Abstract
The HIV-1 transcript is alternatively spliced to over 30 different mRNAs. Whether RNA secondary structure can influence HIV-1 RNA alternative splicing has not previously been examined. Here we have determined the secondary structure of the HIV-1/BRU RNA segment, containing the alternative A3, A4a, A4b, A4c and A5 3′ splice sites. Site A3, required for tat mRNA production, is contained in the terminal loop of a stem–loop structure (SLS2), which is highly conserved in HIV-1 and related SIVcpz strains. The exon splicing silencer (ESS2) acting on site A3 is located in a long irregular stem–loop structure (SLS3). Two SLS3 domains were protected by nuclear components under splicing condition assays. One contains the A4c branch points and a putative SR protein binding site. The other one is adjacent to ESS2. Unexpectedly, only the 3′ A residue of ESS2 was protected. The suboptimal A3 polypyrimidine tract (PPT) is base paired. Using site-directed mutagenesis and transfection of a mini-HIV-1 cDNA into HeLa cells, we found that, in a wild-type PPT context, a mutation of the A3 downstream sequence that reinforced SLS2 stability decreased site A3 utilization. This was not the case with an optimized PPT. Hence, sequence and secondary structure of the PPT may cooperate in limiting site A3 utilization.
INTRODUCTION
Retroviruses transcribe their RNA from an integrated proviral genome. The unspliced primary transcript has two functions after transport to the cytoplasm: (i) as messenger RNA for the production of viral proteins, (ii) as genomic RNA which is packaged into newly synthesized virions. HIV-1 is a complex retrovirus of the lentivirus class. Due to the presence of four splicing donor sites (5′ss) and eight splicing acceptor sites (3′ss) in HIV-1 RNA, the splicing machinery in the infected cell generates at least 40 distinct mRNAs from the unique RNA primary transcript (1). The relative abundance of these mRNAs depends greatly upon the relative efficiencies of splicing at the 3′ss. All of these 3′ss are suboptimal (2–7). Five 3′ss (A3, A4c, A4a, A4b and A5), located in a small central portion of the viral RNA, are in competition with one another. All tat mRNAs are spliced at site A3, the rev mRNAs are spliced at sites A4a, A4b or A4c and most nef and env mRNAs are spliced at site A5 (1; for review see 8). These five central 3′ss are all used in the early phase after infection for production of the Tat, Rev and Nef proteins.
The steady-state level of double-spliced tat mRNA is considerably lower than the levels of double-spliced rev and single-spliced env/vpu mRNAs in both lymphoid and non-lymphoid cells (1). It is possible that the tat mRNA is less stable than the rev and env mRNAs. Alternatively, the A3 3′ss may be less efficient than the other downstream 3′ss. Based on studies of the A3 3′ss (2,3,9), the latter explanation appears to be more likely. Indeed, improvement of the efficiency of the A3 3′ss strongly increases the level of the tat mRNAs in transfected HeLa cells (10). All HIV-1 splicing sites have a suboptimal polypyrimidine tract (PPT). Surprisingly, in spite of the low splicing efficiency at site A3, its PPT is more uridine-rich than any of the other 3′ss in the central region of the HIV-1 genome. Nevertheless, the A3 PPT sequence is one of the parameters that limits A3 3′ss efficiency, since increasing the number of U residues in the A3 PPT increases the use of site A3 in vitro (9). A downstream exon splicing silencer (ESS2) is located 64 nt downstream from site A3 (2,9) and inhibits an early step of spliceosome assembly (3). ESS2 also inhibits splicing when the PPT of the upstream 3′ss is optimized but to a smaller extent (9). This means that the observed low efficiency of the A3 3′ss results from a combined effect of the presence of a suboptimal PPT and ESS2 element. ESS2 shows an inhibitory effect when inserted into a heterologous retroviral context (downstream from Rous sarcoma virus src gene 3′ss). In this heterologous context, the effect of ESS2 was found to be distance dependent (2) and a 10 nt sequence of ESS2 was sufficient for the inhibition. However, the magnitude of ESS2 inhibition in this heterologous context was reduced compared to its effect in its natural context downstream from the A3′ss (9), suggesting that in this natural context, additional sequences or the RNA secondary structure may reinforce ESS2 inhibitory property. The hnRNP A/B proteins were recently proposed to bind ESS2 element and appear to be involved in the observed inhibition (11). However, the precise mechanism of inhibition of spliceosome assembly by ESS2 element is still unknown.
The secondary structure of the HIV-1 RNA region containing site A3 and ESS2 has not been previously studied. It may also contribute to the low efficiency of splicing at site A3. Indeed, several studies have shown an effect of RNA secondary structure on alternative splicing in cellular pre-messenger RNAs and some viral mRNAs. In some cases, RNA secondary structure acts as a splicing enhancer by decreasing physical distances between splice sites (12–15), or by maintaining one or more cis-acting regions of pre-mRNA in a favorable configuration for interaction with trans-acting factors (16–19). However, in several other cases, sequestration of 5′ss or 3′ss was found to inhibit or to limit splicing site utilization (20–22).
To test for a potential role of stem–loop structures in determining the splicing efficiency at the HIV-1 A3 3′ss, we studied the secondary structure of the HIV-1 RNA region containing site A3 and ESS2, for the BRU and the MAL HIV-1 strains. We also developed conditions to investigate RNA secondary structure in the presence of nuclear extract. Because the A3 3′ss was found to be located in a stem–loop structure that is highly conserved in HIV-1 and related SIVcpz strains, we tested whether the stability of this stem–loop structure influences splicing at site A3.
MATERIALS AND METHODS
Plasmid construction
Plasmid pBRU3 (23) was used as the source of cDNA sequences from the HIV-1 BRU/LAI strain (GenBank accession no. K02013). PCR amplifications were done according to Nour et al. (24). Three distinct HIV-1/BRU cDNA fragments, H1, H2 and H3, corresponding to regions 1–385, 5155–5408 and 5155–5626, respectively, were prepared (numbering according to Ratner et al., 25). PCR amplifications of fragments H1, H2 and H3 were performed using pairs of specific DNA primers (O-605/O-606, O-567/O-608, O-567/O-521, respectively) (Table 1).
Table 1. The oligonucleotides used in this study.
| Primer
number |
Sequence (5′-3′) |
Orientation |
Restriction
site |
HIV-1 BRU sequence |
HIV-1 MAL sequence |
Utilization |
| O-605 | TATTCTGGATCCGGTCTCTCTGGT | i | BamHI | 1–12 | PCR | |
| O-606 | TATTCTAGATCTCCCATCGATCTA | c | BglII | 373–385 | PCR | |
| O-567 | AGAAGACCAAGATCTAGAGGGAGCCA | i | BglII | 5155–5165 and 5172–5182 | 5151–5158 and 5165–5175 | PCR |
| O-608 | TATTCTCTGAGTTGCTCTCCTCT | c | PstI | 5397–5408 | PCR primer extension | |
| O-521 | TATTCTCTGCAGTGCTTTGATAGA | c | PstI | 5615–5626 | PCR | |
| O-1458 | TATTCTCTGCAGTTGCTCTTCTCT | c | PstI | 5390–5401 | PCR primer extension | |
| O-1459 | TATTCTCTGCAGATACTACTTACTGCTC | c | 5615–5630 | PCR | ||
| O-998 | TCTAATACGACTCACTATAGGGCGACATAGCAGAATAGGCG | i | 5369–5387 | PCR | ||
| O-954 | TATTCTCTGCAGGATCCGGCAATGAAAGC | c | BamHI | 5503–5514 | PCR | |
| O-825 | AATTCTGCAACAACTttcGTTTATttcTTTCAGAATTGGGTG | i | 5326–5340 and 5353–5367 | Site-directed mutagenesis | ||
| O-826 | CCATTTCAGAATTGGaTaaACAGCAGAATAGGCGTT | i | 5350–5364 and 5375–5389 | Site-directed mutagenesis | ||
| O-994 | ttcTTTCAGAATgaaaTaaACgaaAGAATAGGCGTT | i | 5353–5361 and 5378–5389 | Site-directed mutagenesis | ||
| O-548 | GGCAATGAAAGCAAC | c | 5500–5514 | Primer extension | ||
| O-831 | GAGGTCTTCGTCGCTG | c | 5571–5585 | Primer extension |
The number, sequence and utilization of each oligonucleotide are given. Sequences identical (i) or complementary (c) to HIV-1 regions are underlined. Nucleotide positions of these HIV-1 RNA regions are numbered according to the GenBank accession no. K02013 (HIV-1/BRU) and X04415 (HIV-1/MAL). Restriction sites introduced by the oligonucleotides are indicated and the sequences corresponding to those sites are in italic. Lower cases indicate the mutations introduced by site-directed mutagenesis in the HIV-1/BRU RNA.
