Abstract
Translation of thymidylate synthase (TS) mRNA is controlled by its own protein end-product TS in a negative autoregulatory manner. Disruption of this regulation results in increased synthesis of TS and may lead to the development of cellular drug resistance to TS-directed anticancer agents. As a strategy to inhibit TS expression, antisense 2′-O-methyl RNA oligoribonucleotides (ORNs) were designed to directly target the 5′ upstream cis-acting regulatory element (nucleotides 80–109) of TS mRNA. A 30 nt ORN, HYB0432, inhibited TS expression in human colon cancer RKO cells in a dose-dependent manner but had no effect on the expression of β-actin, α-tubulin or topoisomerase I. TS expression was unaffected by treatment with control sense or mismatched ORNs. HYB0504, an 18 nt ORN targeting the same core sequence, also repressed expression of TS protein. However, further reduction in oligo size resulted in loss of antisense activity. Following HYB0432 treatment, TS protein levels were reduced by 60% within 6 h and were maximally reduced by 24 h. Expression of p53 protein was inversely related to that of TS, suggesting that p53 expression may be directly linked to intracellular levels of TS. Northern blot analysis demonstrated that TS mRNA was unaffected by HYB0432 treatment. The half-life of TS protein was unchanged after antisense treatment suggesting that the mechanism of action of antisense ORNs is mediated through a process of translational arrest. These findings demonstrate that an antisense ORN targeted at a critical cis-acting element on TS mRNA can specifically inhibit expression of TS protein in RKO cells.
INTRODUCTION
Thymidylate synthase (TS) catalyzes the conversion of deoxyuridine monophosphate (dUMP) and 5,10-methylenetetrahydrofolate (CH2THF) to thymidine monophosphate (dTMP) and dihydrofolate (FH2) (1). Because this enzymatic reaction provides for the sole de novo intracellular source of dTMP, TS is a critical therapeutic target in cancer chemotherapy (2–5).
In addition to its role in enzyme catalysis, there is evidence that TS also functions as an RNA binding protein (6,7). Studies from this laboratory have demonstrated that TS binds with relatively high affinity to two different regions on its own mRNA. The first site is a putative stem–loop structure corresponding to nucleotides 80–109 and contains the translational start site (8). The second binding site is located in the protein-coding region within a 70 nt sequence between nucleotides 480–550 (9). Binding of TS to either of these elements results in translational repression. However, when TS is bound by its physiologic nucleotide substrate dUMP or the 5-fluorouracil (5-FU) nucleotide metabolite FdUMP, it is no longer able to bind to its own mRNA, thereby allowing for synthesis of new TS protein (7). This interaction between TS and its cognate TS mRNA represents an efficient mechanism to control the intracellular levels of TS. Disruption of this normal autoregulatory process may result in the acute induction of TS and the rapid development of cellular resistance in response to exposure to inhibitor compounds of TS such as 5-FU and Tomudex (TDX). In support of this model, studies in an in vitro and in vivo setting have observed induction of TS protein with no corresponding change in TS mRNA levels upon exposure to various TS inhibitor agents (10–12).
The ability to increase the expression of TS protein reduces the efficacy of traditional anticancer agents targeting TS protein. Our laboratory has attempted to circumvent the translational induction of TS by designing antisense RNA molecules that inhibit the process of TS mRNA translation (13). Using a rabbit reticulocyte lysate in vitro translation system, we demonstrated that a native 2′-OH antisense oligoribonucleotide (ORN), targeting nucleotides 80–109 on human TS mRNA, was able to effectively repress translation of TS mRNA in a concentration-dependent manner. This effect was specific in that the native antisense ORN was unable to inhibit translation of control mRNAs including Escherichia coli TS and p53. One major drawback of native 2′-OH ORNs is their enhanced sensitivity to nuclease degradation by RNases (14,15). This limitation has been overcome, to a large extent, by chemically modifying the ORN on the ribose ring or the phosphate backbone which stabilizes the ORN against nuclease activity (14). With this in mind, antisense 2′-O-methyl ORNs with either phosphodiester (PO) (HYB0432) or phosphorothioate (PS) (HYB0431) backbones were tested in the in vitro translation system. Using these modified RNA oligos, we observed greater repression of TS mRNA translation than the native 2′-OH ORN. However, non-specific effects on mRNA translation were observed with the 2′-O-methyl PS ORN. In vitro translation experiments showed that TS mRNA, in the presence of antisense ORNs, remained intact suggesting a mechanism of translational arrest (13). These preliminary in vitro experiments revealed that antisense ORNs targeted at the 5′ cis-acting binding element of TS mRNA are effective inhibitors of TS mRNA translation.
In the present study, we demonstrate that antisense HYB0432 inhibits expression of TS protein in the human colon cancer RKO cell line as well as in a TDX-resistant RKO-TDX cell line, which expresses 16-fold higher levels of TS protein. We also show that treatment with HYB0432 does not alter the expression of TS mRNA nor the stability of TS protein. Our studies suggest that an antisense 2′-O-methyl ORN directed against a key cis-acting sequence in human TS mRNA exerts its regulatory effects through a process of translational arrest. The potential therapeutic implications of these antisense ORNs on inhibition of TS expression are discussed.
MATERIALS AND METHODS
Cell culture
The human colon cancer RKO cell line was maintained in 75 cm2 plastic tissue culture flasks (Falcon Labware) in growth medium containing RPMI 1640 medium (Gibco BRL) with 10% dialyzed fetal bovine serum (dFBS, Gibco BRL). The characteristics of the RKO-TDX cell line have been previously characterized (16), and was a kind gift from Dr Peter Danenberg (Norris Cancer Center, USC, Los Angeles, CA). The human colon cancer RKO-TDX cell line was grown under identical conditions and maintained in 10 nM TDX.
Oligonucleotides
All ODNs were complementary to the sequence corresponding to nucleotides 80–109 of TS mRNA. This sequence includes the translational start site, and is as follows: 5′-CCG-CCC-GCC-GCG-CCA-UGC-CUG-UGG-CCG-GCU-3′. A 30 nt DNA antisense PS oligonucleotide (ODN) was synthesized by the Department of Pathology, Yale University. 2′-O-methyl RNA antisense ORNs were synthesized by Hybridon, Inc. TSAS18-MM and TS18 ORNs were synthesized by the Keck Core Facility at Yale University. The purity of the ODNs was confirmed by electrophoresis on a 20% acrylamide/8 M urea gel followed by detection with methylene blue dye. The sequences of the ORNs and ODN are as follows, and the bold nucleotides indicate PS linkages. The mismatched nucleotides are underlined.
