Abstract
Patient registries are important for understanding the causes and origins of rare diseases and estimating their impact; and they may prove critical developing new diagnostics and therapeutics. This paper introduces the [RD] PRISM resource <http://rdprism.org>, an NIH-funded project to develop a library of standardized question and answer sets to support rare disease research. The paper presents a project case-driven plan for creating a new registry using questions from an existing related registry, revising and expanding an existing registry, and showing interoperability of data collected from different registries and data sources. Each of the use cases involves the retrieval of indexed questions for re-use. Successful retrieval of questions can facilitate their re-use in registries, meaning new registries can be implemented more quickly, and the use of “standard” questions can be facilitated. The paper further discusses issues involved in encoding the sets with relevant data standards for interoperability and indexing encoded sets with metadata for optimal retrievability.
Keywords: Patient registries, metadata, rare diseases
Overview
A rare disease is one that affects fewer than 200,000 individuals in the United States at a given time. There are approximately 7,000 rare diseases affecting more than 25 million Americans (NIH, Genetic, 2005). Many of these diseases have little research activity associated with them and no human trials. Often, patient registries are a helpful first step in estimating the impact and understanding the etiology of rare diseases, building a cause for future research, and facilitating enrollment in clinical trials – requisites for the development of new diagnostics and therapeutics. Patient registries use observational study designs that do not specify treatments intended to change patient outcomes (Arts, De Keizer, & Scheffer, 2002; Solomon, Henry, Hogan, Van Amburg, & Taylor, 1991). Although they are not as controlled as other scientific study designs, patient registries play an important role in the development, evaluation, and application of proof for evidence-based-medicine. Registries are an early step in understanding the natural history of disease and the development of clinical endpoints, patient reported outcomes and baseline data to support formal evaluations of therapeutic interventions. They have potential impact on the advancement of rare diseases research and improvement of patient outcomes and quality of life.
Patient registries can include data reported by patients, researchers, or clinicians. Examples of questions might include results of laboratory or genetic tests; requests for information about behaviors or exposures, perception or experience of symptoms or information obtained from family history or physical examination. While patient registries can be sponsored and developed by governments, academic scientists, clinical investigators, or pharmaceutical companies, they are often developed by Patient Advocacy and Support groups (PAGs). For rare diseases in particular, patient groups are often the first to sponsor and develop a registry.
PAGs are generally focused on education and raising public awareness about a disease of interest as well as supporting research efforts to find new treatments and ultimately a cure (NORD, 2009). Patient registries enable PAGs to determine the approximate numbers of people affected by the disease of interest and to gather information about the disease’s natural history. Often the content of these registries (i.e., the questions) change over time as more becomes known about the disease and its clinical variations. Frequently the registries (and the supporting questions) are developed on an ad hoc basis by the PAGs themselves, and there is currently no clear specification for standards and no apparent banks of existing standardized question and answer sets that are appropriate for registry developers, PAGs, or rare disease researchers to access. The challenges and issues noted above need to be addressed. As illustrated in Figure 1, the [RD] PRISM project is addressing these issues by developing a library of rare disease research questions that can be used to build patient registries. This project is guided by five main project demonstration aims that address identifying questions from the rare disease community to include in the library; modifying existing tools used currently in cancer research for storage, retrieval and transformation of questions into web-based forms; developing a metadata indexing model for search and retrieval of questions by the end user; demonstrating the use of the question library in the development of new registries; and identifying a process for expanding the content of the library. This will improve efficiency by enabling researchers and patient advocacy groups to access existing questions, evaluate their relevance for their disease of interest and decrease the amount of time and local resources to develop a registry. The next section provides more details on the specific objectives of the project.
FIGURE 1.
