Abstract
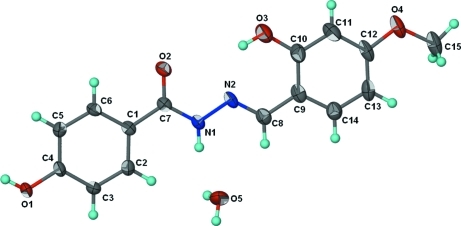
The Schiff base molecule of the title compound, C15H14N2O4·H2O, adopts a trans configuration with respect to the C=N double bond; the Schiff base itself is almost planar (r.m.s. deviation for all non-H atoms = 0.040 Å). The amido N atom is the hydrogen-bond donor to the water molecule, which is the hydrogen-bond donor to the hydroxy groups of two neighboring molecules. One of the hydroxyl groups acts as an intramolecular and the other as an intermolecular hydrogen-bond donor.
Related literature
For the structure of (E)-4-chloro-N′-(2-hydroxy-3-methoxybenzylidene)benzohydrazide, which crystallizes as a monohydrate, see: Cui et al. (2007 ▶). For a series of similar compounds, see: Lu et al. (2008a
▶,b
▶,c
▶). For this and other compounds with antimalarial properties, see: Melnyk et al. (2006 ▶).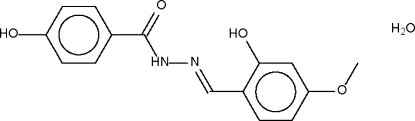
Experimental
Crystal data
C15H14N2O4·H2O
M r = 304.30
Monoclinic,
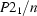
a = 7.1763 (2) Å
b = 16.6507 (5) Å
c = 12.1828 (4) Å
β = 98.022 (2)°
V = 1441.48 (8) Å3
Z = 4
Mo Kα radiation
μ = 0.11 mm−1
T = 100 (2) K
0.16 × 0.04 × 0.04 mm
Data collection
Bruker SMART APEX diffractometer
Absorption correction: none
13327 measured reflections
3315 independent reflections
1903 reflections with I > 2σ(I)
R int = 0.053
Refinement
R[F 2 > 2σ(F 2)] = 0.060
wR(F 2) = 0.180
S = 1.05
3315 reflections
200 parameters
H-atom parameters constrained
Δρmax = 0.66 e Å−3
Δρmin = −0.43 e Å−3
Data collection: APEX2 (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: X-SEED (Barbour, 2001 ▶); software used to prepare material for publication: publCIF (Westrip, 2009 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808042888/bt2836sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808042888/bt2836Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1⋯O2i | 0.84 | 1.86 | 2.621 (3) | 150 |
| O3—H3⋯N2 | 0.84 | 1.94 | 2.575 (3) | 132 |
| O5—H51⋯O1ii | 0.84 | 2.03 | 2.833 (3) | 160 |
| O5—H52⋯O3iii | 0.84 | 2.27 | 3.070 (4) | 160 |
| N1—H11⋯O5 | 0.88 | 2.05 | 2.883 (3) | 158 |
Symmetry codes: (i) 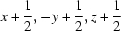 ; (ii)
; (ii) 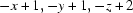 ; (iii)
; (iii) 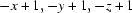 .
.
Acknowledgments
We thank the University of Malaya for funding this study (Science Fund grants 12–02-03–2031, 12–02-03–2051).
supplementary crystallographic information
Experimental
2-Hydroxy-3-methoxybenzaldehyde (0.30 g, 2 mmol) and 4-hydroxybenzohydrazide (0.30 g, 2 mmol) were heated in an ethanol-methanol mixture (50 ml) for 2 h. The solvent was removed and the resulting compound recrystallized from ethanol.
Refinement
Carbon-bound H-atoms were placed in calculated positions (C—H 0.95–0.98 Å) and were included in the refinement in the riding model approximation, with U(H) set to 1.2–1.5U(C). The oxygen- and nitrogen-bound ones were located in a difference Fourier map, and were refined with distance restraints (O–H 0.84±0.01, N–H 0.88±0.01 Å); their isotropic displacement parameters were freely refined.
Figures
Fig. 1.
