Abstract
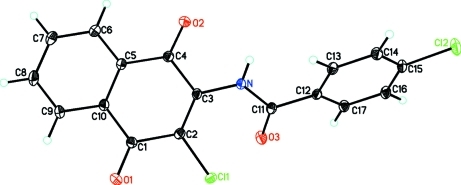
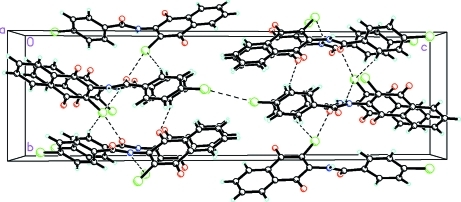
The naphthoquinone ring is almost perpendicular [dihedral angle 71.02 (3)°] to the phenyl group of the title compound, C17H9Cl2NO3, while the dihedral angle between the amide group and the 4-chlorophenyl ring is 21.9 (2)°. The conformation of the N—H and C=O bonds are anti to each other. N—H⋯Cl hydrogen bonds link the molecules into chains in the a-axis direction. In addition, these chains are linked by weak intermolecular C—H⋯O interactions.
Related literature
For similar structures see: Lien et al. (1997 ▶); Huang et al. (2005 ▶); Bakare et al. (2003 ▶); Copeland et al. (2007 ▶); Win et al. (2005 ▶); Rubin-Preminger et al. (2004 ▶). For related literature, see: Gowda, Kožíšek et al. (2008 ▶); Gowda, Tokarčík et al. (2008 ▶); van Oosten et al. (2008 ▶); Shen et al. (2008 ▶).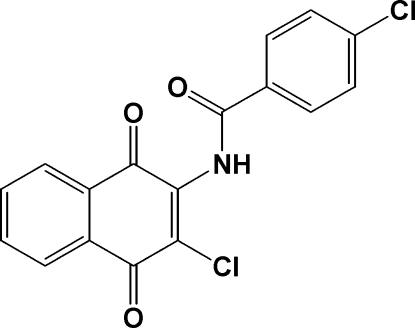
Experimental
Crystal data
C17H9Cl2NO3
M r = 346.15
Monoclinic,
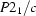
a = 5.6011 (2) Å
b = 8.7237 (3) Å
c = 29.7957 (9) Å
β = 93.504 (3)°
V = 1453.16 (8) Å3
Z = 4
Mo Kα radiation
μ = 0.46 mm−1
T = 200 (2) K
0.49 × 0.41 × 0.12 mm
Data collection
Oxford Diffraction Gemini R diffractometer
Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2007 ▶) T min = 0.887, T max = 1.000 (expected range = 0.839–0.946)
13882 measured reflections
4842 independent reflections
2832 reflections with I > 2σ(I)
R int = 0.035
Refinement
R[F 2 > 2σ(F 2)] = 0.038
wR(F 2) = 0.086
S = 0.93
4842 reflections
208 parameters
H-atom parameters constrained
Δρmax = 0.29 e Å−3
Δρmin = −0.36 e Å−3
Data collection: CrysAlis CCD (Oxford Diffraction, 2007 ▶); cell refinement: CrysAlis RED (Oxford Diffraction, 2007 ▶); data reduction: CrysAlis RED; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808040993/at2690sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808040993/at2690Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N—H0A⋯Cl1i | 0.88 | 2.89 | 3.6491 (12) | 145 |
| C14—H14A⋯O2ii | 0.95 | 2.40 | 3.2517 (19) | 149 |
Symmetry codes: (i) 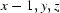 ; (ii)
; (ii) 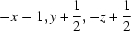 .
.
Acknowledgments
RJB acknowledges the Laboratory for the Structure of Matter at the Naval Research Laboratory for access to their diffractometers.
supplementary crystallographic information
Comment
The amido and imido derivatives of 3-chloro-1,4-naphthoquinone are well known for their anti-inflammatory, antiplatelet, antiallergic and anticancer activities (Lien et al., 1997; Huang et al., 2005; Bakare et al., 2003; Copeland et al., 2007). The title compound, 2-chloro-3-(p-chlorobenzamido)-1,4-naphthoquinone was obtained as an intermediate in the synthesis of some oxazolo-1,4-naphthoquinone and imido-substituted-1,4-naphthoquinone analogs.
