Abstract
Transcription factors control eukaryotic polymerase II function by influencing the recruitment of multiprotein complexes to promoters and their subsequent integrated function. The complexity of the functional ‘transcriptosome’ has necessitated biochemical fractionation and subsequent protein sequencing on a grand scale to identify individual components. As a consequence, much is now known of the basal transcription complex. In contrast, less is known about the complexes formed at distal promoter elements. The c-fos SRE, for example, is known to bind Serum Response Factor (SRF) and ternary complex factors such as Elk-1. Their interaction with other factors at the SRE is implied but, to date, none have been identified. Here we describe the use of mass-spectrometric sequencing to identify six proteins, SRF, Elk-1 and four novel proteins, captured on SRE duplexes linked to magnetic beads. This approach is generally applicable to the characterisation of nucleic acid-bound protein complexes and the post-translational modification of their components.
INTRODUCTION
The unequivocal identification of proteins in DNA-bound multi-component complexes has presented a major problem in the study of transcription mechanisms. The electrophoretic resolution of DNA–protein complexes in native gels has provided an ubiquitous approach (1,2), whereby the identification of proteins relies on complex migration, interference patterns as the result of chemical modification of DNA (3) and the further retardation of complexes by specific antibodies (4). There are numerous shortcomings associated with these approaches: transcription factors belong to families whose members share similar properties; they exist in several closely related isoforms; they are frequently targets for post-translational modifications, all of which can affect their functional behaviour. Most importantly, these approaches are only applicable to protein complexes that remain stable during electrophoresis.
One alternative approach has been taken in which protein complexes have been allowed to form on modified DNA and then captured by precipitation (5,6). A further development of this strategy has allowed the capture of functionally active transcriptional complexes on immobilised DNA (7). In both these instances, proteins were identified by immunoblotting, which presumes some prior knowledge of participating components.
The de novo identification of proteins present in transcriptional complexes has been achieved primarily by biochemical purification on a scale unattainable by DNA-dependent capture and subsequent microsequencing (for examples, see 8,9). Current standards in mass-spectrometry (MS) overcome this quantitative problem. Once isolated in sub-picomole quantities, such complexes can be dissociated, their constituents resolved by SDS–PAGE and revealed by staining. Proteins can then be subjected to limited proteolytic cleavage and analysed by matrix-assisted laser desorption ionisation-time of flight (MALDI-TOF) MS. Peptide mass fingerprinting in this way may allow identification of proteins by comparison with simulated digests of expressed protein databases. However, with nano-electrospray technology, peptide sequence can be generated directly. Such ‘peptide sequence tags’ can be used to identify matching sequences in protein or DNA (including EST) databases or, in the increasingly unlikely event of failure, used as the basis for cDNA cloning strategies (10). In this way the constituents of a multiprotein complex can be identified unequivocally.
Here we have used MS to identify proteins isolated by a DNA pull-down strategy from Human Embryo Kidney (HEK) 293 cells using short oligonucleotide duplexes corresponding to the human c-fos SRE. We have identified several polypeptides bound to this site, including the previously characterised proteins Serum Response Factor (SRF) and Elk-1 (11–13). Furthermore, sequence alterations that increase the palindromic nature of the SRE allow three additional proteins to bind, including the human Dead Box protein DDX1, which is overexpressed in neuroblastoma and retinoblastoma cell lines (14,15), and two novel proteins, one of which bears similarity to the Escherichia coli RtcB gene product (16). From our observations we infer that these proteins may be recruited as a protein complex to cruciform DNA structures.
MATERIALS AND METHODS
Cell culture and extract preparation
HEK 293 and COS1 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% foetal calf serum. Cells were transfected with 3 µg of pCMV5-SRF or pCMV5-SRF-Chis and 6 µg of pCMV5-Elk-1(his) by the standard DNA–calcium phosphate coprecipitation procedure. Eighteen hours after transfection, cells were washed and serum-starved (0.2%) for 24 h. Cells were harvested in phosphate-buffered saline (PBS), after prior stimulation, where appropriate, with EGF (50 ng/ml) for 20 min and lysed in 20 mM Tris–HCl pH 8.0, 1 mM EDTA, 10% glycerol, 1 mM DTT, 0.2% NP-40, 1 mM benzamidine and 0.5 mM Na3VO4. After incubation on ice for 20 min, lysates were cleared by centrifugation.
Plasmids and oligonucleotides
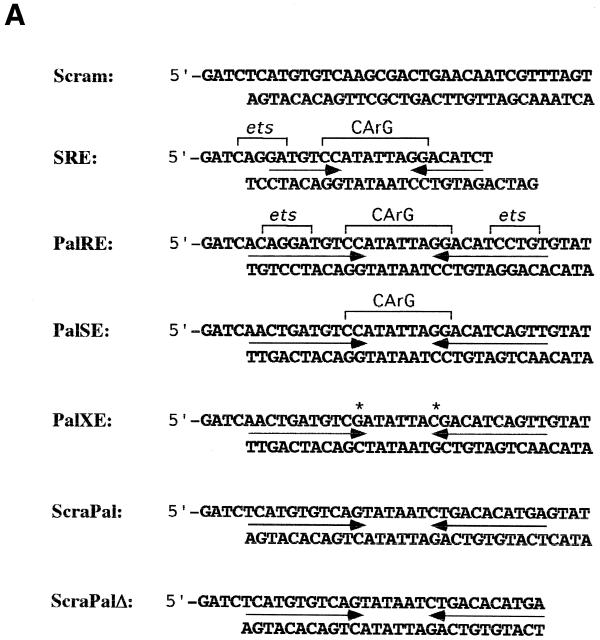
The construction of pBS-Elk-1, pCMV5-Elk-1(his), pCMV5-SRF and pCMV5-SRFChis has been described elsewhere (17; Strahl et al., manuscript submitted). The oligonucleotide pairs used to generate the Scrambled, SRE, PalRE, PalSE, PalXE, ScraPal and ScramPalΔ duplexes for the DNA pull-down assays are presented in Figure 2. All upper strands were 5′ biotinylated.