HYB0432 5′-AGC-CGG-CCA-CAG-GCA-UGG-CGC-GGC-GGG-CGG-3′
ODN 5′-AGC-CGG-CCA-CAG-GCA-UGG-CGC-GGC-GGG-CGG-3′
HYB0848 5′-CCG-CCC-GCC-GCG-CCA-UGC-CUG-UGG-CCG-GCU-3′
HYB0503 5′-CGG-ACA-CAU-GCA-UAG-CGC-GUC-GGG-3′
HYB0504 5′-CCA-CAG-GCA-UGG-CGC-GGC-3′
TSAS18-MM 5′-ACA-CAU-GCA-UAG-CGA-GUC-3′
TS18 5′-GCC-GCG-CCA-UGC-CUG-UGG-3′
Western immunoblot analysis
Human colon cancer RKO cells were plated in 25 cm2 flasks at a density of 2.0 × 105 cells per flask. ODNs were complexed with the cationic lipid Eufectin 7 (JBL Scientific Inc.) in OPTI-MEM medium (Gibco). All flasks were washed with OPTI-MEM medium prior to addition of the oligo–lipid complexes. After 6 h, the medium was replaced with fresh RPMI 1640 medium containing 10% dFBS. After 24 h, RKO cells were trypsinized and washed twice with ice-cold PBS. Cell pellets were stored at –80°C until needed.
RKO cell pellets were resuspended in RIPA buffer (1× PBS pH 7.4, 1.0% IGEPAL, 0.5% deoxycholic acid, 0.1% SDS) containing freshly added PMSF (300 µg/ml) and Protease Inhibitor Cocktail (containing AEBSF, pepstatin A, bestatin, leupeptin, E-64 and aprotinin; Sigma). Suspensions were incubated at 4°C for 15 min and centrifuged for 30 min at 4°C. Protein concentration was determined using the DC Protein Assay (Bio-Rad Laboratories). Equivalent amounts of protein (30 µg) from each cell lysate were resolved on SDS–PAGE (15% acrylamide) using the method of Laemmli (17). Gels were electroblotted onto nitrocellulose membranes (Bio-Rad), and filter membranes were then incubated in blocking solution (1× PBS, 0.2% Tween-20, 5% non-fat dry milk powder) for 2 h at room temperature. Membranes were incubated overnight at 4°C with primary antibodies at the following dilutions: anti-TS106 monoclonal antibody, 1:2000; anti-p53 monoclonal antibody (Santa Cruz), 1:500; anti-β-actin monoclonal antibody (Amersham Life Sciences), 1:30 000; anti-α-tubulin monoclonal antibody (Amersham), 1:60 000; anti-topoisomerase I monoclonal antibody, 1:1000 (a gift from Dr Yung-chi Cheng, Yale University). After four 15 min washes in PBST (1× PBS, 0.2% Tween-20), membranes were incubated with a dilution of 1:2000 of horseradish peroxidase-conjugated secondary antibodies (IgG goat anti-mouse, Bio-Rad) for 1 h at room temperature. After an additional four 15 min PBST washes, membranes were processed by the enhanced chemiluminescence method (SuperSignal Substrate, Pierce) and protein bands were visualized by autoradiography. Quantitation of signal intensities was performed by densitometry on a Hewlett-Packard ScanJet 4p using NIH IMAGE 1.59 software.
TS catalytic assay
RKO cells were treated with HYB0432–Eufectin complexes for 6 h, and allowed to grow for an additional 24 h. RKO cells were harvested as described previously (18), and cell pellets were resuspended in 0.1 M KH2PO4 (pH 7.4). Cell lysis was accomplished by sonication using three 2–3 s bursts. The extracts were centrifuged and the supernatants were assayed immediately. The TS catalytic assay was performed in a total volume of 200 µl containing 10 µM [5-3H]dUMP (specific activity, 20 Ci/mmol), 100 mM 2-mercaptoethanol, 50 mM KH2PO4 (pH 7.4), 150 µM 5,10-methylenetetrahydrofolate and cytosolic extract (30–60 µg total protein) (18). Samples were incubated at 37°C for 30 min, and the reaction was stopped with the addition of 100 µl of 20% trichloroacetic acid. Residual [5-3H]dUMP was removed by the addition of 200 µl of an albumin-coated activated charcoal solution. The charcoal was removed by centrifugation, and a 250 µl sample of the supernatant was assayed for tritium radioactivity.
Isolation of total RNA and northern blot hybridization analysis
After antisense treatment, RKO cells were harvested and washed twice with ice-cold 1× PBS buffer. Total RNA was isolated from cell pellets using the RNeasy kit from Qiagen. RNAs were resolved on a 1% agarose/formaldehyde gel to verify integrity and size. The concentration of RNA was determined by UV spectrophotometry. For northern blot analysis, RNAs (5 µg) were resolved on a 1% agarose/formaldehyde gel and then transferred to a positively charged BrightStar-Plus nylon membrane (Ambion) by capillary transfer. Antisense RNA probes were synthesized in vitro using the MEGAshortscript T7 kit (Ambion, cat. no. 1354). The TS antisense probe was derived from a PCR-generated template complementary to nucleotides 520–1216 of the human TS cDNA. A 28S antisense probe was generated from a template purchased from Ambion. RNA probes were gel-purified and biotin-labeled using the BrightStar Psoralen-Biotin non-isotopic labeling kit (Ambion, cat. no. 1480). The membrane was pre-hybridized for 2 h at 65°C followed by probe hybridization overnight at 65°C. The BrightStar BioDetect kit (Ambion, cat. no. 1930) was then used for detection of cellular mRNAs. Quantitation of signal intensities was performed by densitometry on a Hewlett-Packard ScanJet 4p using NIH IMAGE 1.59 software.
Determination of TS protein half-life
RKO cells were treated for 6 h in the absence or presence of Eufectin–oligo complexes. After the transfection medium was removed, cells were incubated for 30 min with methionine-free RPMI 1640 medium to deplete intracellular pools of methionine. Cells were then incubated with 1 ml of methionine-free medium containing 10% dFBS and 150 µCi/ml 35S-methionine (EasyTag EXPRE35S35S protein labeling mix; specific activity, 1000 Ci/mmol; NEN Life Sciences). Radiolabel was removed after a 2 h incubation, and fresh methionine-containing medium (RPMI 1640) was added to the cell cultures. Cells were then harvested at the following time points: 0, 6, 12, 18 and 24 h, washed twice with ice-cold 1× PBS, lysed in ice-cold RIPA buffer, placed on ice for 15 min and centrifuged for 30 min at 4°C. Immunoprecipitation of TS protein was performed using an anti-TS polyclonal antibody bound to Protein-A agarose beads (Gibco BRL) according to previously described methods (10). Equivalent amounts of trichloroacetic acid-insoluble radioactivity (20 × 106 d.p.m.) from each cell extract were resolved on a 15% SDS–PAGE gel. The relative amounts of 35S-labeled TS protein were determined by autoradiography followed by densitometric scanning.