Anisotropic displacement ellipsoid plot (Barbour, 2001) of C15H14N2O4.H2O at the 70% probability level. Hydrogen atoms are drawn as spheres of arbitrary radius.
Crystal data
| C15H14N2O4·H2O | F(000) = 640 |
| Mr = 304.30 | Dx = 1.402 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2yn | Cell parameters from 1918 reflections |
| a = 7.1763 (2) Å | θ = 2.4–27.3° |
| b = 16.6507 (5) Å | µ = 0.11 mm−1 |
| c = 12.1828 (4) Å | T = 100 K |
| β = 98.022 (2)° | Prism, yellow |
| V = 1441.48 (8) Å3 | 0.16 × 0.04 × 0.04 mm |
| Z = 4 |
Data collection
| Bruker SMART APEX diffractometer | 1903 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.053 |
| graphite | θmax = 27.5°, θmin = 2.1° |
| ω scans | h = −9→9 |
| 13327 measured reflections | k = −21→21 |
| 3315 independent reflections | l = −15→15 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.060 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.180 | H-atom parameters constrained |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.0687P)2 + 1.4764P] where P = (Fo2 + 2Fc2)/3 |
| 3315 reflections | (Δ/σ)max = 0.001 |
| 200 parameters | Δρmax = 0.66 e Å−3 |
| 0 restraints | Δρmin = −0.43 e Å−3 |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.7450 (3) | 0.31702 (12) | 1.09538 (15) | 0.0271 (5) | |
| H1 | 0.7745 | 0.2698 | 1.1150 | 0.041* | |
| O2 | 0.3884 (3) | 0.32742 (11) | 0.58919 (15) | 0.0246 (5) | |
| O3 | 0.1758 (3) | 0.41831 (13) | 0.32215 (18) | 0.0444 (6) | |
| H3 | 0.2262 | 0.4067 | 0.3866 | 0.067* | |
| O4 | −0.1014 (3) | 0.59721 (14) | 0.05179 (16) | 0.0359 (6) | |
| O5 | 0.4249 (4) | 0.61425 (14) | 0.73043 (18) | 0.0495 (7) | |
| H51 | 0.4027 | 0.6363 | 0.7892 | 0.074* | |
| H52 | 0.5420 | 0.6108 | 0.7317 | 0.074* | |
| N1 | 0.3620 (3) | 0.45875 (14) | 0.62786 (18) | 0.0208 (5) | |
| H11 | 0.3800 | 0.4987 | 0.6754 | 0.025* | |
| N2 | 0.2800 (3) | 0.47111 (14) | 0.51985 (17) | 0.0218 (5) | |
| C1 | 0.4997 (3) | 0.36829 (15) | 0.7739 (2) | 0.0174 (5) | |
| C2 | 0.5351 (4) | 0.42869 (16) | 0.8536 (2) | 0.0204 (6) | |
| H2 | 0.5012 | 0.4826 | 0.8345 | 0.024* | |
| C3 | 0.6192 (4) | 0.41082 (16) | 0.9602 (2) | 0.0216 (6) | |
| H3A | 0.6449 | 0.4524 | 1.0135 | 0.026* | |
| C4 | 0.6653 (3) | 0.33229 (16) | 0.9886 (2) | 0.0197 (6) | |
| C5 | 0.6322 (4) | 0.27144 (16) | 0.9108 (2) | 0.0214 (6) | |
| H5A | 0.6655 | 0.2176 | 0.9305 | 0.026* | |
| C6 | 0.5504 (4) | 0.28976 (16) | 0.8043 (2) | 0.0208 (6) | |
| H6 | 0.5284 | 0.2481 | 0.7508 | 0.025* | |
