Abstract
Cytotoxic T lymphocytes (CTL) are hypothesized to play a role in clearance during primary dengue virus (DENV) infections, and contribute to immunopathology during secondary heterologous infections in humans. We previously reported skewed T-cell responses to secondary DENV infection in BALB/c (H-2d) mice, reproducing characteristics of human DENV infection. To set the stage for using widely available transgenic and knockout mice, we extended these studies to identify DENV-specific T-cell responses in C57BL/6 (H-2b) mice. We identified dominant CD8+ T-cell responses to H-2Db-restricted epitopes on the DENV NS4a (aa 249–265) and NS5 (aa 521–537) proteins. High frequencies of IFN-γ- and TNF-α-producing T cells directed at both epitopes were detected following primary infection with all four DENV serotypes, and were augmented by secondary DENV infections. In vivo cytotoxicity assays demonstrated rapid clearance of target cells pulsed with the NS4a peptide; in contrast, NS5 peptide-pulsed target cells were poorly cleared in vivo. These data characterize two H-2b-restricted T-cell epitopes displaying divergent in vivo function. These results should facilitate further studies of the in vivo effects of DENV-specific T cells, including the use of genetically modified mouse strains.
Introduction
Dengue virus (DENV) is a Flavivirus transmitted by the mosquito vector Aedes aegypti. It is the causative agent of dengue fever (1), and the more severe dengue hemorrhagic fever/dengue shock syndrome (DHF/DSS), which are characterized by plasma leakage. DENV infects 50–100 million people worldwide every year, with 500,000 cases of DHF. There are four immunologically distinct serotypes of DENV (DENV-1, DENV-2, DENV-3, and DENV-4). Lifelong homotypic immunity occurs after infection with one of the serotypes; however, heterologous secondary infections can still occur with any of the other three (2,3).
DHF/DSS is seen with significantly increased frequency during heterologous secondary DENV infections (4–6). Multiple immunopathological mechanisms have been proposed to explain these findings. Infection of monocytes and other FcγR-positive cells can be enhanced in the presence of cross-reactive non-neutralizing antibodies by a process known as antibody-dependent enhancement (ADE) (7–9). Cross-reactive memory T cells from the primary DENV infection are hypothesized to be preferentially activated compared to naïve T cells during secondary infection, with more rapid expansion and activation leading to enhanced production of proinflammatory cytokines, but inefficient clearance of the currently-infecting virus, resulting in DHF/DSS (10–12). Pre-existing antibodies that mediate antibody-dependent cellular cytotoxicity (ADCC) and other immune effector cells have also been proposed to contribute to dengue immunopathogenesis (13–18).
The difficulties in discriminating between these mechanisms in observational studies of naturally-infected humans have led researchers to mimic features of DENV infection in experimental animal models, with limited success (19–25). We previously reported that cross-reactive memory T cells were preferentially activated during heterologous secondary DENV infections in BALB/c mice, resulting in augmented IFN-γ and TNF-α responses, and that this effect was dependent on the serotype sequence of infection (26,27). Although this immunological model replicated aspects of human DENV-specific immunity, responses in BALB/c mice are skewed toward Th-2 responses. In contrast, C57BL/6 (H-2b) mice are more skewed to a Th-1-type proinflammatory response (28,29). In addition, many genetically engineered mouse strains are available on the C57BL/6 background, which may be valuable for further elucidating the immune mechanisms occurring during dengue infections.
We performed a comprehensive analysis to identify epitopes that elicited a strong T-cell response in C57BL/6 mice by screening overlapping peptides that spanned all of the DENV proteins. We identified and characterized two H-2b-restricted DENV epitopes. Yauch et al. also recently described one of these epitopes (30). In our studies we observed different T-cell avidity for the two epitopes that resulted in markedly different clearance of antigen-presenting cells. Lastly, we demonstrated serotype-cross-reactive responses, as well as enhanced cytokine responses, to heterologous secondary DENV infections.
Materials and Methods
Viruses
DENV-1 strain Hawaii, DENV-2 strain New Guinea C (31), DENV-3 strain CH53489, and DENV-4 strain 814669 were used. All viruses were propagated in C6/36 mosquito cells, and their titers were determined by plaque assay in Vero cells.
Peptides
Overlapping peptides corresponding to all of the DENV-2 NGC proteins were obtained from the National Institute of Allergy and Infectious Diseases Biodefense and Emerging Infections Research Resources Repository (BEI Resources). Peptides ranged from 14–17 amino acids in length, with 11 amino acid overlaps. Peptide pools for each protein were made with 9–12 overlapping peptides in each pool. Truncated peptides were purchased from Anaspec Inc. (San Jose, CA).
Immunization
C57BL/6 mice (4–6 wk of age) were purchased from Jackson Laboratories (Bar Harbor, ME) and immunized IP with 2 × 105 pfu of DENV, or an equivalent volume of uninfected C6/36 cell culture supernatant. For secondary infections, mice were immunized 28–56 d after the primary infection IP with 2 × 105 pfu of heterologous or homologous DENV serotypes. The time points were determined by previously performed kinetics studies of the post-infection immune response (26). At the indicated time points, the mice were sacrificed and splenectomized and single-cell suspensions were made. To determine the MHC restriction of the epitopes, H2-Db-knockout and H2-Kb-knockout C57BL/6 mice (Taconic Farms, Hudson, NY) were used. The mice were maintained in the Animal Facility at the University of Massachusetts Medical School following AAALAC guidelines.