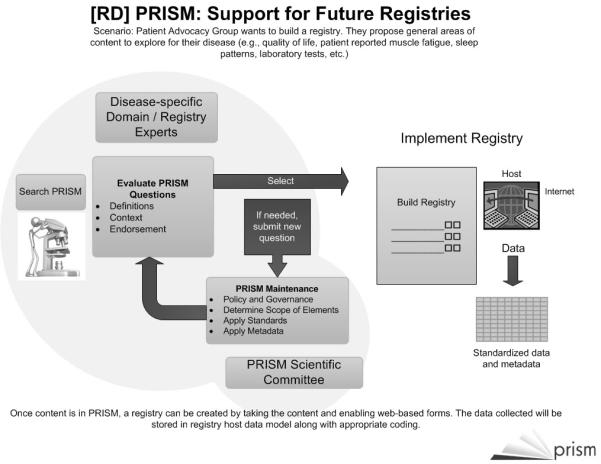
How [RD] PRISM Supports Future Registries
Objectives
The primary objective of the [RD] PRISM project is to develop a library of standardized questions across a broad spectrum of rare diseases that can be used for developing new registries and revising existing ones. This NIH-funded project is a collaborative effort between informatics faculty at the University of South Florida (USF) and the College of American Pathologists (CAP). The project will also receive cooperation and direction from Rare Diseases Clinical Research Network (RDCRN) clinical investigators and representatives of patient advocacy organizations from the rare disease community.
The project is driven by demonstration aims specified previously, that are guided by explicit use cases, i.e., specific and detailed research scenarios. Each use case includes a demonstration objective, a data source (i.e., forms, questions), a data entry source (e.g., patients, physicians), a descriptive scenario of the problem and data flows, and one or more clinical domain experts.
The current scope of the PRISM project includes three use cases—each to guide one of the following demonstrations:
Creating a new registry using questions from an existing related registry
Revising and expanding an existing registry
Interoperability of data collected from different registries and data sources
The final aim of the PRISM project is to expand the content of the library (i.e., add more questions relevant to additional diseases) and identify a process for patient advocacy groups and biomedical researchers to contribute questions to the library, access existing questions from the library, and implement rare disease registries using standardized questions from the library.
Project Plan
To meet our objectives, we have developed a project plan. The plan includes specific aims:
Identify questions from the diverse rare diseases research community that could be included in a standardized question library.
In collaboration with the College of American Pathologists (CAP) <www.capsts.org>, modify the electronic CAP Cancer Checklists tools to facilitate the storage, retrieval, and transformation of questions onto web-based data collection forms that can direct the data to a secure registry data repository.
Develop a metadata model for the questions that will facilitate search and retrieval of questions by various users. This metadata model will embrace current metadata registry standards and use controlled terminology to index library questions by content. Additionally, metadata describing the generalizable/generalized structural features of patient registry questions and answer sets, as well as their context of use, will be included in the PRISM library indexing model.
Demonstrate the use of the question library for the rapid implementation of new registries, the revision or expansion of existing rare disease registries, and the interoperability of data collected from different registries and data sources.
The final aim of the PRISM project is to expand the content of the library (i.e., add more questions relevant to additional diseases) and identify a process for patient advocacy groups and biomedical researchers to contribute questions to the library, access existing questions from the library, and implement rare disease registries using standardized questions from the library.
Using the tools and experience of College of American Pathologists’ (CAP) terminologists and electronic Cancer Checklists (eCC) developers, the project team will adopt and implement the architecture of the CAP eCC structured and standardized data forms to support a library of standardized questions for rare disease registries. Forms created according to this model can be exported into an extensible markup language (XML)-based format, and transformed [via extensible stylesheet language (XSLT)] to generate online registry data-entry forms on-demand.
Similar XML transformations into standardized formats can enable database storage of the completed form data as well as true interoperable transmission of data sets between systems. This section briefly describes the challenges for standard representation of questions and answers, relevant emerging standards efforts, and innovative features of the CAP eCC question/answer coding paradigm and form deployment tools. Our team approach for building a library of standardized registry questions includes plans for addressing various rare disease interests and data standards in the library development and curation process, and for making the library available to the broader rare disease community.