| C7 | 0.4134 (3) | 0.38292 (16) | 0.6579 (2) | 0.0191 (6) | |
| C8 | 0.2221 (4) | 0.54217 (18) | 0.4913 (2) | 0.0231 (6) | |
| H8 | 0.2362 | 0.5848 | 0.5436 | 0.028* | |
| C9 | 0.1350 (4) | 0.55631 (18) | 0.3780 (2) | 0.0235 (6) | |
| C10 | 0.1117 (4) | 0.49462 (18) | 0.2982 (2) | 0.0289 (7) | |
| C11 | 0.0295 (4) | 0.5107 (2) | 0.1905 (2) | 0.0329 (7) | |
| H11A | 0.0125 | 0.4689 | 0.1371 | 0.040* | |
| C12 | −0.0278 (4) | 0.5882 (2) | 0.1614 (2) | 0.0281 (7) | |
| C13 | −0.0097 (4) | 0.64983 (19) | 0.2380 (2) | 0.0290 (7) | |
| H13 | −0.0519 | 0.7025 | 0.2177 | 0.035* | |
| C14 | 0.0714 (4) | 0.63284 (18) | 0.3450 (2) | 0.0270 (6) | |
| H14 | 0.0843 | 0.6749 | 0.3982 | 0.032* | |
| C15 | −0.1581 (4) | 0.6759 (2) | 0.0151 (3) | 0.0385 (8) | |
| H15A | −0.2026 | 0.6749 | −0.0647 | 0.058* | |
| H15B | −0.0508 | 0.7127 | 0.0299 | 0.058* | |
| H15C | −0.2598 | 0.6944 | 0.0549 | 0.058* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0379 (11) | 0.0266 (11) | 0.0146 (9) | 0.0096 (9) | −0.0039 (8) | 0.0015 (8) |
| O2 | 0.0309 (10) | 0.0230 (10) | 0.0188 (10) | −0.0048 (8) | −0.0008 (8) | −0.0005 (8) |
| O3 | 0.0701 (17) | 0.0318 (13) | 0.0264 (12) | −0.0012 (12) | −0.0107 (11) | 0.0029 (10) |
| O4 | 0.0320 (12) | 0.0538 (15) | 0.0203 (10) | 0.0041 (10) | −0.0018 (9) | 0.0118 (10) |
| O5 | 0.0772 (18) | 0.0431 (14) | 0.0259 (12) | 0.0265 (13) | −0.0009 (12) | −0.0119 (11) |
| N1 | 0.0228 (12) | 0.0233 (12) | 0.0152 (11) | 0.0005 (9) | −0.0016 (9) | 0.0003 (9) |
| N2 | 0.0209 (11) | 0.0306 (13) | 0.0132 (11) | −0.0014 (10) | 0.0004 (9) | 0.0039 (9) |
| C1 | 0.0136 (12) | 0.0223 (13) | 0.0164 (13) | −0.0033 (10) | 0.0020 (10) | −0.0001 (11) |
| C2 | 0.0201 (13) | 0.0213 (14) | 0.0197 (13) | 0.0023 (10) | 0.0027 (10) | 0.0031 (11) |
| C3 | 0.0260 (14) | 0.0207 (14) | 0.0172 (13) | 0.0012 (11) | 0.0001 (10) | −0.0035 (11) |
| C4 | 0.0192 (13) | 0.0249 (14) | 0.0144 (12) | 0.0032 (11) | 0.0007 (10) | 0.0034 (11) |
| C5 | 0.0221 (13) | 0.0201 (14) | 0.0214 (13) | 0.0022 (11) | 0.0006 (11) | 0.0011 (11) |
| C6 | 0.0217 (13) | 0.0228 (14) | 0.0170 (13) | −0.0005 (11) | −0.0004 (11) | −0.0033 (11) |
| C7 | 0.0173 (13) | 0.0225 (14) | 0.0176 (13) | −0.0027 (10) | 0.0029 (10) | 0.0000 (11) |
| C8 | 0.0197 (13) | 0.0303 (15) | 0.0195 (14) | 0.0004 (11) | 0.0035 (11) | 0.0013 (12) |
| C9 | 0.0184 (13) | 0.0327 (16) | 0.0198 (14) | −0.0033 (11) | 0.0043 (11) | 0.0066 (12) |
| C10 | 0.0333 (16) | 0.0286 (16) | 0.0242 (14) | −0.0018 (13) | 0.0013 (12) | 0.0074 (13) |