Figure 2.
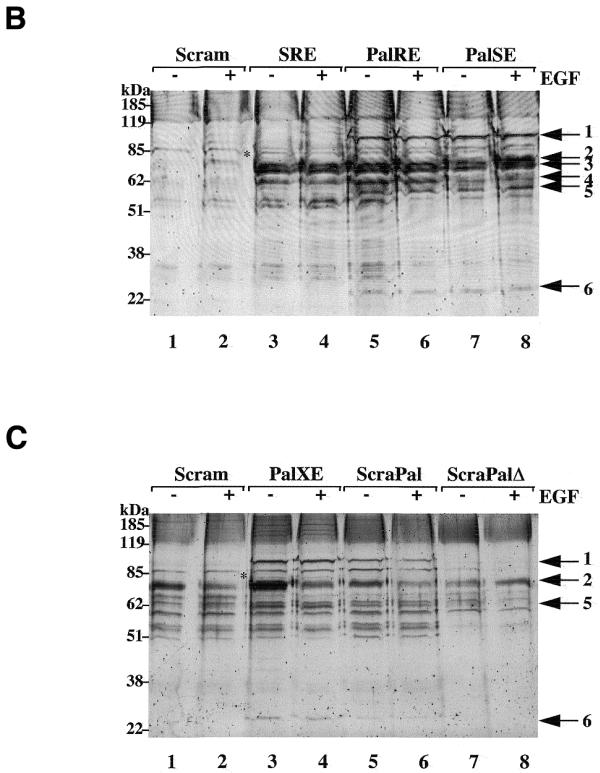
SRE complexes isolated from HEK 293 cell lysates. (A) Oligonucleotide sequences used in this study. Inverted arrows indicate the palindromic sequences while ets binding sites and CArG boxes are bracketed. Asterisks above the PalXE sequence indicate CArG box mutations. (B) Proteins isolated by scrambled (lanes 1 and 2), SRE (lanes 3 and 4), PalRE (lanes 5 and 6) and PalSE (lanes 7 and 8) duplexes from serum-starved (–) or EGF-treated (+) HEK 293 cells resolved by SDS–PAGE and stained with SYPRO ruby. Numbered arrows indicate the proteins analysed in this work. The asterisk between lanes 2 and 3 indicates the 80 kDa protein referred to in the text. (C) Proteins isolated by scrambled (lanes 1 and 2), PalXE (lanes 3 and 4), ScraPal (lanes 5 and 6) and ScraPalΔ (lanes 7 and 8) duplexes from serum-starved (–) or EGF-treated (+) HEK 293 cells. Labelling as in (B).
DNA pull-down assays
Dynabeads Streptavidin (Dynal A.S) were washed three times in buffer A (5 mM Tris pH 8.0, 0.5 mM EDTA, 1 M NaCl). Annealed oligonucleotides were incubated with beads (200 pmol/mg of beads) for 15 min at room temperature in buffer A. Beads were then washed twice with 500 µl buffer A, three times with 500 µl buffer C (20 mM Tris pH 8.0, 1 mM EDTA, 10% glycerol, 1 mM DTT, 50 mM NaCl) and resuspended in buffer C at 10 mg/ml. DNA-bound complexes were isolated as follows: 250 µg beads, bound to the SRE duplex, was incubated in buffer C with 0.1 mg/ml poly(dI-dC), 0.6 mg/ml herring sperm DNA, 1 mM spermidine, 1% BSA, 2 µg coreSRF (18) and various amounts of radiolabelled Elk-1, for 15 min at room temperature. Beads were washed three times in 500 µl buffer C and eluted in 1 M NaCl. Eluted proteins were then analysed by SDS–PAGE and visualised with a Fuji phosphorimaging system.
Purification of proteins from cell lysates was performed essentially as described above, except that 600 µg of whole cell extract was incubated with 1 mg of beads bound to the appropriate oligonucleotides. Samples were analysed by SDS–PAGE and stained with SYPRO Ruby (Bio-Rad) or Coomassie blue.
Mass spectrometry
In-gel tryptic digestion of proteins was performed according to a published protocol (19). Tryptic peptides were micro-purified and sequenced on a QSTAR quadrupole-ToF mass spectrometer (Sciex, Ontario, Canada) equipped with a Protana nanoelectrospray source (Protana, Odense, Denmark) and a Q-ToF2 (micromass UK, Wythenshawe, UK). The data were processed with PPSS2.2 (Protana’s Proteomics Software Suite) or micromass UK software and the peptide sequence tags obtained were queried against non-redundant sequence databases (20).
Protein biotinylation assay
COS1 cells transfected with an expression vector for a C-terminal his-tagged version of SRF were lysed in 50 mM Tris pH 8.0, 150 mM NaCl, 0.1% NP-40, 0.05% sodium deoxycholate, 0.1% SDS and incubated with Nickel-Agarose (Qiagen) for 1 h at 4°C (20 µl slurry per 500 µl lysate). Thereafter the beads were applied to a small column, washed four times with 10 vol of 50 mM imidazole pH 6.8, 500 mM NaCl, 0.5% NP-40 and equilibrated in PBS. Bound proteins were biotinylated as previously described (21) by incubation with NHS-LC biotin (Pierce) for 30 min at 4°C (0.5 mg/ml in PBS). The reaction was stopped with DMEM (10 min, 4°C) and beads were subsequently washed again in 20 vol of 50 mM imidazole pH 6.8, 500 mM NaCl, 0.5% NP-40. Proteins were eluted in 3 column vols of 200 mM imidazole, 150 mM NaCl. After SDS–PAGE and electrotransfer to nitrocellulose, biotinylated proteins were visualised with peroxidase-coupled streptavidin and chemiluminescence (ECL, Amersham).