Challenges
Patient registries are often created by patient advocacy groups as a means for determining the prevalence of a particular disease or to gather information about the natural history of the condition. There are currently no standards for developing questions for use in patient registries (Richesson & Krischer, 2007). This need to identify, codify, and re-use standard questions for data collection is reminiscent of efforts in educational assessment (Sutton, 2008). Despite similar motivations (e.g., to support the administration of appropriate and well-constructed questions) between registry and educational assessment applications, the registry questions need to address many highly specific (even idiosyncratic) questions related to many thousands of questions specific to many thousands of different rare diseases. Because patient registries often are established to support ‘pre-research’ activities, the questions are meant to support exploratory and preliminary data collection. The questions can be changed or refined often, and in many cases are not even thoroughly tested on real patients. In cases where the questions are tested, the very nature of rare diseases severely limits the availability of sufficient populations to establish test metrics for the questions. Depending upon the sponsor of the registry, they might or might not be intended to support research – or even pre-research. They might purely exist to be descriptive of patient populations – providing summary data to affected patients to keep then engaged with the organization and registry until a formal research program can be established. There are also no applications or resources that assist rare disease researchers or patient advocacy organizations to use best practices, emerging standards or existing questions in registry development. The inability to re-use existing standardized registry questions results in time- and resource-intensive registry development efforts and inhibits opportunities for data sharing. A standardized library of data elements will speed the development and deployment of patient registries and allow registries to share and receive data from other registries or data sources. Additionally, a library of standardized registry questions will enable cross-indication and cross-disease analyses of patient populations, facilitate collaboration, and generate more meaningful results for rare disease patients, physicians, and researchers.
A critical and largely unaddressed problem for registries and clinical research data collection in general is the need for tools that allow data collection forms and their component questions and answers to be encoded in such a way that they can be retrieved for re-use (e.g., to support the rapid development of another related rare disease registry) or that the collected data can be interoperable with other data sources (e.g., Personal Health Records or Electronic Medical Records) (Health Information Technology, 2010). Useful standardization of registry forms should enable unambiguous, consistent, and reliable re-use of questions, answers, and groups of question/answer sets among different registries (Andrews, Richesson, & Krischer, 2007). The implementation of common sets of questions and answers derived from other approaches (e.g., Clinical Data Interchange Standards Consortium (CDISC), Cancer Data Standards Registry and Repository (caDSR)/Cancer Biomedical Informatics Grid (caBIG®) between studies is still not common, and the encoding of questions/answers with standard terminologies is not done consistently (Nadkarni & Brandt, 2006; Richesson & Krischer, 2007). Semantic encoding of data elements (i.e., question + answer + definition) is prone to inter-coder variability (Andrews, Richesson, & Krischer, 2007; Richesson, Andrews, & Krischer, 2006), and so makes consistent querying based on these “standard” codes difficult and unreliable, particularly across disparate resources. Further, mechanisms for storing the collected data are also subject to considerable variability, with groups often using fundamentally different data model architectures. In these approaches, data sharing often requires complicated conversions between data models, or the complex and error-prone use of terminological inference approaches (i.e., using computer-based inference methods, such as a transitive closure) to determine the similarity or equivalence of data stored according to different data architectures.
An additional problem in providing standardized questions for rare diseases registry data collection is caused by the context in which questions are presented to the data provider. For example, similar questions may be presented in tabular format versus a linear/vertical format of questions. Discreet answer choices may be presented for a user’s selection in one registry, whereas a write-in section may be presented in a similar form designed for another registry. The set of answer choices presented to the end-user may be subtly different between data-collection instruments, rendering the results not directly comparable. Other data collection instruments may link a series of unrelated answer choices under a single question-like heading, whereas another instrument might break up the answer set into two or more different questions, perhaps with a different set of semantic codes. Some data collection instruments may present mutually-exclusive answer choices, or allow the selection of inconsistent answers, whereas others may have created mutually exclusive, non-overlapping answer sets. Some forms may use constructs, such as “check all that apply” to list all positive findings, whereas others may require that the end-user explicitly state that a given condition is not present, or cannot be answered. Even simple differences such as variations in the use of font size, color, bold-face text, and question placement or order on a form can subtly affect how a form is completed by an end-user(Couper, 2001). The grouping of questions into form sections may also influence the response to certain questions, due to variation in context. The data produced by related non-standardized instruments can be fundamentally incomparable without a standard approach to modeling question/answer sets and registry form construction.