IFN-γ ELISPOT assays
Peptide-specific IFN-γ-secreting T cells were quantified by ELISPOT as previously described (32). Spots were counted either manually or with a CTL Immunospot ELISPOT plate reader. All data shown represent values after subtraction of the media control (median 5.5 spot-forming cells [SFC] per million splenocytes).
Intracellular cytokine staining
Peptide-specific IFN-γ- and TNF-α-producing T cells were quantified by intracellular cytokine staining (ICS) assay as previously described (32). The cells were stimulated for 5 h with the peptides at 10 μg/mL total peptide concentration. The antibodies to surface molecules used were: anti-mouse CD3ɛ (145-2C11; BD Biosciences Pharmingen, Franklin Lakes, NJ), anti-mouse CD4 (GK1.5; BD Biosciences Pharmingen), and anti-mouse CD8α (5.3-6.7; eBioscience, San Diego, CA). Intracellular staining was done using anti-mouse TNF-α (MP6-XT22; BD Biosciences Pharmingen), and IFN-γ (XMG1.2; BD Biosciences Pharmingen). Data were acquired by the Flow Cytometry Core Laboratory at the University of Massachusetts using a FACSCalibur, and analyzed using FlowJo software (Tree Star, Inc., Ashland, OR). A small lymphocyte gate was drawn on forward- and side-scatter low populations, and further gated on either CD3+ CD8+ cells or CD3+ CD4+ cells. The cytokine-positive gate was drawn on the CD4+ or CD8+ and IFN-γ+ or TNF-α+ populations. Background frequencies of IFN-γ+ or TNF-α+ cells (from cells incubated with media) were subtracted from those of peptide-stimulated cells.
In vivo cytotoxicity
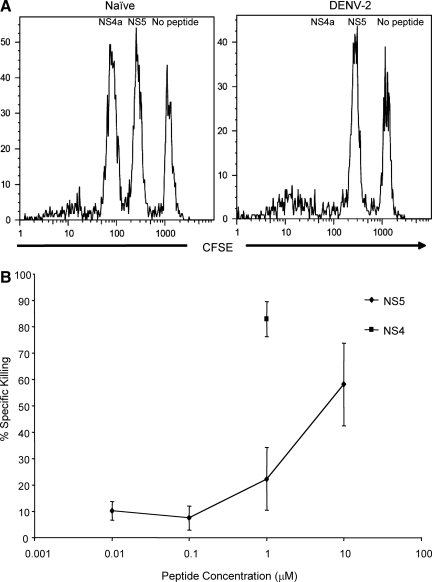
Assays were performed following the method of Jellison et al. (33). Splenocytes from naïve C57BL/6 mice were incubated with peptide (0.01–10 μM) for 45 min at 37°C. The cells were washed twice with phosphate-buffered saline (PBS). Carboxyfluorescein succinimidyl ester (CFSE, 5, 1.5, or 0.4 μM; Molecular Probes, Eugene, OR) was added to the different peptide-pulsed target cells. Dodecyldimethylamine oxide (DDAO, 2 μM; Invitrogen, Carlsbad, CA) was also added to the targets. The cells were incubated for 15 min. in a 37°C water bath. The cells were then washed two times with PBS, and 1 × 107 cells per target per mouse were transferred IV into DENV-2-immune mice in a volume of 200 μL. The mice were sacrificed 16 h later and splenocytes were isolated. The cells were fixed and analyzed by flow cytometry. DDAO+ cells were selected and then analyzed for the presence of CFSE+ cells. Specific killing was calculated as (1 − [ratio immune/ratio naïve]) × 100, with ratio = number of events peptide-coated target/number of events no peptide target (n = 4 mice per group).
Statistical analysis
Medians, confidence intervals, and standard errors were computed using SPSS and Microsoft Excel. ELISPOT and ICS data were compared between groups using the Mann-Whitney U test. p Values <0.05 were considered significant, and all p values <0.10 are shown to illustrate non-statistically significant trends.
Results
Definition of H-2b-restricted dengue viral epitopes
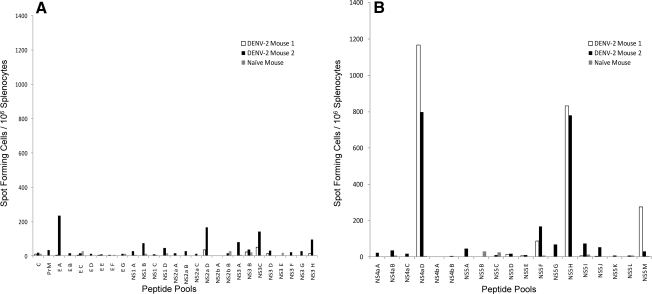
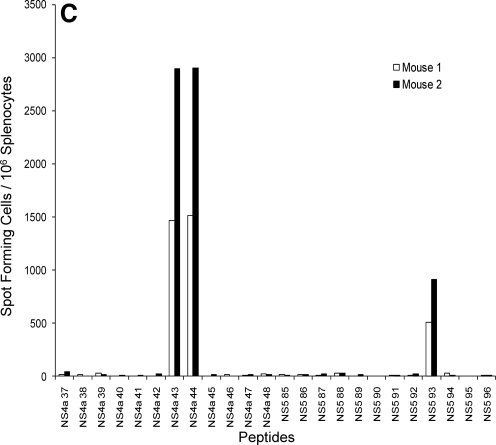
At the time we initiated these studies, bulk culture cytotoxicity assays in DENV-immunized C57BL/6 mice had demonstrated recognition of DENV non-structural proteins, but no epitopes had been identified (34). We immunized C57BL/6 mice with a low dose (2 × 105 pfu) of a laboratory strain of DENV-2, and 7 d later we tested splenocytes from infected mice for responses to overlapping peptide pools (12 peptides per pool) that spanned the entire DENV-2 genome in IFN-γ ELISPOT assays (Fig. 1A and 1B). Two peptide pools, NS4a D and NS5 H, elicited strongly positive IFN-γ responses from DENV-infected, but not naïve, splenocytes (Fig. 1B). Splenocytes from DENV-2-immunized mice were then tested using the individual peptides from positive pools. Two overlapping peptides in the NS4a pool, NS4a 43 (LLAIGCYSQVNPITLTA) and NS4a 44 (YSQVNPITLTAALFLLV), induced IFN-γ production, while a single peptide in NS5 pool H, NS5 93 (DVSKKEGGAMYADDTAG), elicited a positive response (Fig. 1C).