Duplex cleavage by S1 nuclease
Ten picomoles of non-biotinylated oligonucleotide was labelled by incubation in polynucleotide kinase (PNK) buffer with 1.1 MBq [γ-32P]ATP, 4 mM spermidine and 20 U PNK in a final volume of 20 µl at 37°C for 30 min. The complementary oligonucleotide was annealed to this in a final volume of 40 µl and 15 µl was then incubated with an equal volume of 2× buffer C for 15 min at room temperature. Ten volumes of ice-cold S1 buffer containing 10 mg/ml sheared herring sperm DNA was added and samples were incubated with S1 nuclease (25 U) for 1 h at 15°C. Finally, the DNA was precipitated and loaded onto a 15% sequencing gel. Gels were imaged using a Fuji phosphorimaging system.
RESULTS
Ternary complex formation on SRE duplex linked to streptavidin beads
Oligonucleotide duplexes were prepared with a single, covalently-linked biotin moiety and bound to Dynabead Streptavidin (Dynal A.S). Their ability to recruit protein complexes in a sequence-specific manner was initially confirmed with recombinant proteins. As shown in Figure 1, an SRE duplex bound 35S-labelled Elk-1 only in the presence of bacterially-expressed coreSRF (18), indicating that the specificity of ternary complex formation by SRF and Elk-1 observed previously in electrophoretic mobility shift assays is maintained in solution (13,17,18,22).
Figure 1.

Elk-1 and SRF binding to the SRE. A biotinylated SRE duplex bound to streptavidin-conjugated magnetic beads was incubated with increasing amounts of 35S-labelled Elk-1 in the presence (+) or absence (–) of bacterially-expressed coreSRF. Beads were collected, washed and Elk-1 binding was monitored by SDS–PAGE and autoradiography.
To isolate DNA-bound complexes from eukaryotic cells, whole cell extracts were prepared from HEK 293 cells and incubated with beads to which various oligonucleotide duplexes had been attached. As a means of enhancing formation of complexes containing SRF and Elk-1, both proteins were over-expressed in the cells by prior transfection of appropriate expression vectors. After incubation with lysates, beads were collected, washed and the bound proteins eluted and analysed by SDS–PAGE.
Several proteins appear to bind non-specifically to the conjugates, as judged by their presence in samples collected by an oligonucleotide duplex consisting of a scrambled sequence (Fig. 2, lanes 1 and 2). These proteins may have an affinity for double-stranded DNA or may recognise a cryptic sequence element, as streptavidin-coated beads alone yielded a single protein with an estimated molecular weight of 68 kDa (not shown). This protein was subsequently shown to be BSA, with which beads were incubated to suppress other non-specific protein interactions. In contrast, a SRE duplex isolated a number of additional proteins (lanes 3 and 4), which, for the most part, were bound in extracts from both serum-starved and EGF-treated cells. Most prominent among these were three polypeptides with apparent molecular weights in the range 60–70 kDa, which embraces those of BSA (68 kDa), SRF (67 kDa) and Elk-1 (62 kDa).
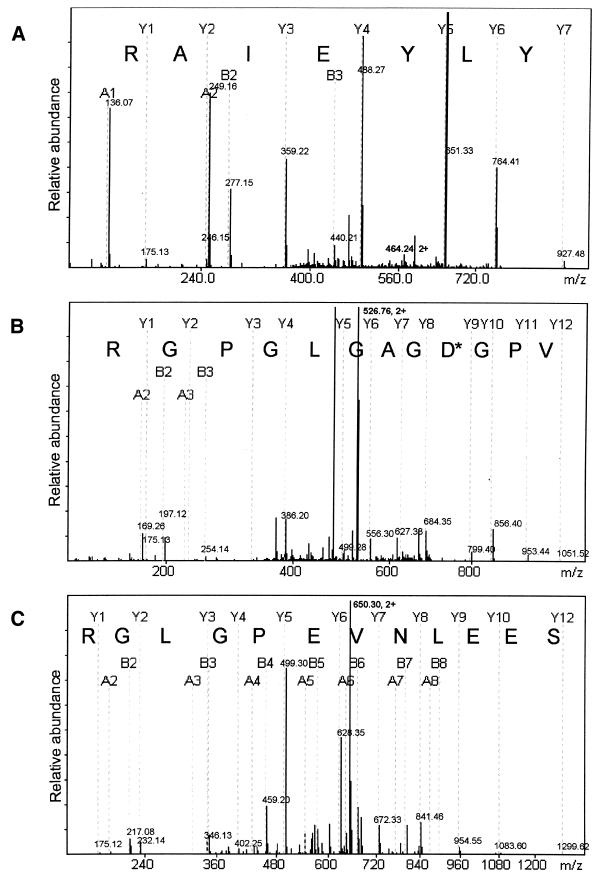
Bands were excised from the gel shown in Figure 2B, subjected to tryptic digestion and analysed by nano-electrospray tandem MS. Bands labelled 2, 3 and 4 each generated several doubly charged ions that generated unambiguous peptide sequence upon analysis by MS-MS (Fig. 3 and Table 1). Moreover, comparison of peptide masses from MS and computer generated tryptic digests confirmed the identify of several other charged peptides. Thus, bands 2, 3 and 4 could be identified as serum albumin, SRF and Elk-1, respectively. The presence of SRF and Elk-1 in the samples collected by SRE duplexes but not in samples collected by the scrambled duplex control underlines the specificity of this approach. As explained above, the presence of serum albumin in the samples is also expected.
Figure 3.
MS-derived sequence for BSA, SRF and Elk-1. The spectra show the derivative ions from doubly charged ions derived from (A) sample 2 (m/z = 464.24), (B) sample 3 (m/z = 525.76) and (C) sample 4 (m/z = 650.30) (see Fig. 2). Peptide sequences are generated from Y ions whereby Y1 corresponds to the C-terminal amino acid, in each case an arginine, consistent with their origin from tryptic digestion. In the case of SRF, the peptide shown differs from the retrieved sequence by one amino acid (asterisk). This difference is interpreted as a deamidation of N47.