Relevant Standards
Three broad areas of standardization must be addressed in a library of standardized registry questions. The first area involves best practices for form styling and question construction. Questions are evaluated according to a process outlined in collaboration with the National Organization for Rare Disorders for vetting rare disease research questions. The second area is metadata model standardization for the library to support the storage and retrieval of the questions, answers and associated data (e.g., definitions, authors, context of use, history). The third area of standardization relevant to a library of standardized registry questions is the ‘binding’ of questions and answers to terminologies to assist retrieval and analysis of questions and data using clinical concepts. The collective experience of all of the key participants supporting this project will certainly guide the development (and adoption of) best practices in registry data collection and question-answer structuring, and will ensure that standards and practice for the [RD] PRISM library will address and leverage any concurrent related efforts. The CAP eCC tools provide a robust architecture based on representing question-answer sets and a suite of tools with demonstrated effectiveness in maintaining and deploying registry questions in cancer pathology. The historical leadership of CAP in SNOMED (Systematized Nomenclature of Medicine-Clinical Terms) creation, maintenance and use is a significant strength of this project, as SNOMED CT will certainly be used to code the clinical content, per Healthcare Information Technology Standards Panel (HITSP) and Office of the National Coordinator for Health Information Technology (ONC) recommendations (CHI, 2004).
There are many modeling and coding practices that must be defined for the semantic encoding of rare disease questions and registry forms. Among the decisions that will be made for this project are:
Coding system to use for questions [SNOMED CT, LOINC (Logical Observation Identifiers Names and Codes) or a combination];
Generic coding (e.g., using common question types and qualifiers as answers) or specific coding (e.g., using form-specific codes);
Assigning codes for entire question/answer set groups vs. discrete codes for questions and answers;
Form change protocol (i.e., changes in questions or changes in answer sets) will require a change in semantic codes;
Procedures to use for code change management;
Use of SNOMED CT core vs. a SNOMED extension (“Ref Set”) mechanism;
Procedures for creating a new mechanism for LOINC code assignment with versioned question/answer set groups;
Alignment of coding paradigm with national and international efforts (e.g., HITSP, Health Level Seven International (HL7), Clinical Document Architecture (CDA), Centers for Disease Control and Prevention (CDC), CDISC/ Clinical Data Acquisition Standards Harmonization (CDASH) research groups, etc.
An optimal, consistent and standards-compliant solution will be developed for the encoding of questions in the rare disease library.
Previous research and the current CHI/ONC standard for assessment instruments have shown that a combination of standards are ideal to represent first the structural and generic features of questions and then the clinical content (CHI, 2006). However, there appears to be no implementation of this that includes the semantics of LOINC axes. Capturing LOINC semantics is ideal in terms of retrieval, but encoding question-answer sets in this manner is time consuming and may not be practical in a short-term, results-driven project. The strategies for the precise use of SNOMED CT and/or LOINC will be decided based upon the nature of the rare disease questions prioritized for the library, the feedback from rare disease investigators regarding objectives of the planned registry implementations, and the availability of HITSP recommendations at the time of implementation.
Other data standards, particularly those that specify data elements for a given domain, will also be incorporated as opportunities arise. For example, family history question groups should include items from the most recent standards recommendations put forth by the American Health Information Community’s (AHIC) Family Health History Multi-Stakeholder Workgroup to the Office of the National Coordinator (ONC) (Feero, Bigley, & Brinner, 2008). Similarly, Office of Management and Budget (OMB) and de facto standards for patient characteristics (e.g., race, ethnicity, and demographics) will be applied, with the goal of limiting unnecessary variations of questions that have relevance across all rare disease registries. New and forthcoming pilot projects sponsored by HL7, caBIG, and CDISC that demonstrate the use of common data elements and shared conceptual domain models for specific therapeutic areas (e.g., cardiovascular, tuberculosis, and diabetes) will also be monitored and explored as a source of standardized questions that could be incorporated into the library and re-used for rare disease registry data collection (Gao, Zhang, Xie, Zhang, & Hu, 2006; Mohanty, et al., 2008; Winget, et al., 2003). Standardized health data from CDC’s PHIN Shared Vocabulary will be searched for value sets standardized for a variety of use cases. Similarly, the most recent CDASH recommendations (under ballot) are promising in terms of standardizing form and section names (e.g., concomitant medication form, maternal family history section), and provide solid guidance on question-answer construction (CDISC CDASH Team, 2008).
