Abstract
The three-ring eudesmanolide, C15H16O3, is a natural product isolated from Dicoma anomala Sond. (Asteraceae). The compound contains an endo–exo cross conjugated methylenecyclohexenone ring with an envelope conformation trans-fused with cyclohexane and trans-annelated with an α-methylene γ-lactone. The absolute structure was assigned by optical rotation measurements compared to those from the synthetic compound with known stereochemistry. The crystal packing is consolidated by C—H⋯O interactions.
Related literature
For NMR studies of this compound, see: Bohlmann & Zdero, (1982 ▶); Grass et al. (2004 ▶). For the chemical synthesis and confirmation of the absolute structure, see: Higuchi et al. (2003 ▶).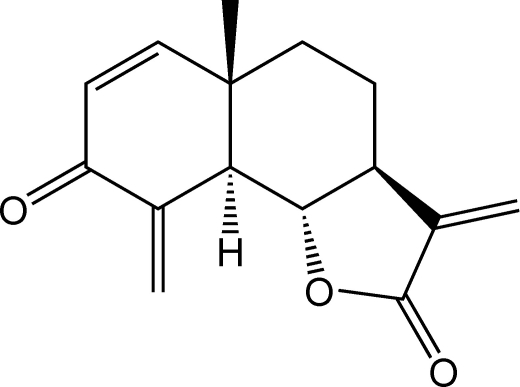
Experimental
Crystal data
C15H16O3
M r = 244.28
Orthorhombic,
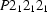
a = 9.5648 (6) Å
b = 11.1631 (6) Å
c = 11.5542 (6) Å
V = 1233.67 (12) Å3
Z = 4
Mo Kα radiation
μ = 0.09 mm−1
T = 150 (2) K
0.50 × 0.50 × 0.40 mm
Data collection
Oxford Diffraction Excalibur2 CCD diffractometer
Absorption correction: multi-scan (Blessing, 1995 ▶) T min = 0.909, T max = 0.963
12604 measured reflections
2294 independent reflections
1988 reflections with I > 2σ(I)
R int = 0.016
Refinement
R[F 2 > 2σ(F 2)] = 0.033
wR(F 2) = 0.095
S = 1.05
2294 reflections
163 parameters
H-atom parameters constrained
Δρmax = 0.31 e Å−3
Δρmin = −0.21 e Å−3
Data collection: CrysAlis CCD (Oxford Diffraction, 2006 ▶); cell refinement: CrysAlis RED (Oxford Diffraction, 2006 ▶); data reduction: CrysAlis RED; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: Mercury (Macrae et al., 2006 ▶); software used to prepare material for publication: PLATON (Spek, 2003 ▶) and WinGX (Farrugia, 1999 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808042402/bi2330sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808042402/bi2330Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C6—H6⋯O1i | 0.98 | 2.39 | 3.360 (2) | 171 |
| C14—H14A⋯O1i | 0.96 | 2.57 | 3.393 (2) | 143 |
Symmetry code: (i) 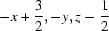 .
.
Acknowledgments
We thank the National Drug Development Platform (NDDP) and the NRF for funding.
supplementary crystallographic information
Comment
The title compound, a sesquiterpene lactone dehydrobrachylaenolide, was isolated from Dicoma anomala Sond (Asteraceae). These bi-functional exo-endo cross conjugated dienones are of importance as synthetic intermediates in the preparation of biologically active natural products (Higuchi et al., 2003). NMR studies of the compound have been reported previously (Bohlmann & Zdero, 1982; Grass et al., 2004) and the absolute stereochemistry has been confirmed as 3-oxoeudesma-1,4(15),11 (13)-triene-12,6a-olide by chemical synthesis (Higuchi et al., 2003). Here we report the crystal structure. Although the absolute structure could not be elucidated by X-ray diffraction, unambiguous assignment of stereochemistry was made on the basis of the value of optical rotation ([α] 24D+68° (c 1/2, CHCl3)) which is identical to that of the synthetic compound ([α] 24D+67.9° (c 0.16, CHCl3)) for which the stereochemistry is known (Higuchi et al., 2003) and very close to the value for the naturally isolated material ([α]24D+67° (c 0.16, CHCl3)) (Bohlmann & Zdero, 1982).