All the constructs were made in plasmid pBluescriptKSII+ cleaved with the BamHI and PstI nucleases. PCR-amplified fragments were digested using restriction sites generated by the primers (Table 1). In plasmid pLD-C3, fragments H1 and H2 have been ligated together using their BglII restriction sites. Similarly, in plasmid pLD-L3, fragments H1 and H3 have been ligated together. Recombinant plasmids pBluescriptKSII+ containing fragment H2 (pA3C) or H3 (pA3L) were prepared.
The HIV-1/MAL region 5148–5401 (fragment H4) and the HIV-1/MAL region 5148–5630 (fragment H5) were PCR amplified with the pairs of primers O-567/O-1458 and O-567/O-1459, respectively (Table 1) using the entire HIV-1/MAL cDNA (26) as the template. Hybrid recombinant plasmids pBluescriptKSII+ (pC3-MAL and pL3-MAL), containing BRU and MAL cDNA sequences, were obtained by insertion of the digestion product of fragment H1 together with the digestion product of fragment H4 or H5, respectively.
The HIV-1/BRU cDNA region encoding the stem–loop 3 RNA fragment containing ESS2 (S3) was PCR amplified from plasmid pLD-L3 with primer O-998 and O-954. Primer O-998 generates a T7 RNA polymerase promoter.
Plasmid pΔPSP, used for the transfection experiments, was constructed in the following way. Infectious HIV-1 plasmid pNL4-3 (GenBank accession no. M19921) was cleaved with SpeI and BalI to generate an 11.9 kb fragment. This was ligated together with oligonucleotides 5′-CTAGACGCGTTTGG-3′ and 5′-CCAAACGCGT-3′, which had been previously annealed, to form a double-stranded linker. This created an HIV-1 plasmid deleted between nt 1511 and 4551.
Site-directed mutagenesis
For site-directed mutagenesis of plasmids pLD-C3 and pLD-L3, the inserted BamHI–PstI fragment of each plasmid was cloned into phage M13mp9. For both M13mp9 recombinants, site-directed mutagenesis was made according to Kramer et al. (27), using the oligonucleotides O-825 and O-826 (Table 1). The mutagenized BamHI–PstI DNA fragments were reinserted into plasmid pBluescriptKSII+ and the resulting plasmids were designated as pSJ-C3.825, pSJ-C3.826, pSJ-L3.825 and pSJ-L3.826, respectively. Constructs pSJ-C3.994 and pSJ-L3.994 were obtained after a second run of site-directed mutagenesis of the BamHI–PstI fragments of plasmids pSJ-C3.825 and pSJ-L3.825. Oligonucleotide O-994 was used to generate the mutation (Table 1). The resulting plasmids were designated as pSJ-C3.994 and pSJ-L3.994, respectively. pΔPSP variants were constructed by replacement of the HIV-1/pNL4-3 EcoRI–Bsu36I fragment (nt 5743–5955) with the corresponding HIV-1/BRU EcoRI–Bsu36I fragment (nt 5325–5537) from plasmids pLD-L3, pSJ-L3.825, pSJ-L3.826 and pSJ-L3.994. All plasmids with mutated DNA were sequenced to confirm the expected base changes.
In vitro transcription
Prior to transcription with T7 RNA polymerase, all the plasmids were linearized with the PstI nuclease. For splicing assays, uniformly labeled capped transcripts were produced as previously described (7). Synthesis of cold transcripts for RNA secondary structure analysis was carried out in a 30 µl mixture, containing 1 µg of linearized plasmid or 1.7 pmol of PCR product, as previously described (15). The RNA was dissolved in 80 µl of sterile water, 1 µl of this solution was used for each chemical reaction or enzymatic digestion.
Chemical and enzymatic probing of RNA in solution
Enzymatic digestions with V1 RNase and S1 nuclease were carried out, as previously described (15), using 1 µl of in vitro synthesized RNA in the presence of 1.25 µg of a commercial yeast tRNA mixture (Roche Diagnostics) for V1 RNase digestion and 2.5 µg of yeast tRNA for S1 nuclease digestion. V1 RNase was prepared from Naja oxiana venom (28) and S1 nuclease was purchased from Amersham Pharmacia Biotech. Prior to enzymatic digestion, the RNA transcript mixed with the tRNA mixture was pre-incubated for 20 min at 20°C, in 50 µl of a 50 mM KCl, 10 mM MgCl2, 10 mM Tris–HCl, pH 7.5 solution for V1 RNase digestion and in 50 µl of a 25 mM KCl, 0.5 mM ZnCl2, 5 mM MgCl2, 12.5 mM AcONa pH 4.5 solution for S1 nuclease digestion. Incubation was for 10 min at 20°C with 0.1 U of V1 RNase or for 12 min at 20°C with 3 U of S1 nuclease. V1 RNase digestion was stopped by addition of 1 µl of 100 mM EDTA, followed by phenol extraction. S1 nuclease digestion was stopped by adding 20 µg of tRNA, chilling on ice and phenol extraction. After phenol extraction, V1 and S1 digested RNAs were ethanol precipitated, dissolved in 3 µl of distilled water and 1 µl was used for reverse transcription analysis.
Prior to 1-cyclohexyl-3-(2-morpholinoethyl)-carbodiimide-metho-p-toluene sulfonate (CMCT) reaction, 1 µl of in vitro synthezised RNA mixed with 10 µg yeast tRNA were pre-incubated for 20 min at 20°C in 50 mM KCl, 10 mM MgCl2, 100 mM sodium borate pH 8, buffer with 3.2 mg of CMCT per 300 µl assay. Incubation was for 30 min at 20°C. At the end of the reaction, RNA was ethanol precipitated and treated as RNA cleaved by enzymes.
Chemical and enzymatic probing of RNA in nuclear extract
Conditions for enzymatic digestion and chemical modification were modified to allow analysis of RNA secondary structure in the presence of the large amounts of protein contained in the nuclear extract. S1 nuclease could not be used under splicing reaction conditions as its optimum pH of action is of 4.5 and splicing reactions are performed at pH 7.9. Because of this, we used T1 and T2 RNases to probe the single-stranded regions. Polyvinylalcohol (PVA) used for splicing assays has an inhibitory effect on V1, T1 and T2 RNase action and therefore enzymatic digestions and chemical modifications were performed in the splicing buffer in the absence of PVA. Secondary structure analysis was performed with 100 ng of an in vitro produced transcript. Prior to enzymatic digestion, the RNA transcript was incubated for 5 min at 0°C, followed by 5 min at 30°C, in 4 µl of HeLa cell nuclear extract in buffer D (0.2 mM EDTA, 0.1 M KCl, 0.5 mM DTT, 0.25 mM PMSF, 20% glycerol, 20 mM HEPES pH 7.9; adapted to Dignam et al., 29) under the conditions used for in vitro splicing assays (2.5 mM MgCl2 and 1 mM ATP), except that PVA was omitted and 5 µg of tRNA were added to the reaction mixture. Incubation was performed for 10 min at 30°C with 0.4 U of V1 RNase, 0.1 U of T1 RNase or 2 U of T2 RNase. V1 RNase digestion was stopped by addition of 2 µl of 100 mM EDTA. T1 and T2 RNase digestions were stopped by cooling the mixture on ice and adding of 20 µg of tRNA. RNA was immediately phenol extracted and ethanol precipitated. It was then dissolved in 4 µl of distilled water and 1 µl was used for primer extension analysis. In parallel, for comparison, V1, T1 and T2 RNase digestions were performed in the same conditions on the same amounts of RNA transcript pre-incubated in the splicing buffer D, instead of the nuclear extract.
We were not able to develop conditions for efficient modification of RNA with CMCT under splicing conditions. To obtain efficient DMS modification, sodium cacodylate had to be added to a 50 mM final concentration. To test for accessible G residues, we used kethoxal. In this case, potassium borate had to be added to a 50 mM final concentration to obtain efficient modification. Incubations with 1 µl of 25% DMS–ethanol solution, or 2 µl of kethoxal (37 mg/ml) were performed for 10 min at 30°C. DMS modifications were stopped by addition of 3 µl of 6 M β-mercaptoethanol. Kethoxal modifications were stopped by addition of 20 µl 0.5 M potassium borate. In both cases, RNA was immediately phenol extracted. RNA modified by kethoxal was dissolved in 4 µl of 25 mM potassium borate, RNA modified by DMS was dissolved in 4 µl of sterile water and 1 µl of each solution was used for reverse transcriptase analysis. Here also, for comparison, DMS and kethoxal modifications were performed in the same conditions on the same amounts of RNA transcript incubated in the splicing buffer D, instead of the nuclear extract.
Identification of modification and cleavage positions and secondary structure prediction
Positions of chemical modifications and enzymatic cleavages were identified by primer extension analysis with the avian myeloblastosis virus reverse transcriptase (Life Science). Four oligonucleotides were used as primers: O-608, O-831, O-548 and O-1458 (Table 1). Oligonucleotide primers were 5′-end labeled with [γ-32P]ATP 3000 Ci/mmol (Amersham Pharmacia Biotech). Annealing of primers and primer extension were performed as previously described (15). To prepare sequencing ladders of unmodified RNA, dideoxynucleotide:deoxynucleotide mixtures at a 1:2 ratio were used. The cDNA products were fractionated by electrophoresis on 7% polyacrylamide sequencing gels.