Checklists
The standardized checklist approach, a format designed to manage the information pathologists provide in reports on cancer specimens to create uniform cancer description and reporting, was pioneered by the CAP, in the form of the internationally-used CAP Cancer Protocols. The CAP Cancer Protocols are a comprehensive set of standardized, evidence-based, “scientifically-validated,” paper-based cancer case forms (the “checklists”) accompanied by detailed background documentation (CAP, 2010). The protocols were created in response to research studies from the 1990’s, which revealed a significant amount of variation in the quality of cancer-related pathology reports. Standardized checklists have been shown repeatedly to result in improved patient care, and to facilitate quality control and surveillance, as well as clinical research (Haynes, et al., 2009). The concept of a computerized checklist based upon standard question/answer sets requires the logical restructuring of existing data collection forms to create question/answer sets consistently, according to domain-specific patterns as expressed in a style guide and following the patterns in a domain-specific “base-template.” It is the team’s experience that the base template provides an outline and library of basic question/answer set structures and organizational patterns that can be used as building blocks for more specific question/answer-based checklist templates.
The template defines the questions, answers, headers, notes, etc. that comprise the lines of a paper checklist form, and specifies the content and general layout of the computerized form. The XML file that represents the question/answer sets structure of the template is called an XML Question/answer set Template (XQT). The basic unit within each XQT is the checklist “item.” The checklist items consist of elements such as Question, Answer, Header or Note. Each item is also able to store metadata (e.g., data type, required status, indentation level, rules, etc.) associated with the particular item in the paper checklist.
Work Flow
The concept of the evidence-based standard checklist can be readily extended to data-collection tools for rare disease registries. For this project, the CAP approach to checklist-based form standardization is used as a model for creating a library of rare disease registry questions and answers, which can be readily assembled to produce electronic forms. Each registry form will be modeled as a checklist, comprised of a series of questions. Most questions are followed by one or more possible answers or responses. Some question/answer sets require that a single answer be selected from a list (e.g. a combo box), whereas others allow multiple selections from a list (e.g. multiple check boxes). In some cases, “fill-in” free text or a numeric value is required. The questions and answers are interspersed with headers and notes that will guide users in the use of the question/answer sets. A collection of questions and answers and their associated descriptive metadata for a given implementation (e.g., a patient registry) are collectively referred to as a (form) template.
The innovative CAP eCC technical architecture is composed of layered XML-based modules which cleanly separate the work roles (e.g. creation of question/answer set designs, semantic encoding, forms customization). The XML-based modular structure is briefly described below:
The Template Editor Module: The CAP eCC template editing environment is essentially a Checklist Authoring Module, which allows a collection of questions and answers to be modeled, encoded, and organized in the context of a specific form (i.e., checklist) deployment.
Checklist/Form Distribution Module: The current release of the eCC technical architecture contains one XML-based XQT file for each current checklist version. The current release contains one XML schema file to constrain all XQT documents.
Checklist XML Transformation Module: This module generates GUI data-entry forms. An XML transformation (XSLT) can convert a template file into any desired format (e.g. XHTML forms). Cascading style sheet(s) (CSS) are used to format the transformed templates.
Checklist Data Output Module: This module handles the data output from the data-entry form. The XML-based output format is based upon the item keys in the Template; the XML output may be transformed into other formats such as HL7-based messages. Semantic codes may be incorporated in this output format.
Checklist Mapping Module: This module is an XML-based mechanism for mapping the Template questions and answers to any coding system for query purposes. In the current eCC release the code-mapping file contains a map to SNOMED CT, which is a controlled clinical terminology and the U.S. recommended standard for encoding many types of clinical data. We are also using coding schemes that are specific for various domains (e.g., Logical Observation Identifiers Names and Codes (LOINC®) http://loinc.org for Laboratory test names and RxNorm for medications). Other mappings can be added as needed to support the indexing of questions in specialized areas.