The naphthoquinone ring is almost perpendicular to the phenyl group of the title compound C17H9Cl2NO3, while the dihedral angle betwen the amide group and the 4-chlorophenyl ring is 21.9 (2)° (Fig. 1). The conformation of the N—H and C=O bonds are anti to each other (Gowda, Kožíšek et al., 2008; Gowda, Tokarčík et al., 2008). N—H···Cl hydrogen bonds link the molecules into chains in the a direction. In addition, these chains are linked by weak intermolecular Ar—H···O interactions (Fig. 2, Table 1).
Experimental
A mixture of 2-amino-3-chloro-1,4-naphthoquinone (213 mg, 1.03 mmol) and 4-chloro-benzoylchloride (2 ml) was refluxed for 2 1/2 h (powerstat setting at 70). The reaction mixture was cooled to room temperature. The precipitate was isolated by vacuum filtration and the yellow-grey solid was washed with diethyl ether. The crude was recrystallized from ethanol (20 ml) to obtain a yellow solid (67 mg, 18.8%). Crystals for x-ray study were obtained by recrystallization from methanol.
Refinement
All H atoms were placed in geometrically idealized positions and constrained to ride on their parent atoms with C—H = 0.95 Å, N—H = 0.88 Å and Uiso(H) = 1.2Ueq(C, N).
Figures
Fig. 1.
View of (I) showing the atom-labelling scheme. Displacement ellipsoids are drawn at the 20% probability level.
Fig. 2.
View of the packing viewed down the a axis. Dashed bonds show weak C—H···O interactions.
Crystal data
| C17H9Cl2NO3 | F(000) = 704 |
| Mr = 346.15 | Dx = 1.582 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 4629 reflections |
| a = 5.6011 (2) Å | θ = 4.6–32.5° |
| b = 8.7237 (3) Å | µ = 0.46 mm−1 |
| c = 29.7957 (9) Å | T = 200 K |
| β = 93.504 (3)° | Plate, pale yellow |
| V = 1453.16 (8) Å3 | 0.49 × 0.41 × 0.12 mm |
| Z = 4 |
Data collection
| Oxford Diffraction Gemini R diffractometer | 4842 independent reflections |
| Radiation source: fine-focus sealed tube | 2832 reflections with I > 2σ(I) |
| graphite | Rint = 0.035 |
| Detector resolution: 10.5081 pixels mm-1 | θmax = 32.6°, θmin = 4.6° |
| φ and ω scans | h = −8→8 |
| Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2007) | k = −12→12 |
| Tmin = 0.887, Tmax = 1.000 | l = −44→44 |
| 13882 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.038 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.086 | H-atom parameters constrained |
| S = 0.93 | w = 1/[σ2(Fo2) + (0.0416P)2] where P = (Fo2 + 2Fc2)/3 |
| 4842 reflections | (Δ/σ)max = 0.001 |