FIG. 1.
Screening of the DENV-2 genome and identification of two immunodominant DENV epitopes. The IFN-γ response in splenocytes of DENV-2-immune mice to peptide pools spanning the entire DENV-2 genome was measured by ELISPOT 8 d post-infection. (A) Responses, reported as spot-forming cells (SFC) per million splenocytes, to the pools corresponding to the DENV-2 C, PrM, E, NS1, NS2a, NSb, and NS3 proteins. (B) Responses to the pools corresponding to the DENV-2 NS4a, NS4b, and NS5 proteins. (C) Responses to individual overlapping peptides.
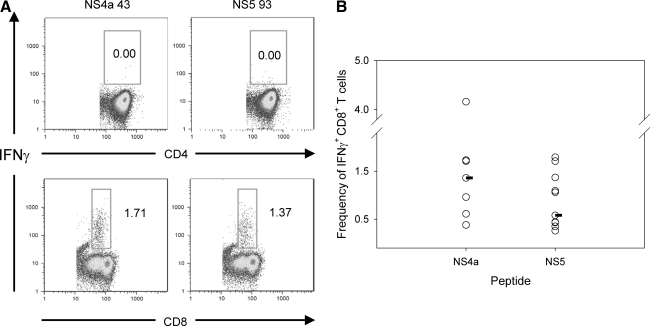
To establish whether these epitopes were recognized by CD4+ or CD8+ T cells, the IFN-γ response from splenocytes of DENV-2-infected mice was measured by flow cytometry. High frequencies of peptide-specific IFN-γ-producing CD8+ T cells were detected, whereas IFN-γ responses among CD4+ T cells were not significantly higher than background (Fig. 2A). These data demonstrate that CD8+ T cells responded to both the NS4a 43 and NS5 93 peptides (Fig. 2A).
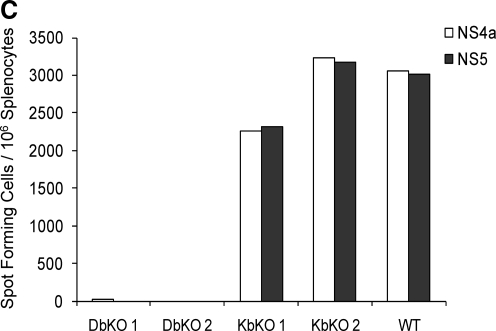
FIG. 2.
Characterization of the MHC restriction of the epitopes 8 d post-infection. (A) Representative frequencies of CD4+ T cells (top panel) or CD8+ T cells (bottom panel) that respond to stimulation with 10 μg/mL of NS4a and NS5 peptides by intracellular cytokine staining. (B) Median IFN-γ frequencies (n = 7–10 mice) of the NS4a and NS5 epitopes in splenocytes of DENV-2 immune mice by intracellular cytokine staining. (C) ELISPOT responses of splenocytes in H2-Db and H2-Kb knockout mice immunized with DENV-2. DbKO 1, DbKO 2, KbKO 1, KbKO 2, and WT indicate Db knockout mouse 1, Db knockout mouse 2, Kb knockout mouse 1, Kb knockout mouse 2, and wild-type mouse, respectively.
To characterize the magnitude of the IFN-γ responses to both the NS4a 43 and NS5 93 epitopes, the frequency of CD8+ IFN-γ-producing cells was determined in DENV-2-immunized mice using intracellular cytokine staining. A median of 1.38% (range 0.38–1.71%) of splenic CD8+ T cells responded to the NS4a 43 peptide by secreting IFN-γ, while frequencies of cells that responded to the NS5 93 peptide were slightly lower (median 0.58% of CD8+ T cells, range 0.26–1.79%; Fig. 2B)
To determine the MHC restriction of these epitopes, H-2Db and H-2Kb knockout mice were immunized with DENV-2, and the IFN-γ responses were measured by ELISPOT. No response to either peptide was seen in the Db knockout mice, while a response equivalent to that seen in wild-type mice was observed in the Kb knockout mice (Fig. 2C). These data indicate that both the NS4a and NS5 epitopes were recognized by CD8+ T cells and were restricted by H-2Db.