The base template is an outline and library of basic question/answer sets structures and organizational patterns that can be used as a building block for more specific question/answer sets-based checklist templates. The base template (prototype version) is shown in Figure 2 and contains subheadings for demographics, medications, etc. Some generic questions exist for each. Others – e.g., “Have you taken methotrexate ever” are disease-sepecific but it is clear where they belong on the template. CAP is adapting their Template Editor to model the question/answer sets structure of a registry data collection form (e.g. a “checklist”). The Template Editor allows the user to view a graphical representation of the question/answer hierarchy in one pane and to edit metadata for each question or answer value in a separate pane, including the editing of semantic codes and related mappings. To support the inclusion of generalizable questions in the library, we have explored research data collection standards and identified general constructs that many registries contain. Examples are Demographics, Medical History, Allergies, Medications.
FIGURE 2.
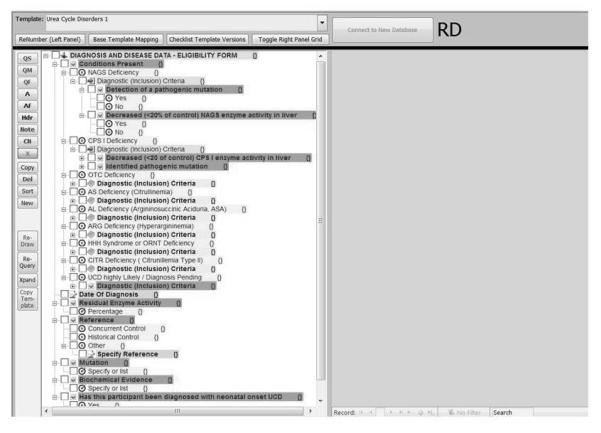
Template Editor
Data entered into the Template Editor can be exported in the XML-based XQT format, and can then be transformed (via XSLT) instantly into an XHTML page (Figure 2). The resulting customizable web page can be used by any rare disease patient or clinical center to submit semantically-encoded form data to a central repository. There are two data formats for the template information: the database representation used in the Template Editor (for editing of checklist line items) and a checklist XML (XQT) distribution format. The XML distribution format may be used to support multiple user implementations derived from a single checklist XML document, such as the generation of online registry data-entry forms, format and storage of the completed form data, and transmission of checklist data sets between systems.
Each form template and component question/answer item is automatically assigned a unique code, which is associated with an institution-specific namespace. Checklists may be released in new versions, while retaining the ability to query the checklist data across any subset of versions. Additionally, descriptive data regarding the authority that produces each checklist or checklist line item and history of checklist changes will be documented, to allow users to understand the domain, history, and context of use of question/answers.
In actual implementations of questions from the Template Editor/Library, certain questions can be designated as required (must be answered) to produce a valid data set. Implementations can specify the order of registry questions and address question dependencies (e.g., “Have you been depressed in past 3 months?” is dependent upon patient response to “Have you ever been depressed?”) and is flexible enough to allow different implementations of the content (e.g., different ordering of questions). All items in the library will be semantically encoded to allow retrieval and organization of the question/answers in the library. Additionally, the CAP Template Editor supports mappings to any number of coding systems.
The question/answer sets model of the CAP eCC currently includes provisions for handling common form metadata issues such as validation, repeating sections, required questions, single-select questions, multi-select questions, common question/answer set “rules,” multi-state checkboxes and single-select (deselecting) checkboxes. The model also includes support for form structural issues, such as activation and deactivation (“skip”) logic, and activation/deactivation of child elements. Also supported in the current model is the derivation of new form templates from a common base template, and derivation from a version of another template. Links to the base template or the parent template may be preserved to produce a chain of provenance. In the future, support is planned for complex rules, custom logic, calculations and algorithmic determination of response, table representations, and clickable images. This innovative approach, combined with semantic encoding, provides for a robust mechanism for inter-template and inter-version querying against sets of forms. Metadata will support all the querying, and identifying, testing, and refining the metadata to optimize question and template retrieval and re-use is an active area of the PRISM project, and a primary research objective.