Predictions of secondary structure were made with the MFold program of the GCG software, version 8.1 Unix (1995), which is based on the thermodynamic values proposed by Jaeger et al. (30) for an RNA in solution in 1 M NaCl buffer at 37°C. The results of the experimental analysis were introduced in the computer search, in order to define the secondary structure that had the best fit with the experimental data.
The GenBank accession numbers of HIV-1 or related HIV-1 sequences used in the comparative analysis are: K02013 (HIV-1/BRU), M19921 (HIV-1/NL4-3), K03455 (HIV-1/HXB2), M38429 (HIV-1/JRCSF), U39362 (HIV-1/P896), M17449 (HIV-1/MN), D10112 (HIV-1/CAM1), M38431 (HIV-1/NY5CG), K02007 (HIV-1/SF2), X04415 (HIV-1/MAL), K03454 (HIV-1/ELI), L20587 (HIV-1/ANT70C), X52154 (SIVcpzGAB) and U42720 (SIVcpzANT).
In vitro splicing assays
In vitro splicing assays were performed with HeLa cell nuclear extracts from the Computer Cell Culture Center S.A. (Belgium), using 105 Cerenkov c.p.m. of RNA transcript per assay, as previously described (7). The reaction mixture was prepared on ice and then incubated at 30°C for 150 min. Spliced products were deproteinized with proteinase K, phenol extracted and analyzed on a 5% polyacrylamide sequencing gel. Splicing efficiency was estimated by scanning the gel with a Molecular Dynamics Phosphorimager using the Image Quant Software, version 3.3. The M:P ratio (moles of mature RNA versus the moles of residual precursor) was determined for each transcript taking into account the estimated radioactivity and the number of uracil residues per molecule.
In vivo splicing assays
HeLa cells were transfected by the modified calcium phosphate coprecipitation technique with 12 µg plasmid DNA as described above (10). Total cellular RNA was isolated from transfected HeLa cells 48 h post-transfection and 3 µg of RNA was reversed transcribed and PCR amplified with forward oligonucleotide primer BSS (5′-GGCTTGCTGAAGCGCGCACGGCAAGAGG-3′; nt 700–727) and reverse primer SJ4.7A, which spans sites D4 and A7 (5′-TTGGGAGGTGGGTTGCTTTGATAGAG-3′; nt 8369–8381 and 6032–6044) as described previously (10). After confirmation of amplified spliced product by PAGE, amplification products (100 ng) were radiolabeled by performing a single round of PCR with the addition of 10 µCi [α-32P]dCTP and the products were analyzed by electrophoresis on a 6% polyacrylamide 7 M urea gel.
RESULTS
Secondary structure analysis of the HIV-1 RNA region containing site A3
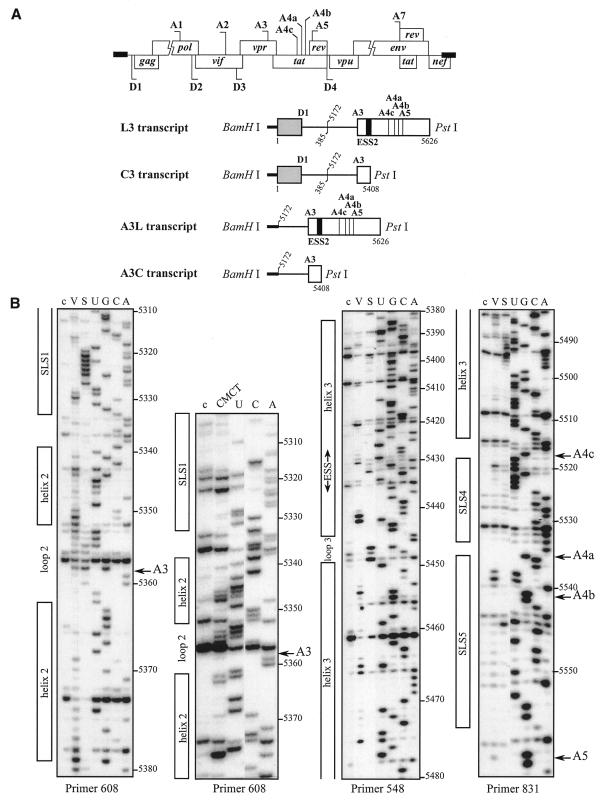
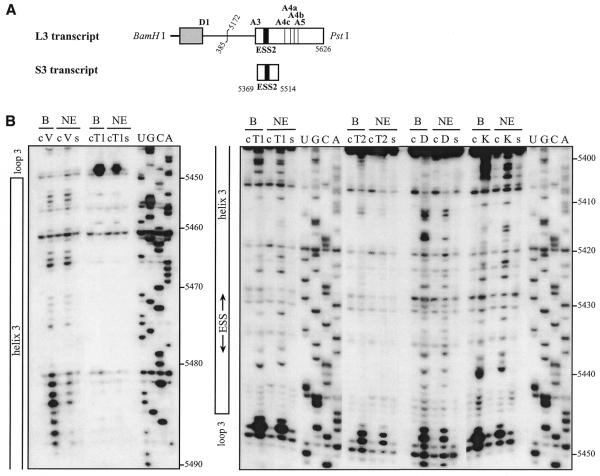
We first studied the secondary structure of the region from positions 5303 to 5559 of the HIV-1/BRU RNA in the L3 transcript (Fig. 1A). Transcript L3 contains the 5′ extremity of the HIV-1/BRU RNA (positions 1–385) fused to a central portion of the HIV-1/BRU RNA (positions 5172–5626). Transcript L3 carries the D1 5′ss and five 3′ss (A3, A4c, A4a, A4b and A5). In order to detect RNA secondary structure artifacts that may be due to the absence of the authentic partners of the sequences located at the extremities of the studied fragments, analyses of a given HIV-1 RNA region were performed on several RNA fragments of different lengths. To this end, in addition to the L3 transcript, the C3 transcript lacking RNA sequence between nt 5409 and 5626 downstream from site A3, and the A3C and A3L transcripts, which do not contain the 5′ part of the HIV-1/BRU RNA, were also subjected to chemical and enzymatic probing experiments (Fig. 1A). The single-stranded specific chemical reagent, CMCT, and two RNases (V1 and S1 with specificities for double- and single-stranded RNA, respectively) were used in the analysis. Positions of chemical modifications and enzymatic cleavages were identified by primer extension analysis, using the oligonucleotide primers given in Table 1. Relevant examples of primer extension analyses of enzymatic cleavages and chemical modifications of transcript L3 are shown in Figure 1B. Identical patterns of chemical modifications and enzymatic cleavages were obtained for the common parts of the four transcripts studied (data not shown). This demonstrated that the presence of ESS2 in the transcript did not alter the secondary structure at the A3 3′ss.
Figure 1.
Secondary structure analysis of the HIV-1/BRU RNA region from positions 5303 to 5559. (A) Schematic representation of the HIV-1 genome and of the RNAs used for secondary structure analysis. In the HIV-1 genome, the 5′ss (D) and 3′ss (A) are shown; boxes, open reading frames. In the four RNA transcripts (L3 to A3C), numbering of HIV-1/BRU RNA sequences is according to Ratner et al. (25), exon sequences are represented by rectangles, introns by thin lines, the thick horizontal lines at the beginning of the transcripts correspond to sequences generated by plasmid pBluescriptKSII+. The 5′ss, 3′ss and ESS2 are indicated. In transcripts C3 and L3, the junction between the two HIV-1 RNA regions within the intron is indicated by a vertical broken line. (B) Examples of primer extension analyses of enzymatically digested and chemically modified L3 transcript. Lanes marked by V, S and CMCT correspond to V1 RNase digestion, S1 nuclease digestion and CMCT modification, respectively. Conditions for digestion and modification are given in Materials and Methods. Lanes marked by c correspond to control experiments with the untreated RNA transcript, lanes UGCA to the sequencing ladders. Numbering of nucleotides in the HIV-1/BRU RNA is on the right of the autoradiogram, as well as the positions of the 3′ss (A3, A4c, A4a, A4b and A5). Positions of the various stem–loop structures identified on the basis of this analysis (Fig. 1C) and of ESS2 are shown on the left. The primers used for extension with reverse transcriptase are indicated below each autoradiogram. (C) The results of enzymatic probing are schematically represented on the proposed secondary structure model. Cleavages by enzymes are shown by arrows surmounted with circles for S1 nuclease and squares for V1 RNase. Three red circles or squares indicate a strong cleavage, two orange circles or squares a medium cleavage, one green circle or square a low cleavage. The free energy of the proposed stem–loop structures at 37°C, in 1 M NaCl were calculated with the MFold software. Positions of nucleotides in the HIV-1/BRU RNA are given. Stem–loop structures are designated as SLS1–5. The various 3′ss are indicated by blue arrows. The ESS2 inhibitory element is squared. Results of CMCT modification of SLS1 and SLS2 are shown in the inset. Red circles indicate a strong, orange a medium and green a low level of modification. Squared nucleotides were not modified.