Differential lysis by epitope-specific CTLs
To assess whether epitope-specific CD8+ T cells could lyse cells presenting peptide-MHC complexes during the acute response to DENV infection, in vivo cytotoxicity assays were performed. Naïve syngeneic splenocytes were pulsed with peptide and adoptively transferred into mice infected with DENV-2 8 d previously. When splenocytes pulsed with 1 μM of peptide were transferred into acutely infected mice, there was efficient killing of NS4a-pulsed target cells, while lysis of NS5 peptide-pulsed cells was no higher than that of unpulsed cells (Fig. 3A). To determine if the failure to clear NS5 peptide-pulsed cells was due to insufficient expression of peptide-MHC, we repeated these experiments using a range of NS5 peptide concentrations (0.01–10 μM). We detected minimal killing at NS5 peptide concentrations ≤1 μM (Fig. 3B); lysis significantly above the negative control targets was only observed at 10 μM of NS5 peptide (Fig. 3B). These results demonstrate a difference in in vivo cytotoxicity responses to the NS5 and NS4a peptides, and suggest a difference in avidity of the T cells for these peptides.
FIG. 3.
Differential in vivo lysis of NS4a and NS5 peptide-pulsed cells in DENV-2-immune mice. (A) Representative examples of loss of CFSE staining in DENV-2-immune mice that received naïve splenocytes pulsed with 1 μM of either NS4a or NS5 peptides or no peptide (n = 4). Splenocytes were first gated on small lymphocytes and donor cells were further identified by gating on DDAO+ cells. (B) Summary of specific lytic activity against NS4a- and NS5-pulsed targets in DENV-2-immune mice. Median values are shown for each concentration. Diamonds represent the cytotoxic response to NS5, and squares represent the response to NS4a. Ninety-five percent confidence intervals were calculated.
Serotype cross-reactive cytokine responses
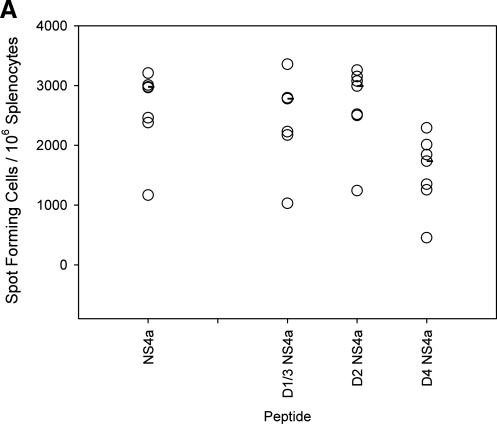
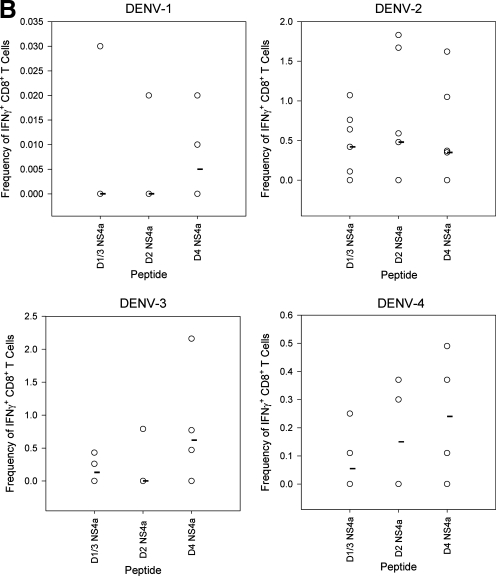
A key assumption of T cell-mediated immunopathogenesis implies that serotype-cross-reactive memory T cells elicited by primary DENV infection will respond to epitope variants of heterologous DENV serotypes. We identified the optimal NS4a epitope for all four serotypes via IFN-γ ELISPOT after primary DENV-2 infection (Fig. 4A). The amino acid sequences for the minimal NS4a epitopes in D1V and D3V were identical (YSQVNPLTL), and are referred to here as D1/3 NS4a. The DENV-2 epitope was also recently reported on by Yauch et al. (30). The peptides corresponding to the NS4a epitope for DENV-2 (YSQVNPITL) and DENV-4 (YSQVNPTTL) are referred to as D2 NS4a and D4 NS4a, respectively. To determine if any cross-reactivity existed in the immune responses to the variant peptides, groups of mice were given a primary infection with each of the four serotypes, and cytokine responses were measured to the peptides corresponding to all of the different serotypes by intracellular cytokine staining (Fig. 4B and C). Responses were detected after infection with all serotypes. Infection with DENV-2 elicited the greatest response, followed by DENV-3 and DENV-4, while infection with DENV-1 elicited the lowest response (Fig. 4B and C). Immunization of mice with DENV-2, DENV-3, and DENV-4 induced T cells capable of responding to homologous and heterologous NS4a peptides (Fig. 4B), indicating the serotype-cross-reactive nature of the T-cell response.