| 208 parameters | Δρmax = 0.29 e Å−3 |
| 0 restraints | Δρmin = −0.36 e Å−3 |
Special details
| Experimental. (CrysAlis RED; Oxford Diffraction, 2007) Empirical absorption correction using spherical harmonics, implemented in SCALE3 ABSPACK scaling algorithm. |
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | 0.42210 (6) | 0.13331 (4) | 0.298854 (11) | 0.02796 (10) | |
| Cl2 | −0.42944 (7) | −0.05032 (6) | 0.057164 (13) | 0.04852 (14) | |
| O1 | 0.54644 (18) | 0.07511 (14) | 0.39281 (4) | 0.0401 (3) | |
| O2 | −0.26101 (17) | −0.20745 (14) | 0.33569 (3) | 0.0378 (3) | |
| O3 | 0.31583 (17) | −0.12832 (14) | 0.24103 (3) | 0.0364 (3) | |
| N | −0.02512 (19) | −0.06128 (15) | 0.27365 (4) | 0.0273 (3) | |
| H0A | −0.1786 | −0.0415 | 0.2689 | 0.033* | |
| C1 | 0.3653 (2) | 0.00688 (18) | 0.38026 (5) | 0.0275 (3) | |
| C2 | 0.2705 (2) | 0.01309 (17) | 0.33244 (4) | 0.0251 (3) | |
| C3 | 0.0712 (2) | −0.06238 (17) | 0.31773 (4) | 0.0240 (3) | |
| C4 | −0.0749 (2) | −0.14777 (18) | 0.34975 (5) | 0.0264 (3) | |
| C5 | 0.0144 (2) | −0.15604 (18) | 0.39758 (5) | 0.0272 (3) | |
| C6 | −0.1138 (3) | −0.2369 (2) | 0.42817 (5) | 0.0391 (4) | |
| H6A | −0.2591 | −0.2864 | 0.4186 | 0.047* | |
| C7 | −0.0289 (3) | −0.2451 (3) | 0.47288 (5) | 0.0477 (5) | |
| H7A | −0.1169 | −0.3002 | 0.4939 | 0.057* | |
| C8 | 0.1823 (3) | −0.1737 (2) | 0.48705 (5) | 0.0477 (5) | |
| H8A | 0.2390 | −0.1794 | 0.5177 | 0.057* | |
| C9 | 0.3111 (3) | −0.0940 (2) | 0.45660 (5) | 0.0391 (4) | |
| H9A | 0.4574 | −0.0459 | 0.4663 | 0.047* | |
| C10 | 0.2273 (2) | −0.08383 (18) | 0.41170 (5) | 0.0287 (3) | |
| C11 | 0.1070 (3) | −0.08964 (17) | 0.23680 (5) | 0.0263 (3) | |
| C12 | −0.0252 (2) | −0.07098 (17) | 0.19218 (4) | 0.0245 (3) | |
| C13 | −0.2316 (2) | 0.01736 (18) | 0.18635 (5) | 0.0277 (3) | |
| H13A | −0.2882 | 0.0726 | 0.2110 | 0.033* | |
| C14 | −0.3550 (3) | 0.02492 (19) | 0.14462 (5) | 0.0315 (3) | |
| H14A | −0.4966 | 0.0846 | 0.1406 | 0.038* | |
| C15 | −0.2697 (3) | −0.05531 (19) | 0.10907 (5) | 0.0307 (3) | |
| C16 | −0.0608 (3) | −0.1395 (2) | 0.11357 (5) | 0.0319 (3) | |
| H16A | −0.0020 | −0.1913 | 0.0885 | 0.038* | |
| C17 | 0.0618 (3) | −0.14706 (19) | 0.15551 (5) | 0.0295 (3) | |
| H17A | 0.2058 | −0.2045 | 0.1592 | 0.035* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.02827 (17) | 0.0286 (2) | 0.02758 (17) | −0.00541 (15) | 0.00608 (13) | 0.00212 (15) |