The primary technical goal of this project is to produce a library of XML-based form representations that can be used to describe all aspects and functions of a computerized registry form, and enable robust, reliable and consistent querying of the accumulated form data. In practice, this means that the Template files must be readily transformable into a completely functional form, with semantic encodings and related mappings intact. Although it is possible to create custom hard-coded (static) forms (e.g., web pages, Windows forms) for each checklist using the XQT metadata, this is not required. The XQT files can be used for metadata-driven creation of forms via on-the-fly form generation. This would enable the user to browse the PRISM library for existing question and answer sets, select ones for re-use and preview in a sample form (Figure 3).
FIGURE 3.
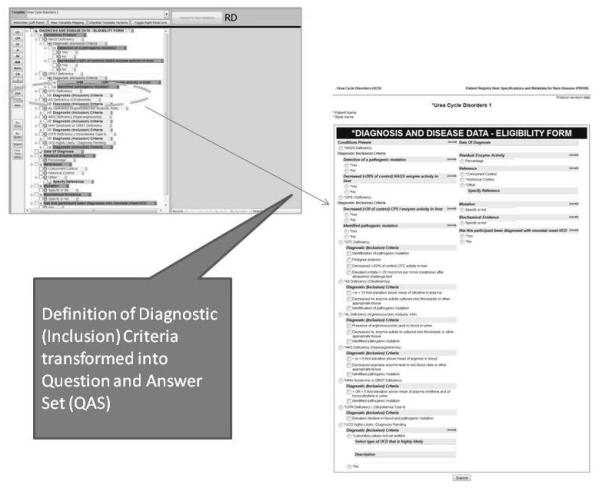
Transformation into Question and Answer Set
Summary
This paper describes the development of a library of standardized questions that can be rapidly implemented in patient registries across a spectrum of rare diseases. Informatics specialists from rare disease research settings guide the adaptation of the existing CAP Cancer Checklist technology for rare disease registry applications. The project is leveraging the existing Rare Disease Clinical Research Network infrastructure and registry to engage rare disease experts, identify valid questions from existing natural history studies, encode them using relevant standard terminologies, import them into a library accessible to the broader research community, and implement standardized questions into patient registry applications. The CAP eCC solution standardizes research questions and answers using XML-based files, which can be immediately transformed into various data-entry forms and can facilitate standardized transmission and interoperability of data sets. This approach enables the rapid generation of online registry forms, and speeds the implementation of registries for an unlimited number of rare diseases. The collaborative team has extensive ties to research and domain experts in rare diseases and informatics, as well as experience with many informatics standards. With this powerful set of skills and resources, project participants can create, implement, analyze and derive maximum benefit from this new, vitally-important interoperable and extensible library of standardized questions for rare diseases within a short period of time. The demonstrations will show the value of the PRISM library as a resource for re-using questions. This facilitates faster implementation of a new registry, a faster extension of an existing registry, and interoperability of data between two different registries. Our demonstrations include various queries of the library by typical users and will be used to refine and evaluate our metadata model and use of controlled vocabulary and data standards for the PRISM Library. We anticipate preliminary results by fall 2010.
FIGURE 4.

Sample Form
Acknowledgement
The Cancer Checklists and the tools described here were developed by our Colleagues at the American College of Pathologists, who are our partners in this work. Specifically we want to thank Rich Moldwin, Debra Konicek, Narciso Albarracin and Debbie Klieman.
Funding
American Reinvestment and Recovery Act (ARRA) <Recovery.gov> The [RD] PRISM Library project is funded by the American Reinvestment and Recovery Act. It is intended to be a fast-moving project with measureable deliverables within two years and demonstrated impact on investigations of rare diseases.
National Library of Medicine Support for the [RD] PRISM Library project is administered through the National Library of Medicine. (NLM Grant Number 1RC1LM010455-01, a component of the NIH, and supported by the Office of Rare Diseases Research. The contents are solely the responsibility of the authors and do not necessarily represent the official views of NLM or ORDR or NIH.)
Contributor Information
Rachel Richesson, Division of Bioinformatics and Biostatistics, University of South Florida (USF) College of Medicine, Tampa, Florida.
Denise Shereff, Division of Bioinformatics and Biostatistics, USF College of Medicine, Tampa, Florida.
James Andrews, School of Library and Information Science, USF, Tampa, Florida.
