg J. Theory of the Phase Transition between Helix and Random Coil in Polypeptide Chains. Journal of Chemical Physics. 1959;31(2):526–535. [Google Scholar]
- Zimm BH. Theory of “Melting” of the Helical Form in Double Chains of the DNA Type. J. Chem. Phys. 1960;33(5):1349–1356. [Google Scholar]