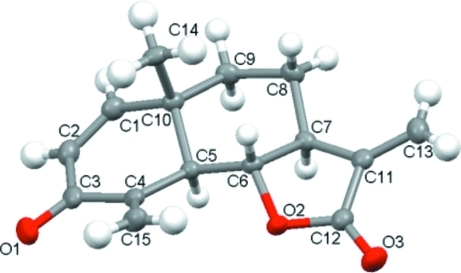
The molecular geometry and labelling scheme are shown in Fig. 1. The methylenecyclohexenone ring adopts an envelope conformation, with the C5 atom out of the plane of the ring by approximately 0.7 Å. The γ-lactone ring is twisted on C6—C7, while the cyclohexane ring adopts a chair conformation. An axial position is occupied by methyl group C14, and the methylene carbon atom C15 is in the equatorial position. A weak intramolecular interaction is formed between C15—H15B···O2. Fig. 2 illustrates the molecular packing viewed down the c axis. Weak intermolecular hydrogen bonds are present between atoms C6—H6···O1i and and C14—H14···O1i [symmetry code (i): 1/2 - x, 1 - y, 1/2 + z].
Experimental
The compound was isolated from Dicoma anomala Sond (Asteraceae), and recrystallized from propanol at room temperature.
Refinement
H atoms were placed geometrically and refined in idealized positions in the riding-model approximation, with C—H = 0.93–0.98 Å with Uiso(H) = 1.2 or 1.5Ueq(C). In the absence of significant anomalous scattering effects, Friedel pairs were merged as equivalent data.
Figures
Fig. 1.
Molecular structure showing displacement ellipsoids at 50% probability for all atoms.
Crystal data
| C15H16O3 | F(000) = 520 |
| Mr = 244.28 | Dx = 1.315 Mg m−3 |
| Orthorhombic, P212121 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P 2ac 2ab | Cell parameters from 8167 reflections |
| a = 9.5648 (6) Å | θ = 4.0–31.8° |
| b = 11.1631 (6) Å | µ = 0.09 mm−1 |
| c = 11.5542 (6) Å | T = 150 K |
| V = 1233.67 (12) Å3 | Block, colourless |
| Z = 4 | 0.50 × 0.50 × 0.40 mm |
Data collection
| Oxford Diffraction Excalibur2 CCD diffractometer | 2294 independent reflections |
| Radiation source: fine-focus sealed tube | 1988 reflections with I > 2σ(I) |
| graphite | Rint = 0.016 |
| ω scans | θmax = 31.9°, θmin = 4.0° |
| Absorption correction: multi-scan (Blessing, 1995) | h = −13→13 |
| Tmin = 0.909, Tmax = 0.963 | k = −15→16 |
| 12604 measured reflections | l = −16→17 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.033 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.095 | H-atom parameters constrained |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.0708P)2] where P = (Fo2 + 2Fc2)/3 |