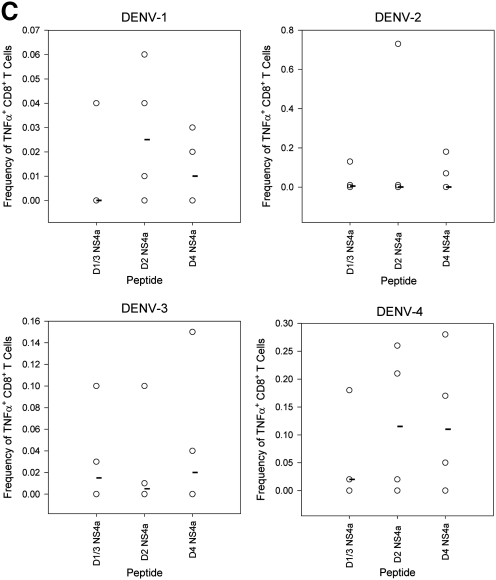
FIG. 4.
Serotype cross-reactive cytokine responses to homologous and heterologous variants of the NS4a epitope 8 d post-primary infection. (A) IFN-γ responses to the 15-mer peptide NS4a 43 and to three variants of the 9-mer epitope were measured by ELISPOT assay (B) IFN-γ responses to the variant 9-mer NS4a peptides by intracellular cytokine staining. Values represent frequencies of IFN-γ-secreting CD8+ T cells in splenocytes of DENV-immune mice. (C) TNF-α responses to the variant NS4a peptides by intracellular cytokine staining. Values represent frequencies of TNF-α-secreting CD8+ T cells in splenocytes of DENV-immune mice. Median values are represented by black bars (n = 4 mice per group for all groups, except n = 7 for mice receiving primary DENV-2).
TNF-α responses were detected in splenocytes after infections with all serotypes, with DENV-2 infections eliciting the largest response, followed by DENV-3, DENV-4, and DENV-1. Cross-reactive TNF-α+ T-cell responses were also observed. Following DENV-1 infection, higher frequencies of cells responded to heterologous peptides compared to the homologous DENV-1 peptide. Splenocytes from DENV-2-infected mice yielded a low TNF-α response to all of the NS4a peptides. For DENV-3 and DENV-4 infections, some heterologous peptide responses were similar in magnitude to homologous peptide responses. Overall the data indicate that cross-reactive TNF-α responses were detected in mice immunized with each of the four serotypes. These data provide evidence supporting the idea that dengue-specific T cells can secrete TNF-α and IFN-γ in response to stimulation with variants of the NS4a epitope.
Augmentation of immune responses by heterologous secondary DENV infection
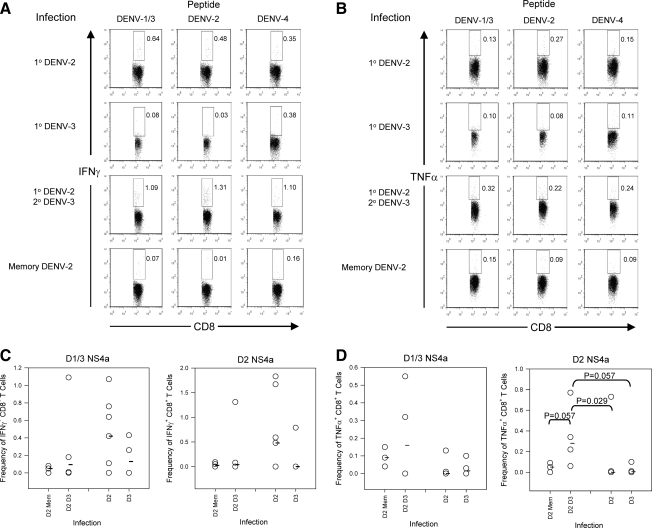
We had previously shown that cross-reactive T cells from a primary DENV infection were preferentially recruited to expand during a heterologous secondary DENV infection in BALB/c mice (26). To determine whether memory T cells from C57BL/6 mice also showed enhanced responses to heterologous secondary DENV infection, we infected mice with DENV-2, and 4 wk later the mice were rechallenged with a secondary DENV-3 infection. T-cell IFN-γ and TNF-α responses to the NS4a variant peptides were measured by intracellular cytokine staining (Fig. 5). Though the IFN-γ response was not significantly augmented (Fig. 5A and C), significant boosting of the TNF-α response was seen to the DENV-2 peptides (Fig. 5B and D).
FIG. 5.
Cytokine responses following heterologous secondary dengue virus infections. Mice were infected with DENV-2 and challenged 4 wk later with DENV-3. The IFN-γ and TNF-α responses to the variant peptides were measured by intracellular cytokine staining on day 9 after secondary infection. (A and B) Representative cytokine staining for the variant NS4a peptides. 1o DENV-2 is staining of splenocytes 8 d post-primary DENV-2 infection. 1o DENV-3 is staining of splenocytes 8 d post-primary DENV-3 infection. 1o DENV-2 2oDENV-3 is staining of splenocytes 9 d post-secondary infection of DENV-2-immune mice with DENV-3. Memory DENV-2 is staining of splenocytes from mice that received a primary DENV-2 infection 28 d previously. (A) Representative IFN-γ staining for the variant NS4a peptides. Numbers represent the frequencies of IFN-γ+ CD8+ T cells. (B) Representative TNF-α staining for the variant NS4a peptides. The numbers represent the frequencies of TNF-α+ CD8+ T cells. (C and D) Total IFN-γ and TNF-α responses. The x-axis represents the infection group. Memory phase post-primary DENV-2 infection (D2 Mem), heterologous secondary infection (D2 D3), primary DENV-2 (D2), and primary DENV-3 (D3). (C) IFN-γ response and (D) TNF-α response. Medians are represented by the black bars. The Mann-Whitney U test was used to calculate p values. Only significant values (p < 0.05) are shown (n = 4 mice per group for all groups, except n = 7 for mice that received primary DENV-2).