Table 1. Synopsis of MS data and proteins identified .
| Sample |
MWa |
Status |
Tryptic peptidesb |
% coveredc |
Protein identified |
| 1 | 90/81 795 | MS-MS | DLGLAFEIPPHMK | 8 | GI:539572 DDX1 DEAD Box Protein 1 |
| FNFGEEEFKFPPK | |||||
| ELAEQTLNNIK* | |||||
| ELLIIGGVAAR | |||||
| MHNQIPQVTSDGKR | |||||
| MDQAIIFCR | |||||
| 2 | 68/71 320 | MS-MS | YLYEIAR* | 6 | GI:3336842 serum albumin (BSA) |
| LGEYGFQNALIVR | |||||
| KVPQVSTPTLVEVSR | |||||
| RPCFSALTPDETYVPK | |||||
| 3 | 67/51 649 | MS-MS | VPGDGAGLGPGR* | 6 | GI:134876 SRF Serum Response Factor |
| IKMEFIDNK | |||||
| ALIQTCLNSPDSPPRS | |||||
| 4 | 62/45 086 | MS-MS | PAVVLPNAAPAGAAAPPSGSRS | 19 | GI:119291 Elk-1 ETS-Domain Protein |
| SEELNVEPGLGR* | |||||
| GFVPETTK | |||||
| AEPEVPPQEGVPAR | |||||
| GFVPETTKAEPEVPPQEGVPAR | |||||
| LPAVVMDTAGQAGGHAASSPEISQPQK | |||||
| DLELPLSPSLLGGPGPE | |||||
| 5 | 55/55 724 | MS-MS | SYNDELQ | 13 | GI:4886424 Hypothet. 55.2 kDa Protein |
| LMFEELR | |||||
| GLGHQVATDALVAMEK | |||||
| IASPEGQDYLK* | |||||
| NLDFQDVLDK | |||||
| NVTDVVNTCHDAGISK | |||||
| 6 | 24/28 182 | MS-MS | NAEPLINLDVNNPDFK* | 20 | GI:4929667 CGI-99 Protein |
| KAGVMALANLLQIQR | (GI:417719 Ribosomal RS3) | ||||
| HDDYLVMLK | (GI:1350997 Ribosomal RS4) | ||||
| EGLPVALDK |
aEstimated/calculated molecular weights.
bSequence data presented for peptides denoted with an asterisk.
cMaximum coverage from a single sample.
In the case of Elk-1, we also detected an unpredicted sequence ion (m/z = 964.0) which, upon MS-MS analysis, was identified as a phosphopeptide corresponding to Elk-1 phosphorylated on serine 324. As the sample in question derived from lysates of EGF-treated cells, this observation confirms previous data showing that Elk-1 phosphorylated on serine 324 is present in mitogen-stimulated cells (17,23,24). However, the presence of the non-phosphorylated peptide in the same sample indicates that phosphorylation of S324 may be sub-stoichiometric.
Novel proteins associated with ternary complex at SRE
SRF and Elk-1 were not the only proteins that interacted specifically with the SRE duplex. An additional protein (molecular weight ∼80 kDa) was found to associate strongly and reproducibly with the SRE (Fig. 2, lanes 3 and 4). This protein could associate either by virtue of direct DNA interactions or through protein–protein contacts with SRF, or possibly Elk-1. In order to distinguish between these two possibilities, we generated oligonucleotide duplexes containing the CArG box to which SRF binds but lacking an ets binding site (PalSE), or lacking the CArG box and ets binding site (PalXE; Fig. 2A). Both duplexes include similar flanking palindromic sequences. The PalSE duplex captured both SRF and the 80 kDa protein (Fig. 2B, lanes 7 and 8), whereas the PalXE bound neither (Fig. 2C, lanes 3 and 4), suggesting that SRF, but not Elk-1, is important for recruitment of the 80 kDa protein.
We also sought some other indication of proteins interacting directly with SRF. To this end, SRF with a C-terminal histidine tag (SRF-Chis) was overexpressed in cells and isolated from lysates by nickel-affinity chromatography. Proteins bound on the column were biotinylated, prior to elution and separation by SDS–PAGE. Biotinylated proteins were then transferred to nitrocellulose and visualised with peroxidase-coupled streptavidin and chemiluminescence. As shown in Figure 4, this approach led to the labelling of a single 80 kDa protein in addition to SRF. This protein was also detected when a his-tagged mutant of SRF (M9) lacking several phosphorylation sites was used (lane 2) (25) but not in the absence of SRF-Chis (lane 3). Taken together, these results suggest that the 80 kDa protein associated with the SRE may have bound via interactions with SRF. The identity of the 80 kDa protein and the exact nature of its interaction with the SRE are currently under further investigation.
Figure 4.
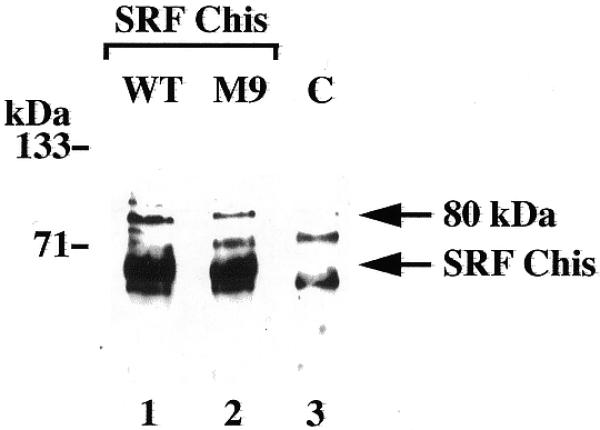
Co-purification of an 80 kDa protein with SRF. Lysates prepared from untransfected COS1 cells (C) or cells expressing a C-terminal his-tagged version of SRF (WT) or SRF-M9 (M9), were incubated with nickel-agarose beads. Bound proteins were labelled with biotin, eluted and visualised with peroxide-coupled streptavidin after SDS–PAGE and electrotransfer to nitrocellulose. Arrows indicate SRF and the co-purified 80 kDa protein.