A unique model of RNA secondary structure was derived from all the experimental data and is shown in Figure 1C. In this model, site A3 is located in the terminal loop of stem–loop structure 2 (SLS2). This stem–loop structure is preceded by a smaller stem–loop structure 1 (SLS1) of lower stability and followed by a long irregular stem–loop structure 3 (SLS3), that contains the ESS2 element (Fig. 1C). Although several internal loops and bulges interrupt the helical structure of SLS3, the calculated stability at 37°C of SLS3 is high (ΔG = –22.3 kcal/mol). Two short stem–loop structures 4 and 5 (SLS4 and SLS5) were detected in the segment containing sites A4c, A4b, A4a and A5 (Fig. 1C).
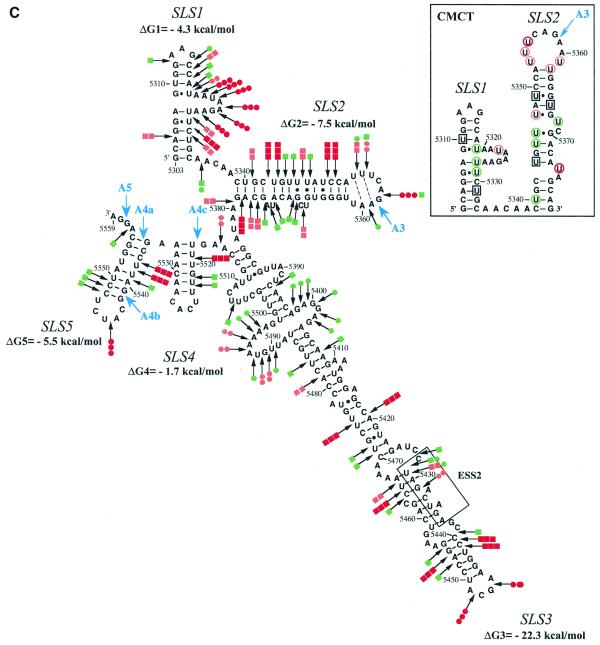
As a test for the biological significance of the proposed structure, we examined the sequences of other HIV-1 and SIV strains for compensatory (replacement of a Watson–Crick base pair by another Watson–Crick base pair) or semi-compensatory (substitution of a Watson–Crick base pair by a G·U or U·G base pair and vice versa) base mutations. The SIVcpzGAB and ANT strains were recently shown to share a common ancestor with virus HIV-1 (31).
As shown in Figure 2A, the possibility to form SLS2 is conserved in clade B and in a recombinant of A, D and I clades (HIV-1/MAL) of group M of HIV-1 strains, as well as in the SIVcpzGAB strain and formation of SLS2 was experimentally verified for the HIV-1/MAL RNA using RNA transcripts L3-MAL and C3-MAL (Fig. 2A, B and C). Although of lower stability, an SLS2 can be formed in the RNA of the ANT70C strain from the group O of HIV-1 strains (Fig. 2A). Such conservation of SLS2 suggested its functional importance in splicing, which was experimentally tested, as described below.
Figure 2.
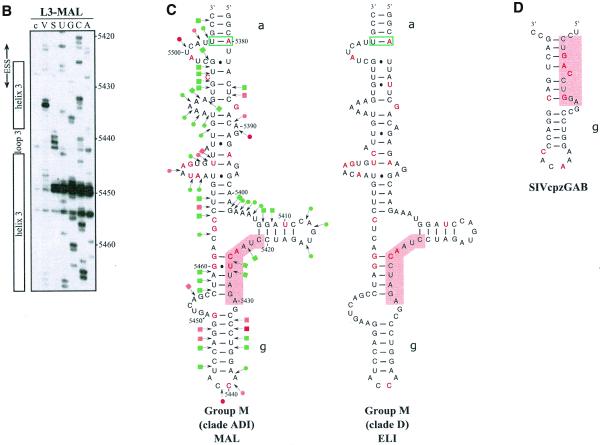
Conservation of SLS2 and SLS1 in the groups M and O of HIV-1 strains and in the SIVcpz strains. Nucleotide sequences in this figure are from the HIV sequence database (http://hiv-web.lanl.gov/, Los Alamos National Laboratory, Los Alamos, NM). They were identified by alignment of the HIV-1/BRU RNA sequence with sequences from HIV-1 RNAs of other strains and the SIVcpz strains using the ClustalW program (43). (A) SLS2s are drawn for various strains of the group M of HIV-1 strains, for the ANT70C strain of group O and for the SIVcpzGAB strain. The name of the strains, the clade and the group are indicated below each structure. The free energy of each SLS2, at 37°C, in 1 M NaCl, as calculated by the MFold program is indicated. Sequence variations as compared to BRU are indicated by red nucleotides. Base pairs within green rectangles are conserved by semi-compensatory mutations. (B) Schematic representation of the transcript used for the experimental study of the HIV-1/MAL RNA region containing site A3. The same symbols were used as in Figure 1A. (C) Probing of the secondary structure of the L3-MAL RNA with V1 RNase (lane V) and S1 nuclease (lane S). Primer 1458 was used for reverse transcriptase analysis of cleaved and intact RNA (lane c) and for generating the sequencing ladders (lanes UGCA). Numbering of the HIV-1/MAL RNA sequence on the right of the autoradiogram is according to the Los Alamos HIV sequence database. Beside the autoradiogram, a schematic representation of the cleavages observed is given on the MAL SLS2 model (squares for V1 cleavages; dots for S1 cleavages; red, orange and green for strong, medium and low cleavages, respectively). Helix and loop positions are indicated on the left of the autoradiogram. (D) Based on the experimentally identified SLS1 of the BRU RNA, ‘SLS1-like’ structures were drawn for the HIV-1/ANT70C, SIVcpzGAB and SIVcpzANT strains. Base pairs in red and green rectangles are conserved by compensatory and semi-compensatory mutations, respectively. The upper part of SLS1, that is not conserved in the SIVcpzANT RNA, is represented by a discontinuous line.
The possibility to form the terminal helix of SLS1 is not as highly conserved in HIV-1 and SIVcpz viruses as SLS2. However, we found that several semi-compensatory and a few compensatory mutations allow formation of the SLS1 bottom helix in the ANT70C strain from group O of HIV-1 strains and in the SIVcpz strains GAB and ANT (Fig. 2D).
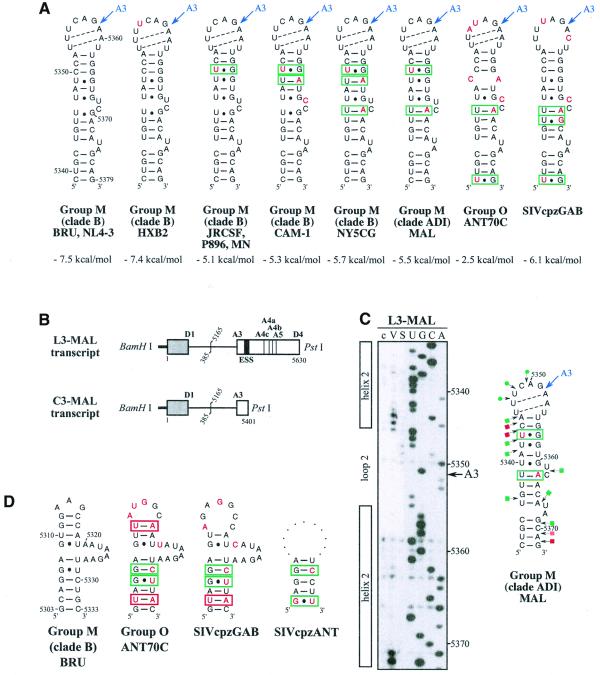
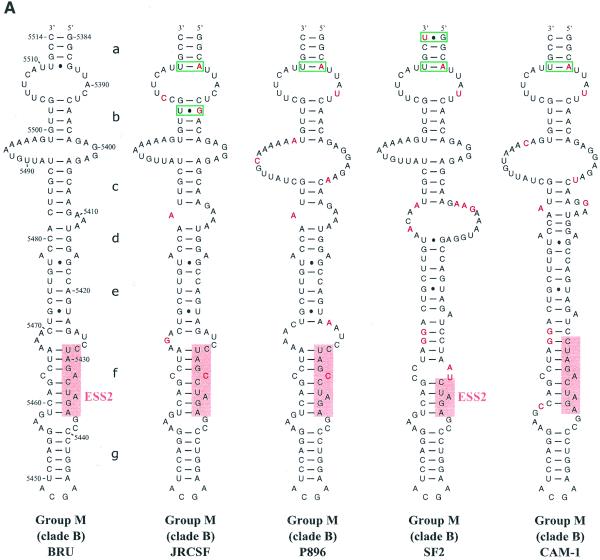
SLS3 is highly conserved in strains from clade B of group M of HIV-1 strains (Fig. 3A). Most of the observed base substitutions and the insertion found in HIV-1/SF2, are located in segments which are single-stranded in the HIV-1/BRU SLS3. The few base substitutions in double-stranded regions are semi-conservative: a C–G pair in helix a, found in almost all studied RNAs, is replaced by a U·G pair in the HIV-1/SF2 RNA; a U·G pair in helix a of the HIV-1/BRU RNA is replaced by a U–A pair in all other strains; finally, a U–A pair in helix b, found in almost all studied RNAs, is replaced by a U·G pair in the HIV-1/JRCSF RNA. Only one region of SLS3 showed a variable number of base pairs, depending on the clade B strain studied, and this is the helical part f containing ESS2 (Fig. 3A). Thus, the phylogenetic study of clade B supports the existence of SLS3.