| C11 | 0.0371 (17) | 0.0391 (18) | 0.0211 (15) | −0.0053 (14) | −0.0013 (12) | 0.0008 (13) |
| C12 | 0.0192 (14) | 0.0471 (19) | 0.0177 (14) | −0.0008 (13) | 0.0011 (11) | 0.0108 (13) |
| C13 | 0.0237 (14) | 0.0358 (17) | 0.0269 (16) | 0.0055 (12) | 0.0018 (12) | 0.0123 (13) |
| C14 | 0.0235 (14) | 0.0327 (16) | 0.0249 (15) | 0.0039 (12) | 0.0043 (12) | 0.0033 (12) |
| C15 | 0.0314 (17) | 0.057 (2) | 0.0263 (16) | 0.0062 (16) | 0.0018 (13) | 0.0173 (16) |
Geometric parameters (Å, °)
| O1—C4 | 1.370 (3) | C3—H3A | 0.9500 |
| O1—H1 | 0.8400 | C4—C5 | 1.385 (4) |
| O2—C7 | 1.243 (3) | C5—C6 | 1.382 (4) |
| O3—C10 | 1.369 (4) | C5—H5A | 0.9500 |
| O3—H3 | 0.8400 | C6—H6 | 0.9500 |
| O4—C12 | 1.374 (3) | C8—C9 | 1.452 (4) |
| O4—C15 | 1.426 (4) | C8—H8 | 0.9500 |
| O5—H51 | 0.8400 | C9—C14 | 1.394 (4) |
| O5—H52 | 0.8400 | C9—C10 | 1.408 (4) |
| N1—C7 | 1.351 (3) | C10—C11 | 1.387 (4) |
| N1—N2 | 1.380 (3) | C11—C12 | 1.385 (4) |
| N1—H11 | 0.8800 | C11—H11A | 0.9500 |
| N2—C8 | 1.286 (4) | C12—C13 | 1.381 (4) |
| C1—C6 | 1.393 (4) | C13—C14 | 1.381 (4) |
| C1—C2 | 1.396 (4) | C13—H13 | 0.9500 |
| C1—C7 | 1.482 (3) | C14—H14 | 0.9500 |
| C2—C3 | 1.386 (4) | C15—H15A | 0.9800 |
| C2—H2 | 0.9500 | C15—H15B | 0.9800 |
| C3—C4 | 1.381 (4) | C15—H15C | 0.9800 |
| C4—O1—H1 | 119.9 | N1—C7—C1 | 118.3 (2) |
| C10—O3—H3 | 120.0 | N2—C8—C9 | 119.1 (3) |
| C12—O4—C15 | 117.3 (3) | N2—C8—H8 | 120.5 |
| H51—O5—H52 | 108.8 | C9—C8—H8 | 120.5 |
| C7—N1—N2 | 117.5 (2) | C14—C9—C10 | 117.7 (3) |
| C7—N1—H11 | 121.3 | C14—C9—C8 | 120.2 (3) |
| N2—N1—H11 | 121.3 | C10—C9—C8 | 122.1 (3) |
| C8—N2—N1 | 118.3 (2) | O3—C10—C11 | 117.8 (3) |
| C6—C1—C2 | 118.4 (2) | O3—C10—C9 | 121.7 (3) |
| C6—C1—C7 | 117.8 (2) | C11—C10—C9 | 120.4 (3) |
| C2—C1—C7 | 123.8 (2) | C12—C11—C10 | 119.6 (3) |
| C3—C2—C1 | 120.7 (2) | C12—C11—H11A | 120.2 |
| C3—C2—H2 | 119.7 | C10—C11—H11A | 120.2 |
| C1—C2—H2 | 119.7 | O4—C12—C13 | 124.3 (3) |
| C4—C3—C2 | 119.8 (2) | O4—C12—C11 | 114.2 (3) |
| C4—C3—H3A | 120.1 | C13—C12—C11 | 121.5 (3) |
| C2—C3—H3A | 120.1 | C12—C13—C14 | 118.3 (3) |
| O1—C4—C3 | 117.9 (2) | C12—C13—H13 | 120.9 |
| O1—C4—C5 | 121.5 (2) | C14—C13—H13 | 120.9 |
| C3—C4—C5 | 120.5 (2) | C13—C14—C9 | 122.5 (3) |
| C6—C5—C4 | 119.4 (2) | C13—C14—H14 | 118.7 |
| C6—C5—H5A | 120.3 | C9—C14—H14 | 118.7 |
| C4—C5—H5A | 120.3 | O4—C15—H15A | 109.5 |
| C5—C6—C1 | 121.2 (2) | O4—C15—H15B | 109.5 |
| C5—C6—H6 | 119.4 | H15A—C15—H15B | 109.5 |