It has previously been shown that the introduction of a second ets binding site at the SRE promotes the formation of a symmetrical quaternary complex containing two Elk-1 molecules and that the formation of this complex, which may involve Elk-1 dimerisation, correlates with SRE activity (26,27). We therefore reasoned that a corresponding oligonucleotide duplex would be more effective than an SRE at recruiting Elk-1 and consequently proteins with which Elk-1 might interact. As shown in Figure 2 (lanes 5 and 6), several additional proteins are detected among those captured by an SRE with two ets sites (PalRE), with molecular weights of 90, 58 and 28 kDa. These three proteins were present in complexes isolated from starved or EGF-treated cell lysates.
The importance of Elk-1 and SRF in the recruitment of the 90, 58 and 28 kDa proteins could also be gauged from their capture by other oligonucleotide duplexes. As can be seen in Figure 2B, lanes 7 and 8, the PalSE captured less Elk-1 than either the SRE (lanes 3 and 4) or PalRE (lanes 5 and 6) although some Elk-1 was found to be present, perhaps reflecting the strength of direct interactions with SRF (28). The PalXE captured neither Elk-1 nor SRF (Fig. 2C, lanes 3 and 4). However, the PalRE, PalSE and PalXE all captured similar levels of the 90, 58 and 28 kDa proteins, suggesting that their presence in the complex is not dependent on SRF or Elk-1 but on some other criterion.
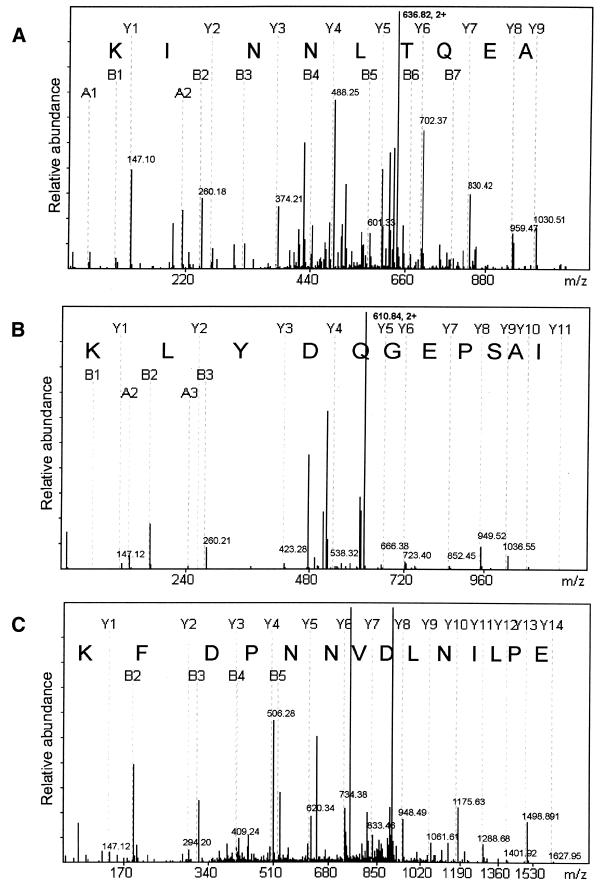
The three proteins were subjected to MS and all gave rise to multiple sequence ions allowing their unambiguous identification (Fig. 5). Of the three, only the 90 kDa protein, a dead box protein called DDX1, has been studied previously (14,15). Both the 58 and 28 kDa proteins correspond to hypothetical proteins of unidentified function, identified by six and five peptide sequences, respectively. However, BLAST searches reveal numerous orthologues of each protein in Caenorhabditis elegans, Drosophila melanogaster, Saccharomyces cerevisiae and several bacteria and archibacteria. It should be noted that the 28 kDa samples also contained three peptides corresponding to tryptic digestion products of human ribosomal proteins RS3 and RS4. These results are summarised in Table 1.
Figure 5.
MS-derived sequences of DDX1, p55.2 and CG1-99. The spectra show derivative ions from doubly charged ions derived from (A) sample 1 (m/z = 636.82), (B) sample 5 (m/z = 610.84) and (C) sample 6 (m/z =906.96) (see Fig. 2). Peptide sequences are generated from Y ions whereby Y1 corresponds to the C-terminal amino acid, in each case a lysine, consistent with their origin from tryptic digestion.
As the binding of these proteins appeared to be indpendent of SRF and Elk-1, we sought an alternative explanation for their presence. In the case of the PalRE duplex, the introduction of a second ets binding site to the SRE increases the length of palindromic sequence from 7 to 11 bp. Similarly, the PalSE, which lacks ets binding sites, and the PalXE, which also lacks a CArG box, both contain extended palindromes of 11 bp. Thus it seemed feasible that the 90, 58 and 28 kDa proteins that bind to the PalRE, PalSE and PalXE recognise a conformational element rather than a sequence motif, the most likely being a stem–loop structure or cruciform.
To test this possibility, palindromic versions of the scrambled duplex were generated. ScramPal is able to form a cruciform similar to those formed by PalRE, PalSE and PalXE. In contrast, ScramPalΔ lacks sequences beyond the palindrome (Fig. 2A) and can yield a biotinylated stem–loop but no cruciform. As shown in Figure 2C, lanes 5–8, the 90, 58 and 28 kDa proteins bind to ScramPal but not to ScramPalΔ, indicating that they recognise a cruciform rather than a stem–loop structure.
To assess cruciform formation by the SRE, PalRE and PalSE, we compared their susceptibility to cleavage by S1 nuclease, a single-strand-specific endonuclease. As shown in Figure 6A, all three duplexes yield S1 cleavage products of 16–19 nt, indicating that in all three duplexes, the A/T-rich core of the CArG box has single-strand character. This suggests that all three have the propensity to form cruciforms. However, the SRE is more resistant to cleavage by S1 nuclease, as judged by the intensity of the bands corresponding to the uncleaved duplexes (Fig. 6B). This result suggests that the PalRE and PalSE duplexes more readily form S1 sensitive structures, such as cruciforms, than the SRE duplex, which could account for their ability to bind the 90, 58 and 28 kDa proteins. Alternatively, as the SRE also appears to have some single-strand character under our experimental conditions, the length of the stem may be the governing factor rather than cruciform formation per se.