Figure 3.
Comparison of SLS3 in clade B of HIV-1 group M strains and of the corresponding structure in the HIV-1/ELI and MAL strains and in the SIVcpzGAB strain. In this figure, the RNA regions from various HIV-1 strains and the SIVcpzGAB strain, corresponding to the HIV-1/BRU RNA region from positions 5407 to 5484, were identified by sequence alignment using the ClustalW program (43). (A) A secondary structure model based on the one established experimentally for the HIV-1/BRU RNA could be drawn for each strain of clade B that was examined. Base substitutions or insertions as compared to the HIV-1/BRU sequence are shown by red letters. Green rectangles indicate semi-compensatory mutations. The experimentally demonstrated ESS from the HIV-1/BRU and SF2 strains and the putative ESSs of the other strains are shown in a pink rectangle. (B) Experimental study of the HIV-1/MAL RNA structure using primer 831 (same legend as in Fig. 2C). (C) Secondary structure model proposed for the HIV-1/MAL RNA region (positions 5377–5507), on the basis of the experimental data in (B). Cleavages are schematically represented with the same symbols as in Figure 2C. Conserved helices a and g as compared to SLS3 are indicated. The putative ESS is indicated in pink. A similar model can be drawn for the ELI RNA. (D) The terminal part of SLS3 can be formed in the SIVcpzGAB RNA.
Using enzymatic probes, we studied experimentally the secondary structure of the HIV-1/MAL region corresponding to the HIV-1/BRU SLS3. As shown in Figure 3B and C, a long irregular stem–loop structure was also observed. However, only helix a and the terminal stem–loop structure g are strictly conserved as compared to the HIV-1/BRU SLS3. As mentioned above, the HIV-1/MAL virus is a recombinant of A, D and I clades (26). A structure similar to that established for the MAL RNA can be proposed for the corresponding region of the ELI RNA (Fig. 3C) (The HIV-1/ELI strain belongs to clade D) (26). Interestingly, the terminal part of the BRU SLS3 can be formed in the SIVcpzGAB RNA (Fig. 3D), which favors the functional importance of this terminal part of SLS3. For the remaining part of SLS3, results suggest some variability depending on the clade of HIV-1 strains.
Whereas an active ESS2 was only demonstrated in three HIV-1 strains from clade B of group M, namely NL4-3 (3,9), SF2 (10) and BRU (present data described below), the presence of an ESS2 element has been proposed to be a specific feature of groups M and N of HIV-1 viruses but not of group O viruses (10). In the various HIV-1 strains of clade B that we studied, ESS2 is always located in the irregular helical part f of SLS3, which precedes the conserved terminal region g (Fig. 3A). ESS2 is composed of two repeats of the CUAGA sequence in the HIV-1/BRU RNA. In some HIV-1 strains, only the 3′-terminal C/UUAGA sequence is conserved [this is the case for the SF2 (Fig. 3A), MAL and ELI strains (Fig. 3C)]. Based on the proposed secondary structure, the conserved C/UUAGA motif of ESS2 is at the border of the highly conserved terminal region g.
SLS3 is formed under splicing assay conditions
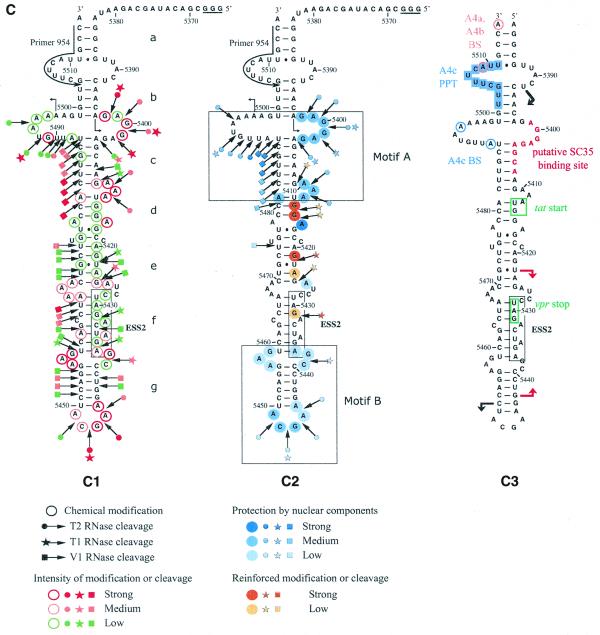
Because our phylogenetic comparisons were less definitive for SLS3 compared to SLS2 and because SLS3 containing ESS2 was determined using the naked RNA, we tested whether this structure was also formed under conditions for in vitro splicing assays. To this end, conditions were developed to probe the RNA secondary structure within a nuclear extract (see Materials and Methods). These conditions were used to analyze the S3 RNA transcript containing all the sequences needed to form SLS3 and part of the SLS2 sequence (Fig. 4A). Three RNases were used in these experiments: (i) V1 RNase to probe accessible double-stranded RNA regions and (ii) T1 and T2 RNases to identify accessible single-stranded regions. Two chemical probes were also used to detect the accessible single-stranded residues, DMS for A and C residues and kethoxal for G residues. To identify the RNA segments protected by the binding of nuclear components, the probing experiments were performed after incubation in a HeLa cell nuclear extract dissolved in buffer D and in buffer D alone.
Figure 4.
The HIV-1/BRU SLS3 is formed in a nuclear extract and two structural motifs A and B are protected by association with nuclear components. (A) The S3 transcript used for the experimental analysis is shown. (B) Examples of primer extension analyses of the S3 transcript cleaved by V1, T1 or T2 nucleases (lanes marked by V, T1, T2, respectively) or modified by kethoxal or DMS (lanes marked by K or D, respectively) are presented. Enzymatic and chemical reactions were either performed in the splicing buffer D (B) or in a nuclear extract (NE). As a control, a primer extension was made with the intact RNA transcript incubated in the absence of reagent, either in buffer D or in the nuclear extract (lanes marked by c). Lanes UGCA correspond to the sequencing ladders. Numbering of the nucleotides in the HIV-1/BRU RNA is indicated on the right. Positions of the loops and helices 3 and of ESS2 are shown on the left. (C) Schematic representation of results of chemical and enzymatic probing of S3, in the splicing buffer D (C1) or in the nuclear extract (C2). In Panel C1, the circled nucleotides were modified by DMS or kethoxal. Cleavages by RNases are shown by arrows, surmounted with a circle for T2 RNase, a star for T1 RNase and a square for V1 RNase. Green, orange and red symbols indicate low, medium and strong modifications or cleavages, respectively. In C2, protection against the action of chemical reagents and nucleases are shown in blue, the intensity of the blue color reflects the level of protection. Increased sensitivity to chemical reagents and nucleases is indicated in yellow (low increase) or orange (medium increase). In both C1 and C2, the oligonucleotide primer 954 used for the reverse transcriptase analysis is indicated. The 3G residues at the 5′-end of the S3 transcript were generated by the T7 RNA polymerase promoter. The portion of the S3 RNA that was analyzed is delimited by the two broken arrows in C1. Due to pause of reverse transcriptase at some of the V1 cleavage sites in the region from positions 5397 to 5438 in the nuclear extract, no estimation of their variation of intensity as compared to naked RNA is given for this part of SLS3. In C3, the HIV-1/BRU functional sequences contained in SLS3 are indicated, namely: the tat start codon and vpr stop codon (squared in green), the putative SC35 binding site (red letters), the branched sites (circled in blue) and PPT (in blue rectangles) of the A4c 3′ss and the branched sites for A4a and b 3′ss (circled in orange). The limits of the RNA fragments used by Caputi et al. (11) and Del Gatto-Konczak et al. (32) for hnRNP A/B crosslinking experiments are indicated by black and red arrows, respectively.