RESULTS
Previous studies from this laboratory have demonstrated in a cell-free in vitro translation system that a 30 nt antisense RNA, HYB0432, targeted to the 5′ upstream binding site of human TS mRNA, is able to repress translation in a concentration- and sequence-specific manner (13). To extend these studies to an intact biological system, we investigated the ability of antisense HYB0432 to repress translation of TS mRNA and subsequent TS expression in the human colon cancer RKO cell line. Given the charged nature of ORNs, a cationic lipid carrier was used to transport the oligo across the cellular membrane. The ability of a lipid carrier to bind and transport the ORN into cells depends on several factors including: (i) the charge of the lipid and the ORN, (ii) ORN base and backbone modifications, (iii) ORN length and (iv) cell line specificity (19). With this in mind, several commercially available lipids including Lipofectin, Lipofectamine, Fugene6 and Eufectin were initially tested for their ability to transfect antisense HYB0432 into RKO cells. While most of these preparations were able to transport HYB0432 to some extent, the intrinsic cytotoxicity of each lipid preparation precluded their subsequent use. Eufectin 7 was selected as the carrier lipid given its lack of intrinsic cytotoxic effects. Various ratios of Eufectin:HYB0432 were then tested as the carrier to oligo ratio has been shown to play a critical role in determining transfection efficiency. A weight ratio (µg:µg) of 5:1 Eufectin:HYB0432 was observed to be the most effective at inhibiting TS protein expression (Fig. 1, lane 4). Eufectin alone, at this concentration, had no effect on the expression of TS (Fig. 1, lane 7). All subsequent experiments with HYB0432 employed a ratio of 5:1 Eufectin:HYB0432.
Figure 1.
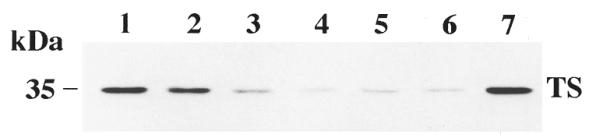
Effect of ratio of Eufectin to HYB0432 on TS expression. RKO cells were incubated with different Eufectin:HYB0432 combinations. After 6 h, complexes were removed and fresh RPMI medium was added. After an additional 24 h, cells were harvested and processed for western blot analysis as described in Materials and Methods. Lane 1 contains cell extracts from untreated RKO cells. Lanes 2–6 contain 190 nM HYB0432 complexed with Eufectin in weight ratios of 1:1 (lane 2), 1:3 (lane 3), 1:5 (lane 4), 1:7 (lane 5) and 1:9 (lane 6). Lane 7 contains cell extracts treated with Eufectin alone (10 µg/ml).
We next investigated the effect of HYB0432 on expression of TS in human colon cancer RKO cells by means of western immunoblot analysis. As seen in Figure 2, treatment with this antisense ORN for 6 h inhibited the expression of TS protein in a dose-dependent manner, with maximal inhibition (>90%) at 250 nM (Fig. 2, lanes 2–5). This reduction in the level of TS protein was accompanied by a significant decrease, of up to 80%, in TS catalytic activity after treatment with 250 nM HYB0432 (data not shown). In contrast, a sense 30 nt, 2′-O-methyl RNA, HYB0848 (Fig. 2, lane 6), and a 4-base mismatch 24 nt RNA, HYB0503 (Fig. 2, lane 7), at a concentration of 250 nM, had absolutely no effect on levels of TS protein. To provide evidence for the specificity of effect of the antisense ORN on TS expression, the levels of several other cellular proteins were analyzed after antisense treatment (Fig. 2). Expression of two housekeeping proteins, α-tubulin and β-actin, was not affected by HYB0432. However, since the half-lives of these housekeeping proteins are significantly longer than that of TS, the effect of antisense RNA treatment on the expression of a protein with a half-life similar to that of TS was also determined. The half-life of topoisomerase I, which is normally 10–16 h (20), remained unchanged with antisense treatment. These experiments demonstrate that an antisense ORN directed at the 5′-upstream cis-acting element of TS mRNA can specifically inhibit the expression of TS protein.
Figure 2.
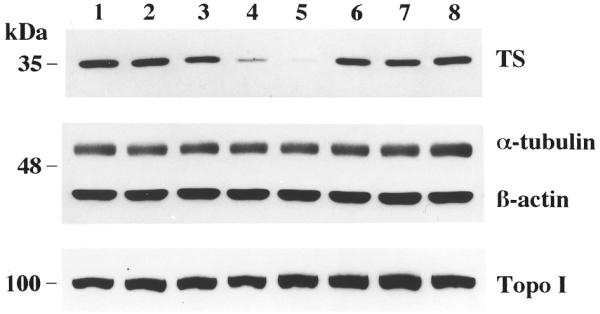
Western blot analysis of RKO cells after treatment with HYB0432. Cells were incubated in the absence (lane 1) or presence (lanes 2–8) of Eufectin–ORN complexes (5:1 ratio; µg:µg) for 6 h, grown for an additional 24 h and then harvested and processed for western blot analysis as described in Materials and Methods. Extracts in lanes 2–5 were treated with complexes containing 63, 126, 190 and 250 nM HYB0432, respectively. Cells were treated with various control ORNs: lane 6, sense ORN HYB0848 (250 nM); lane 7, mismatch ORN HYB0503 (250 nM). Lane 8 contains cell extracts treated with Eufectin alone (13 µg/ml).
To determine the duration of the antisense effect, levels of TS protein were monitored as a function of time following antisense therapy (Fig. 3). HYB0432 effectively decreased levels of TS protein almost immediately upon transfection. During the 6 h incubation period, TS protein levels were reduced by 60% from pre-treatment levels (Fig. 3, lane 2 versus lane 1). Maximal antisense effect occurred by 24 h (Fig. 3, lane 5) with TS protein levels slowly returning to baseline by 72 h. As an important control, the expression of two control proteins, α-tubulin and β-actin, remained unaffected over this same time course. Our laboratory has previously shown that TS protein binds to other cellular mRNAs including p53 (21). Recent studies have demonstrated that TS can control p53 expression at the translational level (22,23). As seen in Figure 3, treatment with HYB0432 resulted in a significant increase in the expression of p53 by up to 7-fold (Fig. 3, lane 3 versus lane 1). As the effect of antisense treatment on TS expression diminished and TS protein levels increased, levels of p53 returned to baseline (Fig. 3, lanes 6–8). These experiments provide further evidence that TS expression is specifically controlled by the antisense ORN. In addition, they lend further support to the theory that p53 expression may be directly linked to the cellular levels of TS protein.
Figure 3.
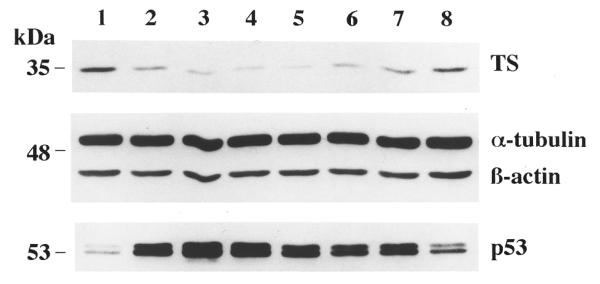
Duration of effect of HYB0432 on expression of TS protein in RKO cells. After a 6 h transfection with HYB0432–Eufectin complexes (190 nM), cells were harvested prior to transfection (lane 1) and at the following times: 0 (after transfection, lane 2), 6 (lane 3), 12 (lane 4), 24 (lane 5), 36 (lane 6), 48 (lane 7) and 72 h (lane 8). Cell extracts were processed for western blot analysis as described in Materials and Methods.