Figure 6.
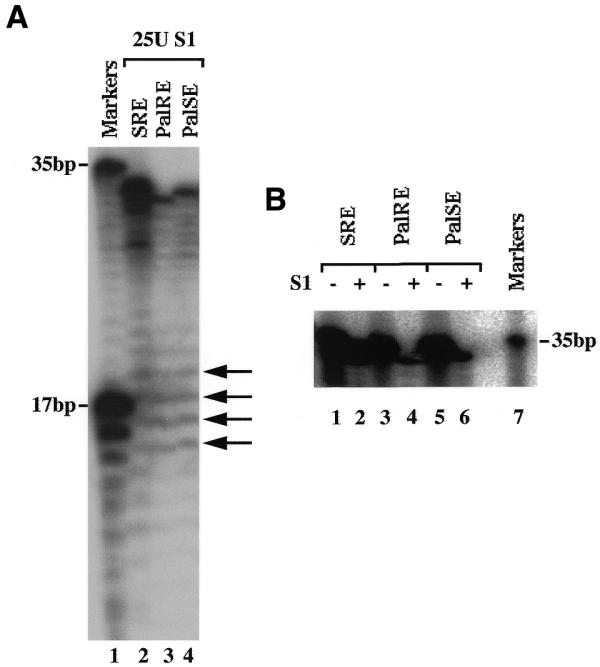
S1 nuclease sensitivity of SRE duplexes. (A) DNA duplexes 32P-labelled at one end were incubated with S1 nuclease and the digestion products resolved on a 15% sequencing gel. Markers correspond to two oligonucleotides of 35 and 17 nt similarly end-labelled. Arrows indicate S1 cleavage within the A/T-rich CArG box. (B) Full-length duplexes before (–) and after (+) S1 digestion.
Notably, in lysates from untransfected cells, the yield of these three proteins bound to the PalRE and PalSE was higher (our unpublished observations), suggesting that increased levels of SRF in the lysates may serve to suppress cruciform formation by these DNA duplexes.
Taken together, our results show that the combined approach of DNA affinity capture and mass-spectrometric protein sequencing allows the unambiguous identification of proteins that participate in the formation of multi-component complexes on DNA through the interaction with specific sequence elements or conformational structures.
DISCUSSION
Eukaryotic transcription involves the establishment of a large, basal transcription factor complex on gene promoters which controls the activity of RNA pol II and is regulated by additional, distally bound transcription factors. While many of the components of the basal transcription machinery have been identified, cloned and studied in varying detail, much less is known about distal, regulatory transcription factor complexes. What is known is that, directly or indirectly, they aid in the recruitment of components such as histone acetyl transferases, deacetylases, chromatin remodelling engines and other activities that modulate transcriptional events (29,30). Here we have used a direct approach towards identifying novel components of such complexes and describe the interaction of six proteins with sequences derived from the human c-fos SRE.
The precipitation of DNA-bound protein complexes for further analysis was first described in the identification of Fos and Jun as components of AP-1 (5,6). Subsequently, an approach has been developed to allow the isolation of pre-initiation complexes on linear DNA templates, which can subsequently be assayed for transcriptional activity (7). Given that the binding of SRF and ternary complex factors such as Elk-1 to the c-fos SRE is well established (12,13,18,22,31), our initial aim was to generate a ternary complex in vitro with purified components and use it to capture additional proteins from cell lysates. Although we were able to achieve the former qualitatively, as demonstrated in Figure 1, we were unable to ensure quantitative formation of ternary complexes on immobilised DNA. We believe this may reflect the highly dynamic interaction of Elk-1 with SRF and the SRE (32). We therefore adopted an alternative approach to enhancing ternary complex formation by overexpressing both SRF and Elk-1 in cells from which lysates were subsequently prepared.
Under our binding conditions we observed that complex capture was not affected by the length of DNA duplex flanking the target element (within the range of 5–10 bp) or by the presence of flush or overhanging duplex ends, but was dependent on DNA sequence and conformation. This suggests that the approach is valid for proteins involved in other aspects of nucleic acid function besides transcription. Indeed, other DNA- and RNA-binding proteins have been isolated by related procedures.
The novelty and analytical power of our approach lies in the subsequent mass-spectrometric analysis of captured complexes. On the basis of their expected complexity, we opted to separate complexes by single dimension SDS–PAGE, rather than on 2-D gels, and analyse individual polypeptides separately. Samples cut from several different gels, stained either with Coomassie blue or SYPRO ruby, gave very similar results, underlying the inherent reproducibility of the approach. In retrospect, having established the efficiency of the procedure, the bulk processing of complexes and chromatographic separation of derivative peptides prior to MS appears to be a realistic option.
Rather than rely on the identification of proteins by tryptic peptide mass fingerprinting, for which single MS (MALDI-TOF) would be appropriate, we predicted that the derivation of peptide sequence information generated by tandem MS-MS would allow us to identify novel proteins and, more importantly, the modification of proteins binding to the SRE. This proved to be the case on both counts. First, two samples yielded several peptide sequences that failed to identify any protein in the currently available databases (data not shown). Secondly, we identified and sequenced one peptide derived from Elk-1 phosphorylated on S324, a site previously found to be modified in mitogen-stimulated cells (17,23,24).
In addition to SRF and Elk-1, we identified an 80 kDa protein that interacted with the SRE, PalRE and PalSE. As Elk-1 binding to the PalSE is compromised, this protein was presumed to bind either to the SRE directly or through interactions with SRF. However, point mutations in the CArG box that abolish SRF binding (PalXE), also prevented capture of the 80 kDa protein, suggesting that its interaction with the SRE required SRF. Moreover, affinity purification and biotinylation of SRF and co-purified proteins (21) revealed a single protein of 80 kDa associated with SRF but absent from control preparations. All these observations are consistent with the notion that the 80 kDa protein associates with SRE duplexes on the basis of its interaction with SRF. The 80 kDa protein is presently the subject of further analysis and will be described in more detail elsewhere.