| 2294 reflections | (Δ/σ)max = 0.001 |
| 163 parameters | Δρmax = 0.31 e Å−3 |
| 0 restraints | Δρmin = −0.21 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C4 | 0.86180 (14) | 0.08184 (11) | 1.24094 (11) | 0.0237 (2) | |
| O2 | 0.87541 (10) | 0.20733 (7) | 1.00141 (7) | 0.0245 (2) | |
| C7 | 1.04877 (12) | 0.07144 (11) | 0.94161 (10) | 0.0210 (2) | |
| H7 | 1.1253 | 0.1211 | 0.9714 | 0.025* | |
| C8 | 1.10043 (14) | −0.05782 (12) | 0.94001 (11) | 0.0246 (3) | |
| H8A | 1.1798 | −0.0657 | 0.8883 | 0.029* | |
| H8B | 1.0268 | −0.1109 | 0.9136 | 0.029* | |
| C9 | 1.14347 (13) | −0.08979 (11) | 1.06503 (11) | 0.0245 (2) | |
| H9A | 1.1717 | −0.1732 | 1.0675 | 0.029* | |
| H9B | 1.2238 | −0.0416 | 1.0866 | 0.029* | |
| O3 | 0.85244 (12) | 0.31953 (9) | 0.84156 (9) | 0.0372 (3) | |
| C14 | 0.90759 (14) | −0.16252 (11) | 1.13671 (12) | 0.0270 (3) | |
| H14A | 0.8694 | −0.1540 | 1.0603 | 0.041* | |
| H14B | 0.8354 | −0.1493 | 1.1930 | 0.041* | |
| H14C | 0.9447 | −0.2419 | 1.1458 | 0.041* | |
| C6 | 0.92565 (13) | 0.08521 (10) | 1.02492 (10) | 0.0197 (2) | |
| H6 | 0.8525 | 0.0278 | 1.0037 | 0.024* | |
| C3 | 0.90807 (15) | 0.04878 (12) | 1.36063 (11) | 0.0274 (3) | |
| O1 | 0.84841 (13) | 0.08529 (10) | 1.44767 (9) | 0.0385 (3) | |
| C11 | 0.98993 (14) | 0.13415 (11) | 0.83767 (11) | 0.0234 (2) | |
| C1 | 1.08111 (15) | −0.08814 (12) | 1.27617 (12) | 0.0279 (3) | |
| H1 | 1.1553 | −0.1409 | 1.2867 | 0.033* | |
| C5 | 0.97025 (12) | 0.06224 (10) | 1.14800 (10) | 0.0197 (2) | |
| H5 | 1.0491 | 0.1156 | 1.1647 | 0.024* | |
| C10 | 1.02589 (13) | −0.06954 (10) | 1.15444 (10) | 0.0213 (2) | |
| C15 | 0.73174 (15) | 0.12080 (12) | 1.22442 (14) | 0.0321 (3) | |
| H15A | 0.6708 | 0.1272 | 1.2868 | 0.039* | |
| H15B | 0.7019 | 0.1416 | 1.1505 | 0.039* | |
| C13 | 1.00061 (16) | 0.11111 (13) | 0.72528 (11) | 0.0303 (3) | |
| H13A | 0.9501 | 0.1564 | 0.6722 | 0.036* | |
| H13B | 1.0586 | 0.0497 | 0.6996 | 0.036* | |
| C12 | 0.89936 (14) | 0.23105 (11) | 0.88715 (11) | 0.0263 (3) | |
| C2 | 1.02875 (16) | −0.03277 (13) | 1.36888 (11) | 0.0304 (3) | |
| H2 | 1.0696 | −0.0460 | 1.4408 | 0.036* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C4 | 0.0291 (6) | 0.0164 (5) | 0.0257 (5) | −0.0021 (5) | 0.0069 (5) | 0.0005 (4) |