| Cl2 | 0.0514 (2) | 0.0680 (4) | 0.02503 (18) | 0.0176 (2) | −0.00744 (17) | −0.0029 (2) |
| O1 | 0.0364 (6) | 0.0500 (8) | 0.0334 (6) | −0.0141 (6) | −0.0034 (5) | −0.0042 (5) |
| O2 | 0.0321 (5) | 0.0490 (8) | 0.0318 (6) | −0.0152 (5) | −0.0010 (5) | 0.0023 (5) |
| O3 | 0.0328 (6) | 0.0476 (8) | 0.0288 (5) | 0.0120 (5) | 0.0007 (4) | −0.0072 (5) |
| N | 0.0240 (6) | 0.0383 (8) | 0.0196 (5) | 0.0012 (5) | 0.0019 (5) | 0.0013 (5) |
| C1 | 0.0266 (7) | 0.0307 (9) | 0.0253 (7) | −0.0004 (6) | 0.0021 (6) | −0.0031 (6) |
| C2 | 0.0272 (7) | 0.0247 (8) | 0.0239 (7) | 0.0006 (6) | 0.0058 (6) | −0.0012 (6) |
| C3 | 0.0252 (6) | 0.0278 (8) | 0.0192 (6) | 0.0026 (6) | 0.0028 (5) | −0.0013 (6) |
| C4 | 0.0278 (7) | 0.0268 (8) | 0.0248 (7) | −0.0014 (6) | 0.0030 (6) | −0.0008 (6) |
| C5 | 0.0294 (7) | 0.0302 (9) | 0.0220 (6) | 0.0000 (6) | 0.0023 (6) | 0.0005 (6) |
| C6 | 0.0374 (8) | 0.0517 (12) | 0.0286 (7) | −0.0087 (8) | 0.0041 (7) | 0.0049 (8) |
| C7 | 0.0502 (10) | 0.0654 (14) | 0.0282 (8) | −0.0078 (9) | 0.0083 (7) | 0.0117 (9) |
| C8 | 0.0532 (10) | 0.0680 (15) | 0.0214 (7) | 0.0003 (10) | −0.0008 (7) | 0.0049 (8) |
| C9 | 0.0374 (8) | 0.0562 (12) | 0.0232 (7) | −0.0047 (8) | −0.0026 (6) | −0.0018 (7) |
| C10 | 0.0296 (7) | 0.0348 (9) | 0.0218 (6) | 0.0004 (6) | 0.0023 (6) | −0.0028 (6) |
| C11 | 0.0306 (7) | 0.0258 (8) | 0.0228 (7) | 0.0018 (6) | 0.0033 (6) | −0.0012 (6) |
| C12 | 0.0275 (7) | 0.0260 (8) | 0.0201 (6) | −0.0020 (6) | 0.0034 (5) | 0.0002 (6) |
| C13 | 0.0321 (7) | 0.0282 (8) | 0.0233 (7) | 0.0034 (6) | 0.0060 (6) | −0.0017 (6) |
| C14 | 0.0293 (7) | 0.0361 (9) | 0.0291 (7) | 0.0071 (7) | 0.0022 (6) | 0.0014 (7) |
| C15 | 0.0359 (8) | 0.0359 (9) | 0.0202 (6) | 0.0015 (7) | −0.0008 (6) | 0.0018 (7) |
| C16 | 0.0347 (8) | 0.0400 (10) | 0.0215 (7) | 0.0064 (7) | 0.0051 (6) | −0.0032 (7) |
| C17 | 0.0292 (7) | 0.0348 (9) | 0.0246 (7) | 0.0052 (6) | 0.0035 (6) | −0.0009 (6) |
Geometric parameters (Å, °)
| Cl1—C2 | 1.7105 (15) | C7—C8 | 1.380 (2) |
| Cl2—C15 | 1.7394 (14) | C7—H7A | 0.9500 |
| O1—C1 | 1.2154 (17) | C8—C9 | 1.381 (2) |
| O2—C4 | 1.2166 (16) | C8—H8A | 0.9500 |
| O3—C11 | 1.2167 (16) | C9—C10 | 1.3932 (19) |
| N—C11 | 1.3834 (18) | C9—H9A | 0.9500 |
| N—C3 | 1.3890 (15) | C11—C12 | 1.4905 (19) |
| N—H0A | 0.8800 | C12—C17 | 1.392 (2) |
| C1—C10 | 1.480 (2) | C12—C13 | 1.392 (2) |
| C1—C2 | 1.4907 (18) | C13—C14 | 1.3867 (19) |
| C2—C3 | 1.3461 (19) | C13—H13A | 0.9500 |
| C3—C4 | 1.494 (2) | C14—C15 | 1.379 (2) |