Examples of primer extension analyses of base modifications and phosphodiester bond cleavages are shown in Figure 4B. Only the probing results showing the larger number of data were selected. Experiments were repeated several times and reproducible results were obtained. For efficient chemical modifications, DMS modifications were performed in the presence of 50 mM sodium cacodylate and kethoxal reactions in the presence of 50 mM potassium borate. However, as shown by the excellent correlation of the results obtained for T1 RNase and kethoxal analysis that both target G residues (for example, see the increased reactivities to both reagents at G residues 5414 and 5415 and the decreased reactivities at G residues 5400 and 5401, Fig. 4B and C), addition of potassium borate in the splicing buffer did not affect the RNA secondary structure and the RNA–protein interactions. Similarly, the very good correlation observed between the T2 RNase and DMS results also revealed that the addition of sodium cacodylate in the splicing buffer did not modify RNA secondary structure and RNA–protein interactions.
Study of the S3 transcript in buffer D, using both enzymatic and chemical probes, confirmed the formation of SLS3 in this buffer. The low level of modification of A and G residues in helical portions d, e and f is the one expected taking into account the expected low thermodynamic stability of these irregular helical portions (Fig. 4C). Using the same probes, SLS3 was also found to be present after incubation of S3 RNA in the nuclear extract (Fig. 4C). In addition, the data suggested that there were two binding sites for nuclear components in SLS3: (i) the structural motif A formed by the segments 5398–5412 and 5480–5499, which includes a large internal loop, the stable helix c and a bulge loop; and (ii) the single-stranded segments of the highly conserved terminal motif B, including the terminal and internal loops (5437–5439 and 5456–5458) (Fig. 4C). A higher degree of protection was observed for motif A as compared to motif B, which suggested a higher affinity of the nuclear components for the former. The accessibility of naked ESS2 to enzymes and chemical reagents is limited in buffer D. However, based on previous studies suggesting the presence of a binding factor (3,11,32), we expected that this limited accessibility would be reduced in a nuclear extract. In fact, protection was only detected for the 3′ terminal A residue and cleavage of the phosphodiester bond between residues 5431 and 5432 by T1 RNase was increased, as well as the modification of G residue 5431 by kethoxal. Hence, SLS3 was formed in a nuclear extract and contained two anchoring motifs, A and B, which were protected in nuclear extract. However, protection of ESS2 was poor in these conditions.
Influence of SLS2 stability on splicing efficiency at site A3
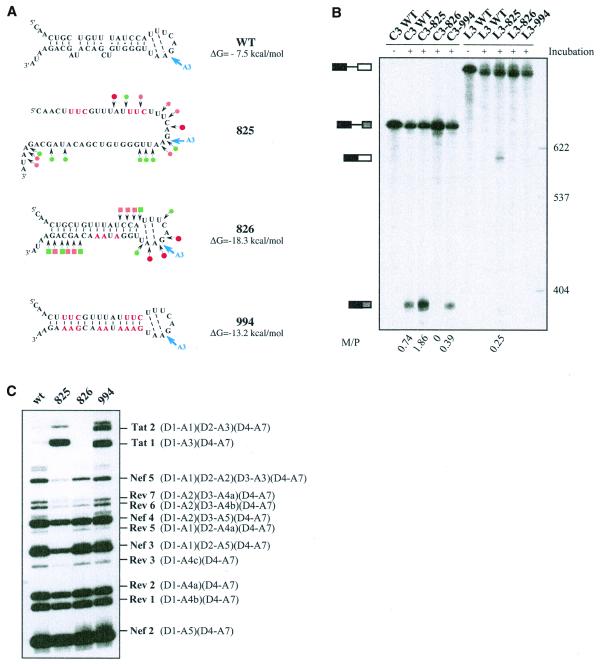
According to our structural model of SLS2, the A3 PPT is base paired (helix 2) (Fig. 1C). As PPTs have an important role in 3′ss activity, we tested for a possible influence of the stability of SLS2 helix on site A3 efficiency. To this end, we produced three RNA variants (825, 826 and 994) (Fig. 5A). Variant 825 was designed to disrupt the helical part of SLS2. However, these mutations also optimized the PPT sequence by conversion of 1G and 3C residues into U residues and 1A and 1U residues into C residues (Fig. 5A). Hence, a cumulative effect of the optimization of the PPT sequence and of the RNA secondary structure destabilization was expected for this mutant. Variant 994 shares the same mutations in the PPT sequence as variant 825 and compensatory mutations were made in the opposite strand to reinforce the RNA secondary structure (Fig. 5A). Hence, variant 994 was expected to have both an optimized PPT sequence and a reinforced SLS2 stability (–13.2 kcal/mol at 37°C versus –7.5 kcal/mol for the wild-type RNA). Finally, variant 826 has a wild-type PPT sequence and was expected to have a reinforced helix 2 (–18.3 kcal/mol at 37°C versus –7.5 kcal/mol for the wild-type RNA). We verified experimentally that the variant RNAs had the expected secondary structure by S1 and V1 nuclease probing (Fig. 5A). In vitro splicing efficiencies of transcripts carrying the various mutations were tested both in a HeLa cell nuclear extract (Fig. 5B) and in a cellular context by transfection of HIV-1 constructs into HeLa cells (Fig. 5C). For this latter purpose, we used a construct containing an HIV-1/pNL4-3 cDNA sequence deleted between nt 1511 and 4550. The deleted sequence is within the D1–A1 intron. Cells transfected with this deleted construct produce an identical pattern of spliced and unspliced HIV-1 mRNAs compared to the wild-type plasmid (P.Bilodeau and C.M.Stoltzfus, unpublished data). Furthermore, using the entire HIV-1 cDNA, it was previously shown that the pattern of HIV-1 mRNAs produced by splicing of the primary transcript is similar in HeLa cells and in infected lymphocytes T (1). Thus, after substitution of the wild-type pNL4-3 sequence from positions 5744 to 5955 with the corresponding sequence of the BRU RNA (wild-type or mutated), we tested the effects of the mutations on the relative use of the central HIV-1 3′ss.
Figure 5.
Effects of alteration of SLS2 on site A3 efficiency in vitro and in vivo. (A) The generated mutations (red nucleotides) and the expected structure of the variant RNAs are shown. For variants C3.825 and C3.826, the secondary structure was probed experimentally, results are schematically represented using the same symbols as in Figure 2C. The free energies of the structures at 37°C in 1 M NaCl, as calculated by the MFold program, are given. (B) PAGE of the in vitro splicing products of the wild-type and variant C3 and L3 transcripts. Uniformly labeled RNAs, prepared as described in Materials and Methods, were incubated for 150 min in a HeLa cell nuclear extract in splicing conditions. Untreated C3 and L3 RNAs were fractionated in parallel, as well as size markers (molecular weights on the right of the panel). Positions of the C3 and L3 transcripts and of their spliced products are indicated on the left of the panel. The M:P ratios (given below the lanes) were calculated by estimation with a phosphorimager of the radioactivity in the bands of gel corresponding to the mature RNA and the residual transcript, respectively. (C) PAGE of the RT–PCR products obtained by amplification of total RNA extracted from HeLa cells transfected with the wild-type or the mutated mini-proviral cDNA pΔPSP, with the oligonucleotide primers BSS and SJ4.7A (see Materials and Methods). Nomenclature of the RT–PCR products is according to Purcell and Martin (1).
Secondary structure had a strong effect on splicing efficiency in vitro as shown by the mature spliced RNA product to precursor (M:P) ratios (Fig. 5B). For the C3 series of RNA transcripts, lacking ESS2, splicing efficiency of variant 825, which had a destabilized SLS2, was increased by a factor of 2.5 as compared to wild-type RNA. Variant 994, which has the optimized PPT sequence and a stabilized SLS2, was spliced somewhat less efficiently than the wild-type RNA. Variant 826 with the wild-type PPT sequence and a reinforced secondary structure was very poorly spliced in vitro (Fig. 5B). As shown in Figure 5B, in the presence of ESS2 (L3 series of RNA transcripts), only variant 825 was spliced. This was a confirmation of the presence of an active ESS2 in the HIV-1/BRU RNA and this showed that in vitro optimization of the PPT sequence accompanied by a destabilization of SLS2 was necessary for splicing to occur in the presence of ESS2. As shown in Figure 5C, the effect of SLS2 stability on the utilization of site A3 in HeLa cells appeared to be dependent upon the sequence of the PPT. Indeed, RT–PCR analysis of the RNAs produced in vivo (Fig. 5C), showed that there was little or no difference in utilization of site A3 when variants 825 and 994 were compared. In contrast, a strong decrease of site A3 utilization was observed for variant 826 with the wild-type PPT and a reinforced SLS2.
DISCUSSION
There has been little information available previously regarding secondary structure of HIV-1 RNA regions containing splicing sites. Only the secondary structure of the MAL region containing the D1 5′ss has previously been investigated (33). Here, we have described the secondary structure of an HIV-1 RNA region containing the central 3′ splice sites: A3, A4a, A4b, A4c and A5 sites. Two main features have emerged from the established structure. Site A3, required for production of the tat mRNAs, is located in a terminal loop and its PPT is involved in the helix. ESS2 within tat exon 2 and acting on site A3 is located in an irregular helical structure in the vicinity of a highly conserved terminal motif. These structural features are conserved in HIV-1 strains as well as the related SIVcpz strains.