To gain insight into the potential mechanism(s) by which the antisense ORN was exerting its effects on TS expression, Northern blot analyses were performed. Figure 4A presents the results of a representative experiment in which the effect of HYB0432 on TS mRNA expression was examined. The corresponding effects of antisense treatment on protein expression are presented in Figure 4B. HYB0432, at a concentration of 190 nM, had no effect on levels of TS mRNA (Fig. 4A, lane 3). However, this same concentration was able to significantly inhibit TS protein expression by nearly 90% (Fig. 4B, lane 3). Sense 30 nt ORN HYB0848, at the same concentration of 190 nM, did not alter the expression of either TS mRNA (Fig. 4A, lane 5) or TS protein (Fig. 4B, lane 5). As an important control, a 30 nt PS DNA oligo targeting the same sequence on TS mRNA was used. Treatment with this antisense DNA oligo significantly reduced TS mRNA expression (Fig. 4A, lane 4) and TS protein expression by 80% (Fig. 4B, lane 4). This finding confirmed that the antisense DNA oligo exerts its effect through degradation of the target mRNA, presumably through activation of RNase H. These experiments, taken together, suggest that the mechanism by which HYB0432 represses TS expression is through a process of translational arrest and not by an RNase-mediated pathway.
Figure 4.
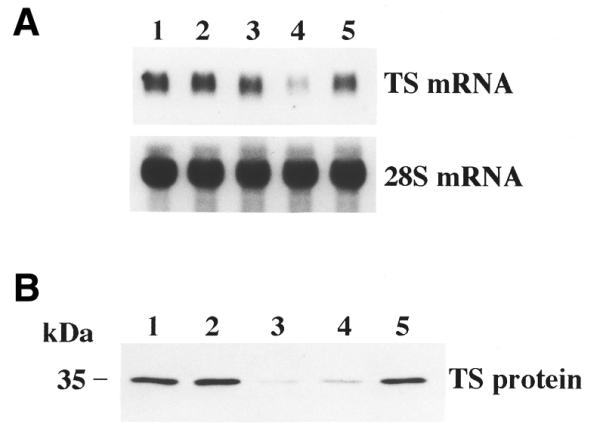
Effect of antisense treatment on expression of TS mRNA and TS protein. RKO cells were treated for 6 h in the absence (lane 1) or presence of oligo–Eufectin complexes (190 nM), grown for an additional 24 h, harvested and processed for northern (A) and western (B) blot analysis as described in Materials and Methods. Lanes 2–5 contain cell extracts treated with Eufectin alone (lane 2) or complexed with either HYB0432 (lane 3), a TS antisense 30mer PS-DNA oligo (lane 4) or TS sense 30mer RNA HYB0848 (lane 5).
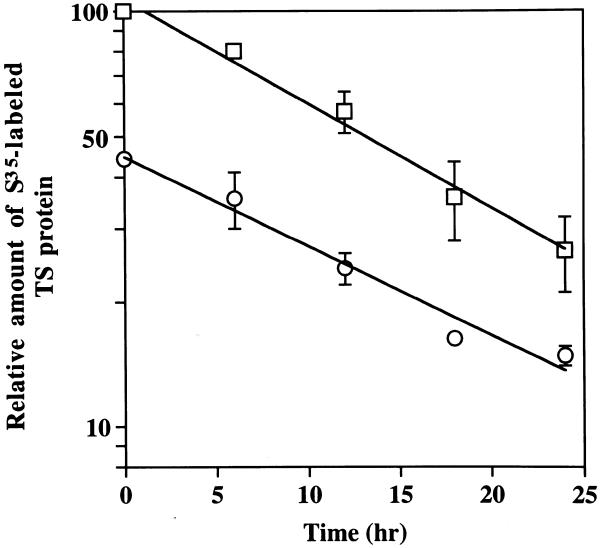
Since TS mRNA levels remained unaffected by HYB0432 treatment, this finding suggested that the regulation of TS expression was controlled at the post-transcriptional level. To rule out the possibility of a post-translational mechanism, pulse-labeling immunoprecipitation studies were performed to determine the half-life of TS protein. As seen in Figure 5, the half-life of TS protein in untreated RKO cells was 12 ± 2 h. While the level of radiolabel incorporated into TS protein was significantly decreased (by 55%) after treatment with HYB0432 (190 nM), the half-life of TS protein remained essentially unchanged (14 ± 1 h). Thus, the decreased levels of TS protein after treatment with HYB0432 did not appear to be due to alterations in half-life of the protein.
Figure 5.
Effect of antisense treatment on the half-life of TS protein in RKO cells. Cells were incubated with Eufectin alone (squares) or Eufectin complexed with HYB0432 (190 nM) (circles) for 6 h, followed by a 2 h incubation with 35S-labeled methionine-containing medium. After removal of the radiolabel, cells were harvested at the indicated times and processed as described in Materials and Methods. The amount of radiolabeled TS protein (treated with Eufectin alone) at 0 h equals 100%. Points represent the mean ± SD from three separate experiments.
We next investigated the minimal length of the ORN required for efficient translational repression of TS mRNA in the RKO cell line. The rationale for this set of experiments was that a shorter ORN should be able to cross the cellular membrane and transport into cells more efficiently. In addition, shorter ORNs should, in theory, display less non-specific interactions with other unrelated cellular mRNAs. Our previous cell-free studies revealed that a 14 nt antisense ORN was as equally effective for inhibition of TS mRNA translation as the 30 nt HYB0432 ORN (13). To address this issue, a series of control experiments were initially performed to determine the optimal Eufectin:ORN ratio for transfecting the 18 nt antisense ORN, HYB0504. In contrast to the 30 nt HYB0432, a ratio of 9:1 Eufectin:HYB0504 was found to be the optimal condition for repressing TS protein expression (data not shown). Using this ratio of 9:1, we observed that the antisense HYB0504 significantly inhibited expression of TS protein in a dose-dependent fashion (Fig. 6). At a concentration of 210 nM, this antisense ORN inhibited TS protein expression by nearly 80% while the expression of control proteins, α-tubulin and β-actin, remained unchanged (Fig. 6, lane 5). Further reduction in oligo size to either 16 or 14 nt, however, resulted in complete loss of in vivo inhibitory activity (data not shown).
Figure 6.