| C4—C5 | 1.4829 (19) | C14—H14A | 0.9500 |
| C5—C6 | 1.387 (2) | C15—C16 | 1.381 (2) |
| C5—C10 | 1.391 (2) | C16—C17 | 1.3903 (19) |
| C6—C7 | 1.389 (2) | C16—H16A | 0.9500 |
| C6—H6A | 0.9500 | C17—H17A | 0.9500 |
| C11—N—C3 | 123.62 (11) | C8—C9—C10 | 120.27 (15) |
| C11—N—H0A | 118.2 | C8—C9—H9A | 119.9 |
| C3—N—H0A | 118.2 | C10—C9—H9A | 119.9 |
| O1—C1—C10 | 121.71 (13) | C5—C10—C9 | 119.60 (14) |
| O1—C1—C2 | 121.20 (14) | C5—C10—C1 | 121.47 (12) |
| C10—C1—C2 | 117.08 (12) | C9—C10—C1 | 118.91 (13) |
| C3—C2—C1 | 122.21 (13) | O3—C11—N | 121.65 (12) |
| C3—C2—Cl1 | 122.74 (11) | O3—C11—C12 | 123.01 (13) |
| C1—C2—Cl1 | 114.90 (10) | N—C11—C12 | 115.34 (12) |
| C2—C3—N | 124.82 (13) | C17—C12—C13 | 119.63 (12) |
| C2—C3—C4 | 120.79 (12) | C17—C12—C11 | 118.00 (12) |
| N—C3—C4 | 114.27 (12) | C13—C12—C11 | 122.36 (12) |
| O2—C4—C5 | 122.80 (13) | C14—C13—C12 | 120.19 (13) |
| O2—C4—C3 | 119.01 (12) | C14—C13—H13A | 119.9 |
| C5—C4—C3 | 118.19 (12) | C12—C13—H13A | 119.9 |
| C6—C5—C10 | 120.03 (13) | C15—C14—C13 | 119.16 (13) |
| C6—C5—C4 | 119.89 (13) | C15—C14—H14A | 120.4 |
| C10—C5—C4 | 120.08 (13) | C13—C14—H14A | 120.4 |
| C5—C6—C7 | 119.71 (15) | C14—C15—C16 | 121.78 (13) |
| C5—C6—H6A | 120.1 | C14—C15—Cl2 | 119.11 (11) |
| C7—C6—H6A | 120.1 | C16—C15—Cl2 | 119.11 (11) |
| C8—C7—C6 | 120.49 (16) | C15—C16—C17 | 118.79 (14) |
| C8—C7—H7A | 119.8 | C15—C16—H16A | 120.6 |
| C6—C7—H7A | 119.8 | C17—C16—H16A | 120.6 |
| C7—C8—C9 | 119.89 (14) | C16—C17—C12 | 120.37 (13) |
| C7—C8—H8A | 120.1 | C16—C17—H17A | 119.8 |
| C9—C8—H8A | 120.1 | C12—C17—H17A | 119.8 |
| O1—C1—C2—C3 | 179.54 (14) | C6—C5—C10—C1 | −178.31 (15) |
| C10—C1—C2—C3 | −1.3 (2) | C4—C5—C10—C1 | 2.1 (2) |
| O1—C1—C2—Cl1 | −4.8 (2) | C8—C9—C10—C5 | −0.8 (3) |
| C10—C1—C2—Cl1 | 174.35 (11) | C8—C9—C10—C1 | 177.93 (16) |
| C1—C2—C3—N | −179.82 (14) | O1—C1—C10—C5 | 177.09 (15) |
| Cl1—C2—C3—N | 4.8 (2) | C2—C1—C10—C5 | −2.0 (2) |
| C1—C2—C3—C4 | 4.4 (2) | O1—C1—C10—C9 | −1.6 (2) |
| Cl1—C2—C3—C4 | −170.99 (11) | C2—C1—C10—C9 | 179.29 (14) |
| C11—N—C3—C2 | 49.6 (2) | C3—N—C11—O3 | 5.2 (2) |
| C11—N—C3—C4 | −134.37 (14) | C3—N—C11—C12 | −175.37 (13) |
| C2—C3—C4—O2 | 175.91 (14) | O3—C11—C12—C17 | 21.9 (2) |
| N—C3—C4—O2 | −0.3 (2) | N—C11—C12—C17 | −157.54 (14) |
| C2—C3—C4—C5 | −4.1 (2) | O3—C11—C12—C13 | −159.00 (15) |
| N—C3—C4—C5 | 179.64 (12) | N—C11—C12—C13 | 21.6 (2) |
| O2—C4—C5—C6 | 1.2 (2) | C17—C12—C13—C14 | 2.4 (2) |