Three proteins, of 90, 55 and 28 kDa, bound specifically to the PalRE, PalSE and PalXE, but not to the SRE. They were identified unequivocally as DDX1, a hypothetical 55.2 kDa protein (p55.2) and CGI-99. DDX1 is a member of the DEAD Box family of proteins, identified as RNA helicases, whereby the eponymous DEAD box (asp-glu-ala-asp) forms a conserved motif in the molecules’ ATPase domains (33). DDX1 is reported to be amplified in some neuroblastoma and retinoblastoma cell lines (14,15) and may play a role in the development of neural tissue. The p55.2 protein has numerous orthologues in eukaryotic and prokaryotic organisms. It is structurally related to E.coli rtcB, a protein of unknown function that forms part of the RNA 3′-terminal phosphate cyclase operon and for which at least one other human relative has been cloned (16). The 28 kDa protein CGI-99 is similar to the chicken protein CLE7 and other related hypothetical proteins have been identified within the genomes of D.melanogaster, C.elegans and S.cerevisiae, although no function has been ascribed to any of them.
As two of these proteins have relatives implicated in RNA catalysis, their interaction with DNA duplexes is intriguing. Although there is no evidence available to suggest that DDX1 or other DEAD Box helicases act on DNA, some DEAD-box proteins do show promiscuity in their substrates, either for helicase activity or for nucleic acid-stimulated ATPase activity (P.Linder, personal communication). Given their specific interaction with some but not all duplexes, the three proteins must recognise a distinct sequence or structural element. By a process of elimination, this was revealed to be an inverted repeat, separated by a 6 bp A/T-rich core corresponding to that of the CArG box to which SRF binds. Moreover, duplexes unable to form cruciforms failed to capture the three proteins. We also found no evidence of sequence specificity.
The cruciform character of the PalRE and PalSE duplexes is indicated by their susceptibility to cleavage and degradation by the single strand-specific S1 nuclease. We infer that they possess stronger cruciform character than the SRE, because they are cleaved by S1 more readily, whereafter they are unstable and digested. An alternative explanation for the failure of the SRE duplex to bind the 90, 55 and 28 kDa proteins may be that the palindrome is only 7 bp in length and too short, whereas in the case of the PalRE and PalSE the stems are 11 bp long and may accommodate protein binding.
When comparing the resolved complexes from Elk-1 and SRF-transfected cell lysates with those from untransfected cells, the only obvious difference, apart from the levels of SRF and Elk-1, was that the yield of the three proteins binding specifically to the PalRE and PalSE was higher from untransfected cell lysates (data not shown). This suggests that increased levels of SRF in the lysates may serve to suppress cruciform formation by these DNA duplexes. As the level of the 80 kDa protein did not appear to change with SRF expression (not shown), it may be expressed at limiting levels in HEK 293 cells.
The consistent and reproducible capture of DDX1, p55.2 and CGI-99 suggests that they may exist together in a functional complex. In the absence of appropriate reagents we have been unable to explore this possibility. At present we also do not know what role this trio of proteins, if any, may play in transcriptional regulation. Given that DDX1 is potentially an ATP-dependent helicase, it may be involved in the resolution of DNA perturbations arising due to changes in superhelical stress during transcription (34). Nonetheless, it remains feasible that their capture in association with a palindromic promoter sequence may be entirely fortuitous. For example, they may recognise and act on Holliday junctions that occur as intermediates during replication (35). However, the binding of many transcription factors as dimers or multimers to palindromic sites suggests that these proteins may play a role in the maintenance of promoter conformation during transcriptional activity.
Acknowledgments
ACKNOWLEDGEMENTS
We thank John Keyte and Kevin Bailey for oligonucleotide synthesis and Jackie Bostock for assistance with the manuscript. This work was supported by a grant to P.E.S. from the The Wellcome Trust.
References
- 1.Fried M. and Crothers,D.M. (1981) Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res., 9, 6505–6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garner M.M. and Revzin,A. (1981) A gel electrophoresis method for quantifying the binding of protein to specific DNA regions: Application to components of the E. coli lactose operon regulatory system. Nucleic Acids Res., 9, 3047–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shaw P.E. and Stewart,A.F. (1993) Identification of Protein-DNA Contacts with Dimethyl Sulphate. Humana, Totowa, NJ.