| O2 | 0.0309 (5) | 0.0174 (4) | 0.0253 (4) | 0.0031 (3) | 0.0029 (4) | 0.0023 (3) |
| C7 | 0.0202 (5) | 0.0206 (5) | 0.0221 (5) | −0.0010 (4) | 0.0013 (4) | −0.0030 (4) |
| C8 | 0.0239 (6) | 0.0254 (6) | 0.0244 (5) | 0.0037 (5) | 0.0003 (5) | −0.0048 (5) |
| C9 | 0.0200 (5) | 0.0245 (6) | 0.0289 (6) | 0.0036 (5) | −0.0014 (5) | −0.0027 (4) |
| O3 | 0.0499 (7) | 0.0262 (5) | 0.0356 (5) | 0.0061 (5) | 0.0000 (5) | 0.0081 (4) |
| C14 | 0.0277 (6) | 0.0168 (5) | 0.0366 (6) | −0.0020 (5) | −0.0027 (5) | 0.0014 (5) |
| C6 | 0.0195 (5) | 0.0148 (5) | 0.0248 (5) | 0.0006 (4) | 0.0012 (4) | −0.0001 (4) |
| C3 | 0.0330 (6) | 0.0233 (6) | 0.0259 (5) | −0.0079 (5) | 0.0076 (5) | 0.0017 (5) |
| O1 | 0.0493 (6) | 0.0366 (6) | 0.0297 (5) | −0.0062 (5) | 0.0165 (5) | −0.0008 (4) |
| C11 | 0.0233 (6) | 0.0207 (5) | 0.0261 (5) | −0.0040 (4) | 0.0015 (5) | 0.0001 (4) |
| C1 | 0.0288 (6) | 0.0252 (6) | 0.0297 (6) | 0.0015 (5) | −0.0043 (5) | 0.0036 (5) |
| C5 | 0.0202 (5) | 0.0171 (5) | 0.0217 (5) | −0.0007 (4) | 0.0021 (4) | −0.0002 (4) |
| C10 | 0.0212 (5) | 0.0186 (5) | 0.0241 (5) | 0.0011 (4) | −0.0022 (4) | 0.0000 (4) |
| C15 | 0.0310 (7) | 0.0263 (6) | 0.0391 (7) | 0.0033 (5) | 0.0129 (6) | 0.0040 (6) |
| C13 | 0.0314 (6) | 0.0337 (7) | 0.0257 (6) | −0.0050 (6) | 0.0028 (6) | 0.0006 (5) |
| C12 | 0.0303 (6) | 0.0219 (6) | 0.0266 (6) | −0.0028 (5) | −0.0002 (5) | 0.0019 (4) |
| C2 | 0.0358 (7) | 0.0301 (6) | 0.0253 (6) | −0.0048 (5) | −0.0011 (5) | 0.0048 (5) |
Geometric parameters (Å, °)
| C4—C15 | 1.332 (2) | C14—H14B | 0.960 |
| C4—C3 | 1.4982 (18) | C14—H14C | 0.960 |
| C4—C5 | 1.5090 (16) | C6—C5 | 1.5067 (16) |
| O2—C12 | 1.3659 (15) | C6—H6 | 0.980 |
| O2—C6 | 1.4708 (14) | C3—O1 | 1.2260 (16) |
| C7—C11 | 1.4997 (17) | C3—C2 | 1.473 (2) |
| C7—C8 | 1.5253 (18) | C11—C13 | 1.3278 (18) |
| C7—C6 | 1.5287 (16) | C11—C12 | 1.4991 (18) |
| C7—H7 | 0.980 | C1—C2 | 1.334 (2) |
| C8—C9 | 1.5439 (18) | C1—C10 | 1.5167 (17) |
| C8—H8A | 0.970 | C1—H1 | 0.930 |
| C8—H8B | 0.970 | C5—C10 | 1.5662 (15) |
| C9—C10 | 1.5437 (17) | C5—H5 | 0.980 |
| C9—H9A | 0.970 | C15—H15A | 0.930 |
| C9—H9B | 0.970 | C15—H15B | 0.930 |