Discussion
We report the identification of two H-2b-restricted T-cell epitopes in the DENV NS4a and NS5 proteins, after screening the entire DENV-2 genome in IFN-γ ELISPOT assays. Both epitopes were recognized by CD8+ T cells and restricted by H-2Db. Yauch et al. also recently described epitopes in DENV recognized by H-2b-restricted T cells, including the one we identified in NS4a, after screening selected candidate MHC-binding peptides (30). Surprisingly, their findings did not include the NS5 epitope that we detected. Significant differences between the design of the two studies may explain the difference in “immunodominant” epitopes identified. We screened the entire DENV polyprotein using overlapping peptide pools, and infected mice with a low dose (2 × 105 pfu) of the prototype DENV-2 strain New Guinea C. In contrast, Yauch et al. screened candidate peptides selected using a computer algorithm, not a comprehensive screen of the entire genome, following infection of mice with a much higher dose (1011 genomic equivalents) of the mouse-adapted DENV-2 strain D2S10. In addition, since they did not report all the peptides that they tested, it was unclear from their study whether or not the NS5 epitope that we describe was tested in their system.
The frequencies of epitope-specific T cells observed in C57BL/6 mice are significantly higher than those we observed earlier in BALB/c mice. This could reflect the greater bias towards Th-1 cytokine production in C57BL/6 mice compared to BALB/c mice (28,29). Alternatively, there may be differences in the expression of DENV antigens in vivo; however, this is difficult to assess, as detectable virus is cleared very rapidly in both mouse strains. The higher frequency of DENV-specific T cells in C57BL/6 mice should be an advantage for further studies in this mouse strain.
Using an in vivo cytotoxicity assay, we detected killing of target cells pulsed with either the NS4a or NS5 epitope in DENV-2-immune mice, suggesting that these T cells would be active at clearing DENV-infected cells in vivo. However, far lower concentrations of peptide were required for killing NS4a peptide-pulsed cells. One possible explanation may be a higher avidity of NS4a-specific T cells for their cognate peptide than the NS5-specific T cells. Yauch et al. found that DENV-specific cytotoxic CD8+ T cells were protective against lethal D2S10 infection in IFN-α/βR−/− mice (30). NS5 peptide-specific T cells might therefore provide less protective immunity against DENV infection in vivo, and instead contribute to immunopathology.
We observed serotype-cross-reactive T-cell cytokine responses to the NS4a peptide in mice immune to any of the four DENV serotypes. Given the cross-reactive nature of the response, we examined the immune response after heterologous secondary DENV infections. Mice infected initially with DENV-2 and subsequently challenged with DENV-3 showed a higher frequency of CD8+ T cells producing TNF-α than animals that had had only primary infection with DENV-2 or DENV-3, suggesting that the primary DENV infection altered the secondary immune response. These data are similar to our previous results in BALB/c mice (26,27), as well as findings in natural secondary DENV infections in humans (4–6). Several studies have associated TNF-α levels with dengue disease severity, and T cells have been suggested to contribute to TNF-α production based on in vitro studies of antigen-stimulated T cells, as well ex vivo staining of PBMCs collected during acute illness (10,32,35–39). The potential for virus-specific T cells to have protective or detrimental effects in vivo, as seen in models of heterologous immunity involving unrelated viruses (40–42), needs further investigation, particularly in the context of the development and assessment of DENV vaccines (43). The identification of H-2b-restricted epitopes should facilitate further studies of the immunological mechanisms of heterologous DENV infection in the mouse model.
Acknowledgments
We would like to thank Dr. Sharone Green for her generous gift of the MHC knockout mice, and also Jurand Janus for the generation of virus stocks. We thank BEI Resources for providing overlapping DENV peptides that spanned all DENV proteins.