| C1—C6—H6 | 119.4 | O4—C15—H15C | 109.5 |
| O2—C7—N1 | 120.3 (2) | H15A—C15—H15C | 109.5 |
| O2—C7—C1 | 121.4 (2) | H15B—C15—H15C | 109.5 |
| C7—N1—N2—C8 | 176.8 (2) | N2—C8—C9—C14 | −179.9 (3) |
| C6—C1—C2—C3 | 0.2 (4) | N2—C8—C9—C10 | 0.0 (4) |
| C7—C1—C2—C3 | −178.9 (2) | C14—C9—C10—O3 | 177.7 (3) |
| C1—C2—C3—C4 | −1.2 (4) | C8—C9—C10—O3 | −2.2 (4) |
| C2—C3—C4—O1 | −178.8 (2) | C14—C9—C10—C11 | 0.6 (4) |
| C2—C3—C4—C5 | 1.5 (4) | C8—C9—C10—C11 | −179.3 (3) |
| O1—C4—C5—C6 | 179.6 (2) | O3—C10—C11—C12 | −176.4 (3) |
| C3—C4—C5—C6 | −0.7 (4) | C9—C10—C11—C12 | 0.8 (4) |
| C4—C5—C6—C1 | −0.3 (4) | C15—O4—C12—C13 | 1.6 (4) |
| C2—C1—C6—C5 | 0.6 (4) | C15—O4—C12—C11 | −178.2 (3) |
| C7—C1—C6—C5 | 179.7 (2) | C10—C11—C12—O4 | 178.0 (3) |
| N2—N1—C7—O2 | 0.7 (4) | C10—C11—C12—C13 | −1.8 (4) |
| N2—N1—C7—C1 | −179.3 (2) | O4—C12—C13—C14 | −178.4 (3) |
| C6—C1—C7—O2 | −0.7 (4) | C11—C12—C13—C14 | 1.4 (4) |
| C2—C1—C7—O2 | 178.4 (2) | C12—C13—C14—C9 | 0.0 (4) |
| C6—C1—C7—N1 | 179.3 (2) | C10—C9—C14—C13 | −1.0 (4) |
| C2—C1—C7—N1 | −1.6 (4) | C8—C9—C14—C13 | 178.9 (3) |
| N1—N2—C8—C9 | −179.8 (2) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···O2i | 0.84 | 1.86 | 2.621 (3) | 150 |
| O3—H3···N2 | 0.84 | 1.94 | 2.575 (3) | 132 |
| O5—H51···O1ii | 0.84 | 2.03 | 2.833 (3) | 160 |
| O5—H52···O3iii | 0.84 | 2.27 | 3.070 (4) | 160 |
| N1—H11···O5 | 0.88 | 2.05 | 2.883 (3) | 158 |
Symmetry codes: (i) x+1/2, −y+1/2, z+1/2; (ii) −x+1, −y+1, −z+2; (iii) −x+1, −y+1, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: BT2836).
References
- Barbour, L. J. (2001). J. Supramol. Chem.1, 189–191.
- Bruker (2007). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Cui, J., Yin, H. & Qiao, Y. (2007). Acta Cryst. E63, o3548.
- Lu, J.-F., Min, S.-T., Ji, X.-H. & Dang, Z.-H. (2008a). Acta Cryst. E64, o1693. [DOI] [PMC free article] [PubMed]
- Lu, J.-F., Min, S.-T., Ji, X.-H. & Dang, Z.-H. (2008b). Acta Cryst. E64, o1694. [DOI] [PMC free article] [PubMed]
- Lu, J.-F., Min, S.-T., Ji, X.-H. & Dang, Z.-H. (2008c). Acta Cryst. E64, o1695. [DOI] [PMC free article] [PubMed]
- Melnyk, P., Leroux, V., Sergheraert, C. & Grellier, P. (2006). Bioorg. & Med. Chem. Lett.16, 31–35. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Westrip, S. P. (2009). publCIF In preparation.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808042888/bt2836sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808042888/bt2836Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report