References
- Andrews JE, Richesson RL, Krischer JP. Variation of SNOMED CT Coding of Clinical Research Concepts among Coding Experts. Journal of the American Medical Informatics Association. 2007;14:497–506. doi: 10.1197/jamia.M2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts DG, De Keizer NF, Scheffer GJ. Defining and improving data quality in medical registries: a literature review, case study, and generic framework. J Am Med Inform Assoc. 2002;9(6):600–611. doi: 10.1197/jamia.M1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- College of American Pathologists (CAP) [Retrieved May 4, 2010];Cancer Protocols and Checklists. 2010 from http://www.cap.org.
- CDISC CDASH Team . Draft version 1.0. Clinical Data Standards Interchange Consortium; 2008. Clinical Data Acquisition Standards Harmonization: Basic Data Collection Fields for Case Report Forms. [Google Scholar]
- CHI . CHI Executive Summaries. Consolidated Health Informatics; 2004. [Google Scholar]
- CHI . Consolidated Health Informatics. Standards Adoption Recommendation. Functioning and Disability. Consolidated Health Informatics; 2006. (No. Disability Public Full.doc) [Google Scholar]
- Couper MP, Traugott ML, Lamais MJ. Web Survey Design and Administration. The Public Opinion Quarterly. 2001;65(2):23. doi: 10.1086/322199. [DOI] [PubMed] [Google Scholar]
- Feero WG, Bigley MB, Brinner KM. New standards and enhanced utility for family health history information in the electronic health record: an update from the American Health Information Community’s Family Health History Multi-Stakeholder Workgroup. J Am Med Inform Assoc. 2008;15(6):723–728. doi: 10.1197/jamia.M2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Zhang YL, Xie ZY, Zhang QP, Hu ZZ. caCORE: core architecture of bioinformation on cancer research in America. Beijing Da Xue Xue Bao. 2006;38(2):218–221. [PubMed] [Google Scholar]
- Haynes AB, Weiser TG, Berry WR, Lipsitz SR, Breizat AH, Dellinger EP, et al. A surgical safety checklist to reduce morbidity and mortality in a global population. N Engl J Med. 2009;360(5):491–499. doi: 10.1056/NEJMsa0810119. [DOI] [PubMed] [Google Scholar]
- Mohanty SK, Mistry AT, Amin W, Parwani AV, Pople AK, Schmandt L, et al. The development and deployment of Common Data Elements for tissue banks for translational research in cancer - an emerging standard based approach for the Mesothelioma Virtual Tissue Bank. BMC Cancer. 2008;8:91. doi: 10.1186/1471-2407-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadkarni PM, Brandt CA. The Common Data Elements for cancer research: remarks on functions and structure. Methods Inf Med. 2006;45(6):594–601. [PMC free article] [PubMed] [Google Scholar]
- National Organization for Rare Disorders (NORD) [Retrieved May 4, 2010];2009 doi: 10.1177/1942602X09352796. from http://rarediseases.org. [DOI] [PubMed]
- Richesson RL, Andrews JE, Krischer JP. Use of SNOMED CT to represent clinical research data: a semantic characterization of data items on case report forms in vasculitis research. J Am Med Inform Assoc. 2006;13(5):536–546. doi: 10.1197/jamia.M2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richesson RL, Krischer J. Data standards in clinical research: gaps, overlaps, challenges and future directions. J Am Med Inform Assoc. 2007;14(6):687–696. doi: 10.1197/jamia.M2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon DJ, Henry RC, Hogan JG, Van Amburg GH, Taylor J. Evaluation and implementation of public health registries. Public Health Rep. 1991;106(2):142–150. [PMC free article] [PubMed] [Google Scholar]
- Sutton SA, Golder D. Achievement Standards Network (ASN): An Application Profile for Mapping K-12 Educational Resources to Achievement Standards. Proceedings of the International Conference on Dublin Core and Metadata Applications.2008. [Google Scholar]
- Winget MD, Baron JA, Spitz MR, Brenner DE, Warzel D, Kincaid H, et al. Development of Common Data Elements: The Experience of and Recommendations from the Early Detection Research Network. International Journal of Medical Informatics. 2003;70:41–48. doi: 10.1016/s1386-5056(03)00005-4. (2003) [DOI] [PubMed] [Google Scholar]