This work was supported by National Institutes of Health grant U19 AI057319, and used core resources supported by the Diabetes Endocrinology Research Center (DERC) grant DK32520 from the National Institute of Diabetes and Digestive and Kidney Diseases. A.L.R. is a member of the University of Massachusetts Medical School DERC (DK32520).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Godfrey DI. Stankovic S. Baxter AG. Developing NKT cells need their calcium. Nat Immunol. 2009;10:231–233. doi: 10.1038/ni0309-231. [DOI] [PubMed] [Google Scholar]
- 2.Burke DS. Nisalak A. Johnson DE. Scott RM. A prospective study of dengue infections in Bangkok. Am J Trop Med Hyg. 1988;38:172–180. doi: 10.4269/ajtmh.1988.38.172. [DOI] [PubMed] [Google Scholar]
- 3.Guzman MG. Kouri G. Martinez E, et al. Clinical and serologic study of Cuban children with dengue hemorrhagic fever/dengue shock syndrome (DHF/DSS) Bull Pan Am Health Organ. 1987;21:270–279. [PubMed] [Google Scholar]
- 4.Sangkawibha N. Rojanasuphot S. Ahandrik S, et al. Risk factors in dengue shock syndrome: a prospective epidemiologic study in Rayong, Thailand. I. The 1980 outbreak. Am J Epidemiol. 1984;120:653–669. doi: 10.1093/oxfordjournals.aje.a113932. [DOI] [PubMed] [Google Scholar]
- 5.Thein S. Aung MM. Shwe TN, et al. Risk factors in dengue shock syndrome. Am J Trop Med Hyg. 1997;56:566–572. doi: 10.4269/ajtmh.1997.56.566. [DOI] [PubMed] [Google Scholar]
- 6.Vaughn DW. Green S. Kalayanarooj S, et al. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J Infect Dis. 2000;181:2–9. doi: 10.1086/315215. [DOI] [PubMed] [Google Scholar]
- 7.Halstead SB. In vivo enhancement of dengue virus infection in rhesus monkeys by passively transferred antibody. J Infect Dis. 1979;140:527–533. doi: 10.1093/infdis/140.4.527. [DOI] [PubMed] [Google Scholar]
- 8.Halstead SB. Shotwell H. Casals J. Studies on the pathogenesis of dengue infection in monkeys. I. Clinical laboratory responses to primary infection. J Infect Dis. 1973;128:7–14. doi: 10.1093/infdis/128.1.7. [DOI] [PubMed] [Google Scholar]
- 9.Halstead SB. Shotwell H. Casals J. Studies on the pathogenesis of dengue infection in monkeys. II. Clinical laboratory responses to heterologous infection. J Infect Dis. 1973;128:15–22. doi: 10.1093/infdis/128.1.15. [DOI] [PubMed] [Google Scholar]
- 10.Bashyam HS. Green S. Rothman AL. Dengue virus-reactive CD8+ T cells display quantitative and qualitative differences in their response to variant epitopes of heterologous viral serotypes. J Immunol. 2006;176:2817–2824. doi: 10.4049/jimmunol.176.5.2817. [DOI] [PubMed] [Google Scholar]
- 11.Kurane I. Innis BL. Nisalak A, et al. Human T cell responses to dengue virus antigens. Proliferative responses and interferon gamma production. J Clin Invest. 1989;83:506–513. doi: 10.1172/JCI113911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathew A. Kurane I. Rothman AL, et al. Dominant recognition by human CD8+ cytotoxic T lymphocytes of dengue virus nonstructural proteins NS3 and NS1.2a. J Clin Invest. 1996;98:1684–1691. doi: 10.1172/JCI118964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Avirutnan P. Malasit P. Seliger B, et al. Dengue virus infection of human endothelial cells leads to chemokine production, complement activation, and apoptosis. J Immunol. 1998;161:6338–6346. [PubMed] [Google Scholar]
- 14.King CA. Anderson R. Marshall JS. Dengue virus selectively induces human mast cell chemokine production. J Virol. 2002;76:8408–8419. doi: 10.1128/JVI.76.16.8408-8419.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laoprasopwattana K. Libraty DH. Endy TP, et al. Antibody-dependent cellular cytotoxicity mediated by plasma obtained before secondary dengue virus infections: potential involvement in early control of viral replication. J Infect Dis. 2007;195:1108–1116. doi: 10.1086/512860. [DOI] [PubMed] [Google Scholar]
- 16.Shresta S. Kyle JL. Robert Beatty P. Harris E. Early activation of natural killer and B cells in response to primary dengue virus infection in A/J mice. Virology. 2004;319:262–273. doi: 10.1016/j.virol.2003.09.048. [DOI] [PubMed] [Google Scholar]
- 17.Shresta S. Kyle JL. Snider HM, et al. Interferon-dependent immunity is essential for resistance to primary dengue virus infection in mice, whereas T- and B-cell-dependent immunity are less critical. J Virol. 2004;78:2701–2710. doi: 10.1128/JVI.78.6.2701-2710.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shresta S. Sharar KL. Prigozhin DM, et al. Critical roles for both STAT1-dependent and STAT1-independent pathways in the control of primary dengue virus infection in mice. J Immunol. 2005;175:3946–3954. doi: 10.4049/jimmunol.175.6.3946. [DOI] [PubMed] [Google Scholar]
- 19.An J. Kimura-Kuroda J. Hirabayashi Y. Yasui K. Development of a novel mouse model for dengue virus infection. Virology. 1999;263:70–77. doi: 10.1006/viro.1999.9887. [DOI] [PubMed] [Google Scholar]
- 20.Bente DA. Melkus MW. Garcia JV. Rico-Hesse R. Dengue fever in humanized NOD/SCID mice. J Virol. 2005;79:13797–13799. doi: 10.1128/JVI.79.21.13797-13799.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang KJ. SY Li. Chen SC, et al. 2000 Manifestation of thrombocytopenia in dengue-2-virus-infected mice. J Gen Virol. 81:2177–2182. doi: 10.1099/0022-1317-81-9-2177. [DOI] [PubMed] [Google Scholar]