RNA secondary structure and the PPT sequence likely both contribute to the low A3 site efficiency
Within L3 (900 nt) as well as C3 (684 nt), A3L (510 nt) and A3C (294 nt) transcripts, the HIV-1/BRU RNA region, which is located between positions 5339 and 5379 and contains site A3, always folds into the same stem–loop structure (SLS2). When various strains were compared, we found that all the base substitutions found in SLS2 are either in the single-stranded segments or are such that secondary structure is preserved (semi-compensatory base mutations). Taking into account the strong constraints on evolution exerted by overlapping and juxtaposed ORFs in HIV-1 RNA, the presence of semi-compensatory mutations in an HIV-1 secondary structure motif reflects an important functional role of this motif. Interestingly, conservation of SLS2 is even observed in the SIVcpzGAB virus from the chimpanzee subspecies Pan troglodytes troglodytes, the primary reservoir of HIV-1 (31). One possible reason for the conservation of SLS2 is its contribution to the suboptimal property of the A3 PPT. This may limit the access of the A3 PPT to splicing factors, such as U2AF (34). We found a stronger effect of the RNA secondary structure at site A3 using in vitro splicing assays as compared to transfection assays in HeLa cells. Such a difference between in vitro and ex vivo assays has already been reported for the regulation of the alternative splicing of the chicken β-tropomyosin pre-mRNA (22).
In vitro, a reinforcement of SLS2 stability had a negative effect on site A3 utilization with a wild-type as well as with an optimized PPT. However, in HeLa cells, using an HIV-1 genomic construct, we showed the dominant effect of the PPT sequence versus the RNA secondary structure. Indeed, reinforcement of the stability of the helix (ΔG difference of –5.5 kcal/mol) had no deleterious effect on site A3 utilization when the PPT was optimized (conversion of three Cs and one G into Us and one A and one U into Cs, variant 994). However, site A3 utilization was decreased for variant 826 with a wild-type PPT and a stabilized SLS2 (ΔG difference of –10.8 kcal/mol). In addition to secondary structure reinforcement, disruption of an exonic splicing enhancer (ESE) element in the wild-type sequence may participate in the decreased utilization of site A3 in variant 826. As efficient splicing at site A3 was observed for variant 994, which carried the same three mutations downstream from site A3, as variant 826 together with additional mutations, if an ESE is present downstream from site A3, it is not required in the presence of an optimized PPT.
Our data should also be examined in the light of data from Wu et al. (35), Guth et al. (36) and Reed (37), who showed that mammalian nuclear introns are AG dependent when their PPT is shorter than 14 nt. The A3 wild-type PPT contains 14 pyrimidine residues but is interrupted by four purine residues. Consequently, splicing at a wild-type A3 site is likely to be AG dependent, while splicing at an A3 site with an optimized PPT is likely to be AG independent. This may, therefore, increase the sensitivity of spliceosome assembly to RNA secondary structure.
Hence, although we cannot discard the possibility of an ESE element downstream from site A3, a reasonable explanation for our in vivo and in vitro observations is that the secondary structure and the suboptimal sequence of the PPT both participate to the low efficiency of site A3. This may be the reason for the high conservation of SLS2 in HIV-1 and SIVcpz strains.
ESS2 element is located in an irregular flexible structure that binds several nuclear components
Because the secondary structure at site A3 was identical in transcript C3 lacking ESS2 and in transcript L3 containing ESS2, the data clearly demonstrate that ESS2 does not act by a direct RNA–RNA interaction with a functional sequence of site A3. Instead, as previously proposed, it is more likely that the inhibition occurs by binding of inhibitory component(s) acting in trans (3,11,32).
In the HIV-1/BRU RNA, ESS2 is located in a long irregular stem–loop structure SLS3, that is formed independently of the length of the studied fragment, the buffer conditions (pH, mono and divalent cation concentrations) and in the presence or the absence of nuclear proteins. Only the terminal loop and helix of SLS3 are strictly conserved in HIV-1 strains. However, a common property of this part of HIV-1 RNAs is the presence of a flexible irregular structure, interrupted by several internal loops and bulges and by a lateral helix in the MAL and ELI RNAs. During the RNA folding process, the conserved stable terminal element may be the nucleation center of this irregular structure.
Based on its location in a region of reduced base-pairing (f), the BRU ESS2 would be expected to be available for RNA–protein interaction. However, its protection in a nuclear extract was limited to the 3′ terminal A residue of the downstream CUAGA motif and an increased reactivity of the upstream CUAGA motif to kethoxal and T1 RNase was observed. Interestingly, this upstream CUAGA motif of ESS2 is the less conserved of the two CUAGA motifs. For instance, in the SF2 strain where an active ESS2 was demonstrated, only the downstream CUAGA motif is present (Fig. 3A). ESS2 is adjacent to the highly conserved terminal motif B, which showed a partial protection in a nuclear extract. It should be noted that the protected A residue of ESS2 and the adjacent protected GC dinucleotide belong to a UAGAGC sequence that is similar to the reported protein hnRNP A1 binding site consensus sequence [UAGGG(A/U)] (38). Therefore, it is possible that binding of hnRNP A1 to HIV-1 RNA segments containing ESS2, detected by cross-linking experiments (11,32) (Fig. 4C), occurs at the UAGAGC sequence overlapping the last CUAGA repeat of ESS2 and the internal loop. In support of this hypothesis, a similar sequence (UAGAGU), which regulates 5′ss utilization in the hnRNP A1 pre-mRNA, was found to be a binding site for protein hnRNP A1 (39), and binding of a UAGAG sequence to hnRNP A1 RRMs is compatible with recent crystallographic data (40). The low protection observed for the core part of ESS2 may be due to the fact that stable binding of the repressor factor(s) to ESS2 needs association with splicing components bound at site A3 (the S3 transcript, that we used for the assays, did not contain the A3 site, Fig. 4A). The presence of numerous internal loops in SLS3 may ensure a greater flexibility allowing free or bound ESS2 to interact with distant components, for instance components bound at the A3 site. Conservation of the irregular architecture of SLS3 may be required for ESS2 mechanism of action.
In contrast to ESS2, the SLS3 motif B was highly protected in a nuclear extract. Interestingly, one of the highly protected sequences in motif A contains the two adenosine residues, used as branch residues for site A4c (Fig. 4C) (41). Protection may be due to the binding of splicing components involved in branch site recognition. Another highly protected single-stranded sequence in motif A (positions 5398–5407) shows homology with one of the consensus sequence established for the binding sites of the SR protein SC35 (Fig. 4C) [HIV-1 sequence 5′-AGGAGAGCA-3′, SC35 binding site consensus sequence 5′-AG(C/G)AGAGUA-3′; 42]. It should be noted that, in the proposed secondary structure, the A4c PPT is largely single stranded.
In addition to its importance for the splicing efficiency at site A3, which determines the yield of tat mRNA production, the SLS3 architecture may also be important for the regulation of tat mRNA translation. Indeed, SLS3 is likely to be present in the spliced tat mRNAs. In SLS3 from strains of clade B, the tat start codon is located in a short helical portion (d) of low stability (Fig. 4C). However, it is preceded by the stable helix portion (c), which may limit scanning of the tat mRNA 5′ UTR by the translation initiation complex.
In summary, it is likely that the RNA secondary structure of the HIV-1 RNA region that we determined play important roles in the regulation of RNA splicing and of mRNA translation, as well as increasing HIV-1 RNA resistance to cellular nucleases.
Acknowledgments
ACKNOWLEDGEMENTS
F. Clavel and J. L. Darlix are thanked for providing plasmid pBRU3 and MAL cDNA, respectively. S.J., D.R. and L.D. were fellows of the French Ministère de la Recherche et de l’Enseignement Supérieur and L.D. was also a fellow of the SIDACTION foundation. This work was supported by the Agence Nationale de Recherche sur le SIDA (ANRS), the Centre National de la Recherche Scientifique (CNRS), the French Ministère de la Recherche et de l’Enseignement Supérieur and by PHS grant AI36073 to C.M.S. from the National Institute of Allergy and Infectious Disease.