| C3—C4—C5—C6 | −178.73 (15) | C11—C12—C13—C14 | −176.72 (14) |
| O2—C4—C5—C10 | −179.23 (15) | C12—C13—C14—C15 | −0.4 (2) |
| C3—C4—C5—C10 | 0.8 (2) | C13—C14—C15—C16 | −1.9 (2) |
| C10—C5—C6—C7 | 0.1 (3) | C13—C14—C15—Cl2 | 178.10 (12) |
| C4—C5—C6—C7 | 179.64 (16) | C14—C15—C16—C17 | 2.1 (2) |
| C5—C6—C7—C8 | −0.2 (3) | Cl2—C15—C16—C17 | −177.92 (13) |
| C6—C7—C8—C9 | −0.3 (3) | C15—C16—C17—C12 | 0.0 (2) |
| C7—C8—C9—C10 | 0.7 (3) | C13—C12—C17—C16 | −2.2 (2) |
| C6—C5—C10—C9 | 0.4 (2) | C11—C12—C17—C16 | 176.94 (14) |
| C4—C5—C10—C9 | −179.17 (15) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N—H0A···Cl1i | 0.88 | 2.89 | 3.6491 (12) | 145 |
| C14—H14A···O2ii | 0.95 | 2.40 | 3.2517 (19) | 149 |
Symmetry codes: (i) x−1, y, z; (ii) −x−1, y+1/2, −z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: AT2690).
References
- Bakare, O., Ashendel, C. L., Peng, H., Zalkow, L. H. & Burgess, E. M. (2003). Bioorg. Med. Chem.11, 3165–3170. [DOI] [PubMed]
- Copeland, R. L., Das, J. R., Bakare, O., Enwerem, N. M., Berhe, S., Hillaire, K., White, D., Beyene, D., Kassim, O. O. & Kanaan, Y. M. (2007). Anticancer Res.27, 1537–1546. [PubMed]
- Gowda, B. T., Kožíšek, J., Tokarčík, M. & Fuess, H. (2008). Acta Cryst. E64, o987. [DOI] [PMC free article] [PubMed]
- Gowda, B. T., Tokarčík, M., Kožíšek, J., Sowmya, B. P. & Fuess, H. (2008). Acta Cryst. E64, o950. [DOI] [PMC free article] [PubMed]
- Huang, L., Chang, F., Lee, K., Wang, J., Teng, C. & Kuo, S. (2005). Bioorg. Med. Chem.6, 2261–2269. [DOI] [PubMed]
- Lien, J., Huang, L., Wang, J., Teng, C., Lee, K. & Kuo, S. (1997). Bioorg. Med. Chem.5, 2111–2120. [DOI] [PubMed]
- Oosten, E. M. van, Lough, A. J. & Vasdev, N. (2008). Acta Cryst. E64, o1005. [DOI] [PMC free article] [PubMed]
- Oxford Diffraction (2007). CrysAlis CCD and CrysAlis RED (including SCALE3 ABSPACK). Oxford Diffraction Ltd, Abingdon, England.
- Rubin-Preminger, J. M., Win, T., Granot, Y. & Bittner, S. (2004). Z. Kristallogr. New Cryst. Struct.219, 323–324.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Shen, Q., Yu, S.-Q., Hu, B.-B. & Lu, P. (2008). Acta Cryst. E64, o996. [DOI] [PMC free article] [PubMed]
- Win, T., Yerushalmi, S. & Bittner, S. (2005). Synthesis, pp. 1631–1634.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808040993/at2690sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808040993/at2690Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report