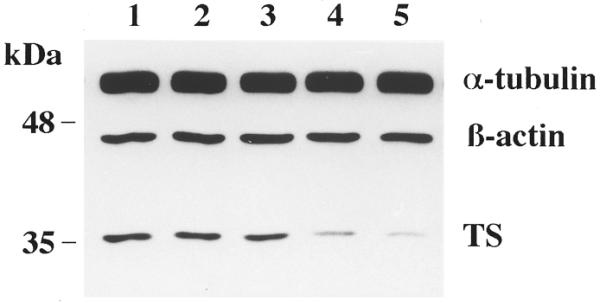
Effect of 18 nt HYB0504 on expression of TS protein in RKO cells. Cells were incubated in the absence (lane 1) or presence of Eufectin–ORN complexes (9:1 ratio, µg:µg) for 3 h, grown for an additional 24 h and then harvested and processed for western blot analysis as described in Materials and Methods. Lanes 2–5 were treated with complexes containing 53, 106, 159 and 212 nM HYB0504, respectively.
In addition to the parent RKO cell line, we investigated the activity of the 18 nt HYB0504 in the TDX-resistant RKO cell line, RKO-TDX. This cell line was established by exposing parent RKO cells to a gradual stepwise increase in TDX concentration in the cellular growth medium resulting in a cell line 5000-fold resistant to TDX (16). Western blot analysis revealed that RKO-TDX cells express 16-fold more TS protein than parent RKO cells (Fig. 7, lane 2 versus lane 1). Treatment of RKO-TDX cells with HYB0504 for 12 h resulted in a dose-dependent inhibition of TS protein expression (Fig. 7, lanes 3–6). At the highest concentration of HYB0504 tested (318 nM), TS protein levels were reduced by 85% when compared to control, untreated cells (Fig. 7, lane 6 versus lane 2). In contrast, treatment with a 5-base mismatch 2′-O-methyl RNA, TSAS18-MM (Fig. 7, lane 7), and a sense 18 nt RNA, TS18 (Fig. 7, lane 8), had absolutely no effect on levels of TS protein. Furthermore, expression of two housekeeping proteins, α-tubulin and β-actin, as well as topoisomerase I, was not affected by HYB0504 or any of the RNA control oligos (data not shown). These findings provide evidence that antisense ORNs can inhibit expression of TS even in cells that significantly overexpress TS protein. Further studies will be necessary to determine whether repeat or prolonged treatments with this antisense ORN can reduce the levels of TS protein in RKO-TDX cells to those expressed in the parent RKO cell line.
Figure 7.
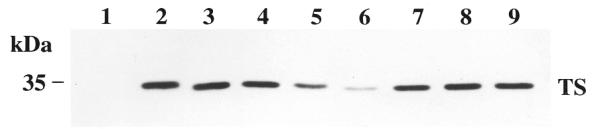
Effect of HYB0504 on expression of TS protein in TDX-resistant, RKO-TDX cells. Cells were treated for 12 h with increasing concentrations of HYB0504–Eufectin complexes: untreated (lane 2), 53 nM (lane 3), 105 nM (lane 4), 158 nM (lane 5), 315 nM (lane 6). Lanes 7 and 8 contain extracts from cells treated with 315 nM TSAS18-MM, a 5-base mismatch 18mer RNA and 210 nM TS18, a sense 18mer RNA, respectively. Lane 9 contains cell extracts treated with Eufectin alone (18 µg/ml). Lane 1 contains protein extract from the parent RKO cell line. After 24 h, cells were harvested and processed for western blot analysis as described in Materials and Methods.
DISCUSSION
In the present study, we investigated the effect of a 30 nt antisense ORN (HYB0432), targeted to the 5′ upstream cis-acting element on TS mRNA, on expression of TS protein in the human colon cancer RKO cell line. Based on our studies, expression of TS was repressed by this antisense RNA in both a dose- and time-dependent manner. This inhibitory effect on TS expression appeared to be specific as no alterations were observed in the expression of several important housekeeping proteins such as β-actin, α-tubulin and topoisomerase I. Treatment with a shorter 18 nt ORN (HYB0504) also resulted in significant repression of TS expression with no effect on control proteins.
With regard to the mechanism of action of the ORN, the levels of TS mRNA remained unaffected by ORN treatment suggesting a post-transcriptional mechanism. Further studies revealed that the reduction in TS protein expression was not associated with alterations in protein half-life, indicating that the effect of HYB0432 on TS expression was not at the post-translational level. It should be noted that the RKO cells were treated with antisense ORNs for 6 h prior to incubation with 35S-methionine. Thus, after antisense treatment, our labeling studies did not suggest an effect of the antisense RNA on the half-life of TS protein. However, during the 6 h antisense treatment, the western blot analysis (Fig. 3) revealed a decrease by 60% in the levels of TS protein. This reduction in TS expression is significantly greater than that expected based on the calculated half-life of the protein (14 h). During the 6 h treatment, a maximal decrease of 20–25% in the levels of TS protein would have been predicted. This finding suggests then that an effect of the antisense molecule on the stability of the TS protein cannot be ruled out entirely. The effect on protein stability appears to occur during antisense treatment since decreased expression of TS protein at later time points does not appear to be due to enhanced protein degradation based on the determined half-life of TS. Since TS is an RNA binding protein, it is conceivable that TS directly binds to the antisense ORN itself or to the TS mRNA–ORN complex resulting in activation of a protein degradation pathway. At this time, the mechanism by which this interaction occurs remains unclear and studies are currently on-going to determine the mechanism by which the antisense ORN might result in enhanced degradation of TS. Taken together, these experiments suggest that the antisense ORN controls the expression of TS at the post-transcriptional level, most likely through a combination of translational arrest of TS mRNA and post-translational processes that may involve enhanced degradation of TS protein.
These findings are consistent with our previous work in which we demonstrated, in an in vitro rabbit reticulocyte lysate translation system, that 2′-O-methyl ORNs with a natural PO backbone were able to effectively suppress TS mRNA translation (13). In both studies, we observed that ORNs inhibited TS mRNA expression more effectively than ODNs. However, treatment with ORNs containing PS backbones resulted in non-specific effects on unrelated mRNAs. There are several theoretic advantages of ORNs over their corresponding ODNs. First, ORNs bind to their target RNA with markedly greater affinity, resulting in a more thermally stable ORN–RNA complex than the corresponding ODN–RNA (15,24). Second, RNA–RNA duplexes do not serve as substrates for RNase H. For this reason, the antisense ORN approach does not utilize an RNase H-dependent mechanism. As presented in the Results, HYB0432 did not affect the levels of its target mRNA in contrast to the PS ODN which caused significant degradation of TS mRNA. Thus, the antisense ORN mediates its effect on TS mRNA most likely through translational arrest by inhibiting the process of ribosomal scanning. Finally, because ORNs do not induce cleavage of their target sequence, non-specific effects may be limited, as interaction of the ORNs with an unrelated cellular mRNA would result in only temporary inhibition of translation. In contrast, non-specific binding of ODNs may result in irreversible degradation of critical cellular mRNAs. In fact, it was recently suggested that 5 bp are sufficient to induce RNase H activity (25). Thus, the potential is quite high for an ODN to possess non-specific effects on expression of cellular genes involved in metabolism and proliferation.