References
- 1.Purcell D.F. and Martin,M.A. (1993) Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication and infectivity. J. Virol., 67, 6365–6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amendt B.A., Hesslein,D., Chang,L.J. and Stoltzfus,C.M. (1994) Presence of negative and positive cis-acting RNA splicing elements within and flanking the first tat coding exon of human immunodeficiency virus type 1. Mol. Cell. Biol., 14, 3960–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amendt B.A., Si,Z.H. and Stoltzfus,C.M. (1995) Presence of exon splicing silencers within human immunodeficiency virus type 1 tat exon 2 and tat-rev exon 3: evidence for inhibition mediated by cellular factors. Mol. Cell. Biol., 15, 6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Reilly M.M., McNally,M.T. and Beemon,K.L. (1995) Two strong 5’ splice sites and competing, suboptimal 3’ splice sites involved in alternative splicing of human immunodeficiency virus type 1 RNA. Virology, 213, 373–385. [DOI] [PubMed] [Google Scholar]
- 5.Staffa A. and Cochrane,A. (1994) The tat/rev intron of human immunodeficiency virus type 1 is inefficiently spliced because of suboptimal signals in the 3’ splice site. J. Virol., 68, 3071–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Staffa A. and Cochrane,A. (1995) Identification of positive and negative splicing regulatory elements within the terminal tat-rev exon of human immunodeficiency virus type 1. Mol. Cell. Biol., 15, 4597–4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Damier L., Domenjoud,L. and Branlant,C. (1997) The D1-A2 and D2-A2 pairs of splice sites from human immunodeficiency virus type 1 are highly efficient in vitro, in spite of an unusual branch site. Biochem. Biophys. Res. Commun., 237, 182–187. [DOI] [PubMed] [Google Scholar]
- 8.Frankel A.D. and Young,J.A. (1998) HIV-1: fifteen proteins and an RNA. Annu. Rev. Biochem., 67, 1–25. [DOI] [PubMed] [Google Scholar]
- 9.Si Z., Amendt,B.A. and Stoltzfus,C.M. (1997) Splicing efficiency of human immunodeficiency virus type 1 tat RNA is determined by both a suboptimal 3’ splice site and a 10 nucleotide exon splicing silencer element located within tat exon 2. Nucleic Acids Res., 25, 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bilodeau P.S., Domsic,J.K. and Stoltzfus,C.M. (1999) Splicing regulatory elements within tat exon 2 of human immunodeficiency virus type 1 (HIV-1) are characteristic of group M but not group O HIV-1 strains. J. Virol., 73, 9764–9772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caputi M., Mayeda,A., Krainer,A.R. and Zahler,A.M. (1999) hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J., 18, 4060–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chebli K., Gattoni,R., Schmitt,P., Hildwein,G. and Stevenin,J. (1989) The 216-nucleotide intron of the E1A pre-mRNA contains a hairpin structure that permits utilization of unusually distant branch acceptors. Mol. Cell. Biol., 9, 4852–4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deshler J.O. and Rossi,J.J. (1991) Unexpected point mutations activate cryptic 3’ splice sites by perturbing a natural secondary structure within a yeast intron. Genes Dev., 5, 1252–1263. [DOI] [PubMed] [Google Scholar]
- 14.Charpentier B. and Rosbash,M. (1996) Intramolecular structure in yeast introns aids the early steps of in vitro spliceosome assembly. RNA, 2, 509–522. [PMC free article] [PubMed] [Google Scholar]
- 15.Mougin A., Grégoire,A., Banroques,J., Ségault,V., Fournier,R., Brulé,F., Chevrier-Miller,M. and Branlant,C. (1996) Secondary structure of the yeast Saccharomyces cerevisiae pre-U3A snoRNA and its implication for splicing efficiency. RNA, 2, 1079–1093. [PMC free article] [PubMed] [Google Scholar]
- 16.Muro A.F., Caputi,M., Pariyarath,R., Pagani,F., Buratti,E. and Baralle,F.E. (1999) Regulation of fibronectin EDA exon alternative splicing: possible role of RNA secondary structure for enhancer display. Mol. Cell. Biol., 19, 2657–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muesing M.A., Smith,D.H. and Capon,D.J. (1987) Regulation of mRNA accumulation by a human immunodeficiency virus trans-activator protein. Cell, 48, 691–701. [DOI] [PubMed] [Google Scholar]
- 18.Shi H., Hoffman,B.E. and Lis,J.T. (1997) A specific RNA hairpin loop structure binds the RNA recognition motifs of the Drosophila SR protein B52. Mol. Cell. Biol., 17, 2649–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Handa N., Nureki,O., Kurimoto,K., Kim,I., Sakamoto,H., Shimura,Y., Muto,Y. and Yokoyama,S. (1999) Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature, 398, 579–585. [DOI] [PubMed] [Google Scholar]
- 20.Watakabe A., Inoue,K., Sakamoto,H. and Shimura,Y. (1989) A secondary structure at the 3’ splice site affects the in vitro splicing reaction of mouse immunoglobulin m chain pre-mRNAs. Nucleic Acids Res., 17, 8159–8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clouet d’Orval B., d’Aubenton-Carafa,Y., Brody,J.M. and Brody,E. (1991) Determination of an RNA structure involved in splicing inhibition of a muscle-specific exon. J. Mol. Biol., 221, 837–856. [DOI] [PubMed] [Google Scholar]
- 22.Libri D., Piseri,A. and Fiszman,M.Y. (1991) Tissue-specific splicing in vivo of the beta-tropomyosin gene: dependence on an RNA secondary structure. Science, 252, 1842–1845. [DOI] [PubMed] [Google Scholar]
- 23.Charneau P., Alizon,M. and Clavel,F. (1992) A second origin of DNA plus-strand synthesis is required for optimal human immunodeficiency virus replication. J. Virol., 66, 2814–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nour M., Naimi,A., Beck,G. and Branlant,C. (1995) 16S-23S and 23S-5S intergenic spacer regions of Streptococcus thermophilus and Streptococcus salivarius, primary and secondary structure. Curr. Microbiol., 31, 270–278. [DOI] [PubMed]
- 25.Ratner L., Haseltine,W., Patarca,R., Livak,K.J., Starcich,B., Josephs,S.F., Doran,E.R., Rafalski,J.A., Whitehorn,E.A., Baumeister,K., et al. (1985) Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature, 313, 277–283. [DOI] [PubMed] [Google Scholar]
- 26.Alizon M., Wain-Hobson,S., Montagnier,L. and Sonigo,P. (1986) Genetic variability of the AIDS virus: nucleotide sequence analysis of two isolates from African patients. Cell, 46, 63–74. [DOI] [PubMed] [Google Scholar]
- 27.Kramer W., Drutsa,V., Jansen,H.W., Kramer,B., Pflugfelder,M. and Fritz,H.J. (1984) The gapped duplex DNA approach to oligonucleotide-directed mutation construction. Nucleic Acids Res., 12, 9441–9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vassilenko S.K. and Babkina,G.T. (1965) Isolation and properties of the cobra venom nuclease. Biokhymiya, 30, 705–712. [PubMed]
- 29.Dignam J.D., Lebovitz,R.M. and Roeder,R.G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res., 11, 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaeger J.A., Turner,D.H. and Zuker,M. (1989) Improved predictions of secondary structures for RNA. Proc. Natl Acad. Sci. USA, 86, 7706–7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao F., Bailes,E., Robertson,D.L., Chen,Y., Rodenburg,C.M., Michael,S.F., Cummins,L.B., Arthur,L.O., Peeters,M., Shaw,G.M. et al. (1999) Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature, 397, 436–441. [DOI] [PubMed] [Google Scholar]
- 32.Del Gatto-Konczak F., Olive,M., Gesnel,M.C. and Breathnach,R. (1999) hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol. Cell. Biol., 19, 251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baudin F., Marquet,R., Isel,C., Darlix,J.L., Ehresmann,B. and Ehresmann,C. (1993) Functional sites in the 5′ region of human immunodeficiency virus type 1 RNA form defined structural domains. J. Mol. Biol., 229, 382–397. [DOI] [PubMed] [Google Scholar]
- 34.Zamore P.D., Patton,J.G. and Green,M.R. (1992) Cloning and domain structure of the mammalian splicing factor U2AF. Nature, 355, 609–614. [DOI] [PubMed] [Google Scholar]
- 35.Wu S., Romfo,C.M., Nilsen,T.W. and Green,M.R. (1999) Functional recognition of the 3’ splice site AG by the splicing factor U2AF35. Nature, 402, 832–835. [DOI] [PubMed] [Google Scholar]
- 36.Guth S., Martinez,C., Gaur,R.K. and Valcarcel,J. (1999) Evidence for substrate-specific requirement of the splicing factor U2AF35 and for its function after polypyrimidine tract recognition by U2AF65. Mol. Cell. Biol., 19, 8263–8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reed R. (1989) The organization of 3’ splice-site sequences in mammalian introns. Genes Dev., 3, 2113–2123. [DOI] [PubMed] [Google Scholar]
- 38.Burd C.G. and Dreyfuss,G. (1994) RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J., 13, 1197–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chabot B., Blanchette,M., Lapierre,I. and La Branche,H. (1997) An intron element modulating 5’ splice site selection in the hnRNP A1 pre-mRNA interacts with hnRNP A1. Mol. Cell. Biol., 17, 1776–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding J., Hayashi,M.K., Zhang,Y., Manche,L., Krainer,A.R. and Xu,R.M. (1999) Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev., 13, 1102–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swanson A.K. and Stoltzfus,C.M. (1998) Overlapping cis sites used for splicing of HIV-1 env/nef and rev mRNAs. J. Biol. Chem., 273, 34551–34557. [DOI] [PubMed] [Google Scholar]
- 42.Tacke R. and Manley,J.L. (1995) The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J., 14, 3540–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]