- 4.Roberts M.S., Boundy,A., O’Hare,P., Pizzorno,M.C., Ciufo,D.M. and Hayward,G.S. (1988) Direct correlation between a negative autoregulatory response element at the cap site of the herpes simplex virus type 1 IE175 (alpha 4) promoter and a specific binding site for the IE175 (ICP4) protein. J. Virol., 62, 4307–4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franza B.R., Rauscher,F.J.,III, Josephs,S.F. and Curran,T. (1988) The Fos complex and Fos-related antigens recognize sequence elements that contain AP-1 binding sites. Science, 239, 1150–1153. [DOI] [PubMed] [Google Scholar]
- 6.Rauscher F.J.,III, Cohen,D.R., Curren,T., Bos,T.J., Vogt,P.K., Bohmann,D., Tjian,R. and Franza,R.J. (1988) Fos-associated protein p39 is the product of the jun proto-oncogene. Science, 240, 1010–1016. [DOI] [PubMed] [Google Scholar]
- 7.Roberts S.G. and Green,M.R. (1996) Purification and analysis of functional preinitiation complexes. Methods Enzymol., 273, 110–118. [DOI] [PubMed] [Google Scholar]
- 8.Dynlacht B.D., Hoey,T. and Tjian,R. (1991) Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell, 66, 563–576. [DOI] [PubMed] [Google Scholar]
- 9.Kretzschmar M., Kaiser,K., Lottspeich,F. and Meisterernst,M. (1994) A Novel Mediator of Class II Gene Transcription with Homology to Viral Immediate-Early Transcriptinal Regulators. Cell, 78, 525–534. [DOI] [PubMed] [Google Scholar]
- 10.Blackstock W.P. and Weir,M.P. (1999) Proteomics: quantitative and physical mapping of cellular proteins. Tibtech, 17, 121–127. [DOI] [PubMed] [Google Scholar]
- 11.Norman C., Runswick,M., Pollock,R. and Treisman,R. (1988) Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds the c-fos serum response element. Cell, 55, 989–1003. [DOI] [PubMed] [Google Scholar]
- 12.Herrera R.E., Shaw,P.E. and Nordheim,A. (1989) Occupation of the c-fos serum response element in vivo by a multi-protein complex is unaltered by growth factor induction. Nature, 340, 68–70. [DOI] [PubMed] [Google Scholar]
- 13.Hipskind R.A., Rao,V.N., Mueller,C.G.F., Reddy,E.S.P. and Nordheim,A. (1991) Ets-related protein Elk-1 is homologous to the c-fos regulatory factor p62TCF. Nature, 354, 531–534. [DOI] [PubMed] [Google Scholar]
- 14.Godbout R. and Squire,J. (1993) Amplification of a DEAD box protein gene in retinoblastoma cell lines. Proc. Natl Acad. Sci. USA, 90, 7578–7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Godbout R., Packer,M. and Wenjun,B. (1998) Overexpression of a DEAD box protein (DDX1) in neuroblastoma and retinoblastoma cell lines. J. Biol. Chem., 273, 21161–21168. [DOI] [PubMed] [Google Scholar]
- 16.Genschik P., Drabikowski,K. and Filipowicz,W. (1998) Characterisation of the Escherichia coli RNA 3′-Terminal Phosphate Cyclase and its σ54-Regulated Operon. J. Biol. Chem., 273, 25516–25526. [DOI] [PubMed] [Google Scholar]
- 17.Gille H., Kortenjann,M., Thomae,O., Moomaw,C., Slaughter,C., Cobb,M.H. and Shaw,P.E. (1995) ERK phosphorylation potentiates ELK-1-mediated ternary complex formation and transactivation. EMBO J., 14, 951–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gille H., Sharrocks,A.D. and Shaw,P.E. (1992) Phosphorylation of transcription factor p62TCF by MAP kinase stimulates ternary complex formation at c-fos promoter. Nature, 358, 414–417. [DOI] [PubMed] [Google Scholar]
- 19.Shevchenko A., Wilm,M., Vorm,O. and Mann,M. (1996) Mass Spectrometric Sequencing of Proteins from Silver-Stained Polyacrylamide Gels. Anal. Chem., 68, 850–858. [DOI] [PubMed] [Google Scholar]
- 20.Mann M. and Wilm,M.S. (1994) Error Tolerant Identification of Peptides in Sequence Databases by Peptide Sequence Tags. Anal. Chem., 66, 4390–4399. [DOI] [PubMed] [Google Scholar]
- 21.Kim K.-M., Adachi,T., Nielsen,P.J., Terashima,M., Lamers,M.C., Köhler,G. and Reth,M. (1994) Two new proteins preferentially associated with membrane immunoglobulin D. EMBO J., 13, 3793–3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaw P.E., Schröter,H. and Nordheim,A. (1989) The ability of a ternary complex to form over the serum response element correlates with serum inducibility of the human c- fos promoter. Cell, 56, 563–572. [DOI] [PubMed] [Google Scholar]
- 23.Marais R., Wynne,J. and Treisman,R. (1993) The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell, 73, 381–393. [DOI] [PubMed] [Google Scholar]
- 24.Janknecht R., Ernst,W.H., Pingoud,V. and Nordheim,A. (1993) Activation of ternary complex factor ELK-1 by MAP kinases. EMBO J., 12, 5097–5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janknecht R., Hipskind,R.A., Houthaeve,T., Nordheim,A. and Stunnenberg,H.G. (1992) Identification of multiple SRF N-terminal phosphorylation sites affecting DNA binding properties. EMBO J., 11, 1045–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gille H., Kortenjann,M., Strahl,T. and Shaw,P.E. (1996) Phosphorylation-Dependent Formation of a Quaternary Complex at the c-fos SRE. Mol. Cell. Biol., 16, 1094–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drewett V., Muller,S., Goodall,J. and Shaw,P.E. (2000) Dimer formation by ternary complex factor Elk-1. J. Biol. Chem., 275, 1757–1762. [DOI] [PubMed] [Google Scholar]
- 28.Shore P. and Sharrocks,A.D. (1994) The transcription factors Elk-1 and serum response factor interact by direct protein-protein contacts mediated by a short region of Elk-1. Mol. Cell. Biol., 14, 3283–3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Struhl K. (1998) Histone acetylation and transcriptional regulatory mechanisms. Genes Dev., 12, 599–606. [DOI] [PubMed] [Google Scholar]
- 30.Kadonaga J.T. (1998) Eukaryotic Transcription: An Interlaced Network of Transcription Factors and Chromatin-Modifying Machines. Cell, 92, 307–313. [DOI] [PubMed] [Google Scholar]
- 31.Treisman R. (1987) Identification and purification of a polypeptide that binds to the c-fos serum response element. EMBO J., 6, 2711–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janknecht R. and Nordheim,A. (1992) Elk-1 protein domains required for direct and SRF-assisted DNA-binding. Nucleic Acids Res., 20, 3317–3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Linder P. and Daugeron,M.-C. (2000) Are DEAD-box proteins becoming respectable helicases? Nature Struct. Biol., 7, 97–99. [DOI] [PubMed] [Google Scholar]
- 34.Liu L.F. and Wang,J.C. (1987) Supercoiling of the DNA Template During Transcription. Proc. Natl Acad. Sci. USA, 84, 7024–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGlynn P. and Lloyd,R.G. (2000) Modulation of RNA Polymerase by (p)ppGpp Reveals a RecG-Dependent Mechanism for Replication Fork Progression. Cell, 101, 35–45. [DOI] [PubMed] [Google Scholar]