- 22.Johnson AJ. Roehrig JT. New mouse model for dengue virus vaccine testing. J Virol. 1999;73:783–786. doi: 10.1128/jvi.73.1.783-786.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mota J. Rico-Hesse R. Humanized mice show clinical signs of dengue fever according to infecting virus genotype. J Virol. 2009;83:8638–8645. doi: 10.1128/JVI.00581-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin YL. Liao CL. Chen LK, et al. Study of Dengue virus infection in SCID mice engrafted with human K562 cells. J Virol. 1998;72:9729–9737. doi: 10.1128/jvi.72.12.9729-9737.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Der Most RG. Murali-Krishna K. Ahmed R. Strauss JH. Chimeric yellow fever/dengue virus as a candidate dengue vaccine: quantitation of the dengue virus-specific CD8 T-cell response. J Virol. 2000;74:8094–8101. doi: 10.1128/jvi.74.17.8094-8101.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beaumier CM. Mathew A. Bashyam HS. Rothman AL. Cross-reactive memory CD8(+) T cells alter the immune response to heterologous secondary dengue virus infections in mice in a sequence-specific manner. J Infect Dis. 2008;197:608–617. doi: 10.1086/526790. [DOI] [PubMed] [Google Scholar]
- 27.Beaumier CM. Rothman AL. Cross-reactive memory CD4+ T cells alter the CD8+ T-cell response to heterologous secondary dengue virus infections in mice in a sequence-specific manner. Viral Immunol. 2009;22:215–219. doi: 10.1089/vim.2008.0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gessner A. Blum H. Rollinghoff M. Differential regulation of IL-9-expression after infection with Leishmania major in susceptible and resistant mice. Immunobiology. 1993;189:419–435. doi: 10.1016/S0171-2985(11)80414-6. [DOI] [PubMed] [Google Scholar]
- 29.Locksley RM. Heinzel FP. Sadick MD, et al. Murine cutaneous leishmaniasis: susceptibility correlates with differential expansion of helper T-cell subsets. Ann Inst Pasteur Immunol. 1987;138:744–749. doi: 10.1016/s0769-2625(87)80030-2. [DOI] [PubMed] [Google Scholar]
- 30.Yauch LE. Zellweger RM. Kotturi MF, et al. A protective role for dengue virus-specific CD8+ T cells. J Immunol. 2009;182:4865–4873. doi: 10.4049/jimmunol.0801974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mongkolsapaya J. Dejnirattisai W. Xu XN, et al. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat Med. 2003;9:921–927. doi: 10.1038/nm887. [DOI] [PubMed] [Google Scholar]
- 32.Mangada MM. Rothman AL. Altered cytokine responses of dengue-specific CD4+ T cells to heterologous serotypes. J Immunol. 2005;175:2676–2683. doi: 10.4049/jimmunol.175.4.2676. [DOI] [PubMed] [Google Scholar]
- 33.Jellison ER. Kim SK. Welsh RM. Cutting edge: MHC class II-restricted killing in vivo during viral infection. J Immunol. 2005;174:614–618. doi: 10.4049/jimmunol.174.2.614. [DOI] [PubMed] [Google Scholar]
- 34.Rothman AL. Kurane I. Lai CJ, et al. Dengue virus protein recognition by virus-specific murine CD8+ cytotoxic T lymphocytes. J Virol. 1993;67:801–806. doi: 10.1128/jvi.67.2.801-806.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Green S. Vaughn DW. Kalayanarooj S, et al. Early immune activation in acute dengue illness is related to development of plasma leakage and disease severity. J Infect Dis. 1999;179:755–762. doi: 10.1086/314680. [DOI] [PubMed] [Google Scholar]
- 36.Hober D. Poli L. Roblin B, et al. Serum levels of tumor necrosis factor-alpha (TNF-alpha), interleukin-6 (IL-6), and interleukin-1 beta (IL-1 beta) in dengue-infected patients. Am J Trop Med Hyg. 1993;48:324–331. doi: 10.4269/ajtmh.1993.48.324. [DOI] [PubMed] [Google Scholar]
- 37.Mangada MM. Endy TP. Nisalak A, et al. Dengue-specific T cell responses in peripheral blood mononuclear cells obtained prior to secondary dengue virus infections in Thai schoolchildren. J Infect Dis. 2002;185:1697–1703. doi: 10.1086/340822. [DOI] [PubMed] [Google Scholar]
- 38.Gagnon SJ. Ennis FA. Rothman AL. Bystander target cell lysis and cytokine production by dengue virus-specific human CD4(+) cytotoxic T-lymphocyte clones. J Virol. 1999;73:3623–3629. doi: 10.1128/jvi.73.5.3623-3629.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gagnon SJ. Mori M. Kurane I, et al. Cytokine gene expression and protein production in peripheral blood mononuclear cells of children with acute dengue virus infections. J Med Virol. 2002;67:41–46. doi: 10.1002/jmv.2190. [DOI] [PubMed] [Google Scholar]
- 40.Brehm MA. Pinto AK. Daniels KA, et al. T cell immunodominance and maintenance of memory regulated by unexpectedly cross-reactive pathogens. Nat Immunol. 2002;3:627–634. doi: 10.1038/ni806. [DOI] [PubMed] [Google Scholar]
- 41.Chen HD. Fraire AE. Joris I, et al. Memory CD8+ T cells in heterologous antiviral immunity and immunopathology in the lung. Nat Immunol. 2001;2:1067–1076. doi: 10.1038/ni727. [DOI] [PubMed] [Google Scholar]
- 42.Selin LK. Varga SM. Wong IC. Welsh RM. Protective heterologous antiviral immunity and enhanced immunopathogenesis mediated by memory T cell populations. J Exp Med. 1998;188:1705–1715. doi: 10.1084/jem.188.9.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Welsh RM. Fujinami RS. Pathogenic epitopes, heterologous immunity and vaccine design. Nat Rev Microbiol. 2007;5:555–563. doi: 10.1038/nrmicro1709. [DOI] [PMC free article] [PubMed] [Google Scholar]