Several groups have developed an antisense approach with ODNs to inhibit the expression of TS. Mader et al. used PS ODNs to target the 3′-UTR of TS mRNA, but they were unable to demonstrate a cytotoxic effect in eight different colon tumor cell lines (26). Ju et al. designed several PO ODNs targeting various sequences on the human TS mRNA (27). ODNs directed at the 5′-UTR had inhibitory activity in an in vitro rabbit reticulocyte lysate translation system. However, none demonstrated biological activity in the human colon cancer HT-29 cell line. While in situ TS catalytic activity was significantly repressed within 4 h of treatment with ODN AS-1, the total amount of TS protein, as determined by the [3H]FdUMP binding assay, remained unchanged and actually increased >2-fold by 36 h. Increased levels of TS may explain why the sensitivity of the cells to FdUrd was decreased 12-fold after AS-1 treatment. DeMoor et al. used PS ODNs containing 6 nt methoxyethoxylated ends to target either the translational start site or stop site (28). While neither the levels of expression of TS mRNA nor TS protein were affected by antisense therapy, treatment with ODNs targeting the translational start site resulted in increased transcription of TS whereas ODNs targeting the stop site had no effect on transcription. Ferguson et al. targeted the 3′-UTR of human TS mRNA with PS ODNs containing 6 nt methoxyethoxylated ends (29). Treatment of human cervical cancer HeLa cells with these ODNs resulted in decreased expression of both TS mRNA and TS protein levels, with resultant inhibition of cell proliferation. They also observed that antisense treatment sensitized cells to FUdR. Clearly, significant differences have been observed regarding the efficacy of these various antisense molecules. This finding may be explained by several factors including transfection reagent, cell type and intracellular accessibility to the target sequence. In addition, the specific chemistry of the ODNs is a critical factor that affects intracellular stability, affinity for its complementary mRNA sequence and non-specific interactions. One further limitation of these previous studies is that, in each case, the expression of other cellular proteins was not investigated to confirm that these ODNs were indeed specific for controlling TS expression. This is an especially important issue as ODNs with PS backbones are well-documented to have a variety of non-specific interactions with unrelated mRNAs and proteins (25).
Previous studies from our laboratory have documented that, besides its role in enzyme catalysis, TS also functions as an RNA binding protein (6,7). In addition to interacting with its own mRNA, there is now evidence that TS is complexed in intact human colon cancer cells with other cellular mRNAs, such as the p53 tumor suppressor gene (21). Recent studies from our laboratory, using both in vitro and in vivo model systems, have demonstrated that TS regulates p53 expression by binding directly to a sequence in the coding region of p53 mRNA and repressing translation (22,23). In this report, we provide further evidence that p53 expression may be linked to the cellular levels of TS protein. Within 6 h of treatment with antisense HYB0432, the level of TS protein was significantly reduced with a rapid rise in the levels of p53 protein. As the antisense effect diminished, TS protein levels returned to baseline levels with a corresponding reduction in p53 levels. Experiments are ongoing to determine whether elevation of p53 is directly related to reduction in TS or whether increased levels of p53 arise as a result of an adaptive response to depletion of essential deoxynucleotides.
The p53 tumor suppressor protein plays an essential role for preserving the integrity of the genome and for maintaining regulation of cell-cycle progression. Expression of p53 has been associated with arrest of the G1 and G2 checkpoints. This arrest of the cell cycle allows sufficient time for cells to repair DNA damage before DNA synthesis and replication can continue. Several investigators have shown that the levels of p53 are acutely increased in both normal and malignant cells in response to DNA-damaging agents (30–33). It appears that induced expression of p53 after DNA damage is regulated by both translational and post-translational regulatory processes (34–36). Recent studies from our laboratory have demonstrated that overexpression of human His-tag TS protein results in a 5-fold reduction in the level of p53 protein (23). Our findings suggest that the synthesis of p53 in cells overexpressing TS is regulated at the translational level and mediated by direct binding of TS protein to p53 mRNA. Following treatment with γ-irradiation or various anticancer agents, TS-overexpressing cells, which express considerably reduced p53 protein levels, are significantly impaired in their ability to arrest in G1 and to undergo apoptosis when compared to parent cells expressing normal levels of TS and wild-type p53 protein. In the absence of intact checkpoint controls and an intact apoptotic mechanism, these TS overexpressing cells are less sensitive to the cytotoxic effects of DNA-damaging agents and thus develop resistance to such agents. Since p53 expression appears to be controlled, in part, by the cellular levels of TS protein, antisense ORNs, as described in the present report, may provide a novel strategy to circumvent drug resistance in TS-overexpressing cells. The goal would be to use antisense ORNs to reduce intracellular levels of TS protein below an apparent ‘threshold’ value in which TS inhibitors have been found to be clinically effective (37,38). We have shown that HYB0504 can inhibit expression of TS by 85% in the TS-overexpressing cell line, RKO-TDX. Reduction in TS protein would allow restoration of p53 levels which, in turn, would aid in facilitating an apoptotic response to TS inhibitors. Furthermore, the induction of p53 may enhance the cytotoxic effects of other non-TS targeted DNA damaging drugs such as the platinum analogs and the topoisomerase I inhibitors.
One final issue to address is whether treatment with antisense ORNs has a specific inhibitory effect on cell proliferation. We focused our initial efforts on determining the specificity of effect as well as the mechanism of action of these ORNs against TS expression. Our preliminary cytotoxicity experiments demonstrate that the 18 nt antisense ORN HYB0504 can effectively inhibit cell proliferation with an IC50 of 200 nM. Surprisingly, when control oligos were tested including sense and mismatch ORNs, IC50 values in the submicromolar range were also achieved. As noted previously, none of the control ORNs directly affected TS expression. A complete GenBank search revealed that each of these ORNs show at least 66% homology with a variety of other cellular mRNAs. For example, the antisense RNA HYB0504 has sequence homology with tumor protein p63 mRNA (83%), Shaw-type potassium channel 3 mRNA (72%) and arachidonate 5-lipoxygenase mRNA (66%). Thus, it is conceivable that antisense and control RNAs may target other cellular mRNAs. For this reason, the cytotoxic effects of HYB0504 may not be due entirely to inhibition of TS expression. It is difficult to predict whether partial homology with a target sequence will cause repression of translation. However, treatment with HYB0503, a 24mer with 4 bases mismatched against TS mRNA (83% homology), had no effect on TS expression. Furthermore, most of these ORNs target sequences downstream from the translational start site of these putative alternative targets. It remains to be proven whether ORNs targeting other sites on a mRNA are effective repressors of translation. Thus, studies are ongoing to determine whether the cytotoxic effects of the ORNs are due to (i) repression of other cellular targets, (ii) non-specific interactions with cellular proteins, and/or (iii) non-specific effects due to free 2′-O-methyl ribonucleotides as a result of ORN degradation.
Several limitations remain for the antisense strategy to find direct application in the clinical setting. Because of their size and charge, antisense oligos display poor entry into cells. Molecules such as cholesterol have been conjugated to oligos to aid in their cellular uptake (39). Other investigators have encapsulated oligos in cationic liposomes to aid in their availability (40). To address tissue-specific uptake of the oligos, Wang et al. conjugated a folic acid molecule on the outside of a cationic liposome in order to enhance intracellular delivery of the encapsulated oligo via a process of folate receptor-mediated endocytosis (41). In addition, chemical modifications to the nucleotides have helped to address the issue of stability against nucleases, but it remains unknown whether such modified nucleotides, once they are eventually degraded, will affect various normal pathways of cellular metabolism.
Given these limitations, the antisense approach may be viewed as an important tool to identify potential new lead compounds for clinical therapy. Once a lead antisense ORN with biological activity has been identified, structural information from NMR or X-ray crystallography studies may allow modern drug design applications to identify small molecules that mimic the structural features of the antisense ORN. Such molecules could then be designed, synthesized and tested. The theoretical advantage of such an approach is that small ‘designer’ compounds, in contrast to nucleic acids, would have enhanced spatial, biophysical and biochemical properties, which might then be delivered with far greater efficiency and at higher concentrations into the target cells.
In the present report, we show that an RNA antisense strategy targeted against the 5′-upstream cis-acting element of TS mRNA can specifically inhibit TS expression in human colon cancer RKO cells. Our findings demonstrate that these ORNs exert their effect on TS expression through translational and post-translational processes. This approach may help to prevent and/or overcome the acute induction of TS and the subsequent development of cellular resistance observed with TS inhibitor compounds now being used in the clinical setting. Antisense ORNs may be useful as a tool to aid our understanding of the translational regulation of TS as well as to provide insights into the potential downstream effects of such a regulatory process. Finally, this antisense strategy may have a therapeutic application to be used alone or in combination with other established anticancer agents for the treatment of human cancer.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by grants from the Department of Veterans Affairs (VA Merit Review to E.C.) and the National Cancer Institute (CA16359 and CA75712 to E.C.).
References
- 1.Carreras C.W. and Santi,D.V. (1995) The catalytic mechanism and structure of thymidylate synthase. Annu. Rev. Biochem., 64, 721–762. [DOI] [PubMed] [Google Scholar]
- 2.Cohen S.S., Flaks,J.G., Barner,H.D. and Lichtenstein,J. (1958) The mode of action of fluorouracil and its derivatives. Proc. Natl Acad. Sci. USA, 44, 1004–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Danenberg P.V. (1977) Thymidylate synthetase – a target enzyme in cancer chemotherapy. Biochim. Biophys. Acta, 473, 73–92. [DOI] [PubMed] [Google Scholar]
- 4.Hardy L.W., Finer-Moore,J.S., Montfort,W.R., Jones,M.O., Santi,D.V. and Stroud,R.M. (1987) Atomic structure of thymidylate synthase: target for rational drug design. Science, 235, 448–455. [DOI] [PubMed] [Google Scholar]
- 5.Shoichet B.K., Stroud,R.M., Santi,D.V., Kuntz,I.D. and Perry,K.M. (1993) Structure-based discovery of inhibitors of thymidylate synthase. Science, 259, 1445–1450. [DOI] [PubMed] [Google Scholar]
- 6.Chu E. and Allegra,C.J. (1996) The role of thymidylate synthase as an RNA binding protein. Bioessays, 18, 191–198. [DOI] [PubMed] [Google Scholar]
- 7.Chu E., Koeller,D.M., Casey,J.L., Drake,J.C., Chabner,B.A., Elwood,P.C., Zinn,S. and Allegra,C.J. (1991) Autoregulation of human thymidylate synthase messenger RNA translation by thymidylate synthase. Proc. Natl Acad. Sci. USA, 88, 8977–8981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu E., Voeller,D., Koeller,D.M., Drake,J.C., Takimoto,C.H., Maley,G.F., Maley,F. and Allegra,C.J. (1993) Identification of an RNA binding site for human thymidylate synthase. Proc. Natl Acad. Sci. USA, 90, 517–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin X., Parsels,L.A., Voeller,D.M., Allegra,C.J., Maley,G.F., Maley,F. and Chu,E. (2000) Characterization of a cis-acting regulatory element in the protein coding region of thymidylate synthase mRNA. Nucleic Acids Res., 28, 1381–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu E. and Allegra,C.J. (1993) Regulation of thymidylate synthase in human colon cancer cells treated with 5-fluorouracil and interferon-gamma. Adv. Exp. Med. Biol., 339, 143–151. [DOI] [PubMed] [Google Scholar]
- 11.Keyomarsi K., Samet,J., Molnar,G. and Pardee,A.B. (1993) The thymidylate synthase inhibitor, ICI D1694, overcomes translational detainment of the enzyme. J. Biol. Chem., 268, 15142–15149. [PubMed] [Google Scholar]
- 12.Van der Wilt C.L., Pinedo,H.M., Smid,K. and Peters,G.J. (1992) Elevation of thymidylate synthase following 5-fluorouracil treatment is prevented by the addition of leucovorin in murine colon tumors. Cancer Res., 52, 4922–4928. [PubMed] [Google Scholar]
- 13.Schmitz J.C., Agrawal,S. and Chu,E. (1998) Repression of human thymidylate synthase mRNA translation by antisense 2′-O-methyl oligoribonucleotides. Antisense Nucleic Acid Drug Dev., 8, 371–378. [DOI] [PubMed] [Google Scholar]
- 14.Morvan F., Porumb,H., Degols,G., Lefebvre,I., Pompon,A., Sproat,B.S., Rayner,B., Malvy,C., Lebleu,B. and Imbach,J.L. (1993) Comparative evaluation of seven oligonucleotide analogues as potential antisense agents. J. Med. Chem., 36, 280–287. [DOI] [PubMed] [Google Scholar]
- 15.Stein D., Foster,E., Huang,S.B., Weller,D. and Summerton,J. (1997) A specificity comparison of four antisense types: morpholino, 2′-O-methyl RNA, DNA and phosphorothioate DNA. Antisense Nucleic Acid Drug Dev., 7, 151–157. [DOI] [PubMed] [Google Scholar]
- 16.Johnston P.G., Behan,K.A., Allegra,C.J. and Drake,J.C. (1995) Fluorouracil: active in ZD1694 (tomudex)-resistant cell lines with markedly elevated thymidylate synthase levels. J. Natl Cancer Inst., 87, 1558–1559. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 18.Chu E., Zinn,S., Boarman,D. and Allegra,C.J. (1990) Interaction of gamma interferon and 5-fluorouracil in the H630 human colon carcinoma cell line. Cancer Res., 50, 5834–5840. [PubMed] [Google Scholar]
- 19.Conrad A.H., Behlke,M.A., Jaffredo,T. and Conrad,G.W. (1998) Optimal lipofection reagent varies with the molecular modifications of the DNA. Antisense Nucleic Acid Drug Dev., 8, 427–434. [DOI] [PubMed] [Google Scholar]
- 20.Desai S.D., Liu,L.F., Vazquez-Abad,D. and D’Arpa,P. (1997) Ubiquitin-dependent destruction of topoisomerase I is stimulated by the antitumor drug camptothecin. J. Biol. Chem., 272, 24159–24164. [DOI] [PubMed] [Google Scholar]
- 21.Chu E., Cogliati,T., Copur,S.M., Borre,A., Voeller,D.M., Allegra,C.J. and Segal,S. (1996) Identification of in vivo target RNA sequences bound by thymidylate synthase. Nucleic Acids Res., 24, 3222–3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chu E., Copur,S.M., Ju,J., Chen,T.M., Khleif,S., Voeller,D.M., Mizunuma,N., Patel,M., Maley,G.F., Maley,F. and Allegra,C.J. (1999) Thymidylate synthase protein and p53 mRNA form an in vivo ribonucleoprotein complex. Mol. Cell. Biol., 19, 1582–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ju J., Pedersen-Lane,J., Maley,F. and Chu,E. (1999) Regulation of p53 expression by thymidylate synthase. Proc. Natl Acad. Sci. USA, 96, 3769–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inoue H., Hayase,Y., Imura,A., Iwai,S., Miura,K. and Ohtsuka,E. (1987) Synthesis and hybridization studies on two complementary nona(2′-O-methyl) ribonucleotides. Nucleic Acids Res., 15, 6131–6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stein C.A. (2000) Is irrelevant cleavage the price of antisense efficacy? Pharmacol. Ther., 85, 231–236. [DOI] [PubMed] [Google Scholar]
- 26.Mader R.M., Sieder,A.E., Braun,J., Rizovski,B., Kalipciyan,M., Mueller,M.W., Jakesz,R., Rainer,H. and Steger,G.G. (1997) Transcription and activity of 5-fluorouracil converting enzymes in fluoropyrimidine resistance in colon cancer in vitro. Biochem. Pharmacol., 54, 1233–1242. [DOI] [PubMed] [Google Scholar]
- 27.Ju J., Kane,S.E., Lenz,H.J., Danenberg,K.D., Chu,E. and Danenberg,P.V. (1998) Desensitization and sensitization of cells to fluoropyrimidines with different antisenses directed against thymidylate synthase messenger RNA. Clin. Cancer Res., 4, 2229–2236. [PubMed] [Google Scholar]
- 28.DeMoor J.M., Vincent,M.D., Collins,O.M. and Koropatnick,J. (1998) Antisense nucleic acids targeted to the thymidylate synthase (TS) mRNA translation start site stimulate TS gene transcription. Exp. Cell Res., 243, 11–21. [DOI] [PubMed] [Google Scholar]
- 29.Ferguson P.J., Collins,O., Dean,N.M., DeMoor,J., Li,C.S., Vincent,M.D. and Koropatnick,J. (1999) Antisense down-regulation of thymidylate synthase to suppress growth and enhance cytotoxicity of 5-FUdR, 5-FU and Tomudex in HeLa cells. Br. J. Pharmacol., 127, 1777–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kastan M.B., Onyekwere,O., Sidransky,D., Vogelstein,B. and Craig,R.W. (1991) Participation of p53 protein in the cellular response to DNA damage. Cancer Res., 51, 6304–6311. [PubMed] [Google Scholar]
- 31.Linke S.P., Clarkin,K.C., Di Leonardo,A., Tsou,A. and Wahl,G.M. (1996) A reversible, p53-dependent G0/G1 cell cycle arrest induced by ribonucleotide depletion in the absence of detectable DNA damage. Genes Dev., 10, 934–947. [DOI] [PubMed] [Google Scholar]
- 32.Maltzman W. and Czyzyk,L. (1984) UV irradiation stimulates levels of p53 cellular tumor antigen in nontransformed mouse cells. Mol. Cell. Biol., 4, 1689–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhan Q., Carrier,F. and Fornace,A.J.,Jr (1993) Induction of cellular p53 activity by DNA-damaging agents and growth arrest. Mol. Cell. Biol., 13, 4242–4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mosner J., Mummenbrauer,T., Bauer,C., Sczakiel,G., Grosse,F. and Deppert,W. (1995) Negative feedback regulation of wild-type p53 biosynthesis. EMBO J., 14, 4442–4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fu L., Ma,W. and Benchimol,S. (1999) A translation repressor element resides in the 3′ untranslated region of human p53 mRNA. Oncogene, 18, 6419–6424. [DOI] [PubMed] [Google Scholar]
- 36.Fu L., Minden,M.D. and Benchimol,S. (1996) Translational regulation of human p53 gene expression. EMBO J., 15, 4392–4401. [PMC free article] [PubMed] [Google Scholar]
- 37.Johnston P.G., Lenz,H.J., Leichman,C.G., Danenberg,K.D., Allegra,C.J., Danenberg,P.V. and Leichman,L. (1995) Thymidylate synthase gene and protein expression correlate and are associated with response to 5-fluorouracil in human colorectal and gastric tumors. Cancer Res., 55, 1407–1412. [PubMed] [Google Scholar]
- 38.Horikoshi T., Danenberg,K.D., Stadlbauer,T.H., Volkenandt,M., Shea,L.C., Aigner,K., Gustavsson,B., Leichman,L., Frosing,R., Ray,M. et al. (1992) Quantitation of thymidylate synthase, dihydrofolate reductase and DT-diaphorase gene expression in human tumors using the polymerase chain reaction. Cancer Res., 52, 108–116. [PubMed] [Google Scholar]
- 39.LeDoan T., Etore,F., Tenu,J.P., Letourneux,Y. and Agrawal,S. (1999) Cell binding, uptake and cytosolic partition of HIV anti-gag phosphodiester oligonucleotides 3′-linked to cholesterol derivatives in macrophages. Bioorg. Med. Chem., 7, 2263–2269. [DOI] [PubMed] [Google Scholar]
- 40.Stuart D.D., Kao,G.Y. and Allen,T.M. (2000) A novel, long-circulating and functional liposomal formulation of antisense oligodeoxynucleotides targeted against MDR1. Cancer Gene Ther., 7, 466–475. [DOI] [PubMed] [Google Scholar]
- 41.Wang S., Lee,R.J., Cauchon,G., Gorenstein,D.G. and Low,P.S. (1995) Delivery of antisense oligodeoxyribonucleotides against the human epidermal growth factor receptor into cultured KB cells with liposomes conjugated to folate via polyethylene glycol. Proc. Natl Acad. Sci. USA, 92, 3318–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]