| O3—C12 | 1.2059 (16) | C13—H13A | 0.930 |
| C14—C10 | 1.5490 (17) | C13—H13B | 0.930 |
| C14—H14A | 0.960 | C2—H2 | 0.930 |
| C15—C4—C3 | 119.26 (12) | O1—C3—C2 | 121.14 (13) |
| C15—C4—C5 | 125.99 (12) | O1—C3—C4 | 122.53 (13) |
| C3—C4—C5 | 114.71 (11) | C2—C3—C4 | 116.32 (11) |
| C12—O2—C6 | 107.66 (9) | C13—C11—C12 | 123.86 (13) |
| C11—C7—C8 | 123.60 (10) | C13—C11—C7 | 131.60 (13) |
| C11—C7—C6 | 99.69 (10) | C12—C11—C7 | 104.38 (10) |
| C8—C7—C6 | 110.63 (10) | C2—C1—C10 | 123.39 (12) |
| C11—C7—H7 | 107.3 | C2—C1—H1 | 118.3 |
| C8—C7—H7 | 107.3 | C10—C1—H1 | 118.3 |
| C6—C7—H7 | 107.3 | C6—C5—C4 | 116.89 (10) |
| C7—C8—C9 | 107.08 (10) | C6—C5—C10 | 107.51 (9) |
| C7—C8—H8A | 110.3 | C4—C5—C10 | 109.63 (9) |
| C9—C8—H8A | 110.3 | C6—C5—H5 | 107.5 |
| C7—C8—H8B | 110.3 | C4—C5—H5 | 107.5 |
| C9—C8—H8B | 110.3 | C10—C5—H5 | 107.5 |
| H8A—C8—H8B | 108.6 | C1—C10—C9 | 110.29 (10) |
| C10—C9—C8 | 113.46 (10) | C1—C10—C14 | 106.58 (10) |
| C10—C9—H9A | 108.9 | C9—C10—C14 | 110.22 (10) |
| C8—C9—H9A | 108.9 | C1—C10—C5 | 106.91 (10) |
| C10—C9—H9B | 108.9 | C9—C10—C5 | 110.69 (9) |
| C8—C9—H9B | 108.9 | C14—C10—C5 | 112.02 (9) |
| H9A—C9—H9B | 107.7 | C4—C15—H15A | 120.0 |
| C10—C14—H14A | 109.5 | C4—C15—H15B | 120.0 |
| C10—C14—H14B | 109.5 | H15A—C15—H15B | 120.0 |
| H14A—C14—H14B | 109.5 | C11—C13—H13A | 120.0 |
| C10—C14—H14C | 109.5 | C11—C13—H13B | 120.0 |
| H14A—C14—H14C | 109.5 | H13A—C13—H13B | 120.0 |
| H14B—C14—H14C | 109.5 | O3—C12—O2 | 121.23 (12) |
| O2—C6—C5 | 115.13 (9) | O3—C12—C11 | 129.75 (13) |
| O2—C6—C7 | 103.21 (9) | O2—C12—C11 | 109.00 (10) |
| C5—C6—C7 | 111.05 (10) | C1—C2—C3 | 121.90 (12) |
| O2—C6—H6 | 109.1 | C1—C2—H2 | 119.1 |
| C5—C6—H6 | 109.1 | C3—C2—H2 | 119.1 |
| C7—C6—H6 | 109.1 | ||
| C11—C7—C8—C9 | −176.53 (11) | C15—C4—C5—C10 | −124.67 (13) |
| C6—C7—C8—C9 | −58.75 (13) | C3—C4—C5—C10 | 52.90 (13) |
| C7—C8—C9—C10 | 55.27 (14) | C2—C1—C10—C9 | 151.41 (13) |
| C12—O2—C6—C5 | 153.05 (11) | C2—C1—C10—C14 | −88.95 (15) |
| C12—O2—C6—C7 | 31.88 (12) | C2—C1—C10—C5 | 31.01 (17) |
| C11—C7—C6—O2 | −39.39 (11) | C8—C9—C10—C1 | −173.06 (11) |
| C8—C7—C6—O2 | −171.01 (9) | C8—C9—C10—C14 | 69.52 (13) |
| C11—C7—C6—C5 | −163.29 (9) | C8—C9—C10—C5 | −54.96 (13) |
| C8—C7—C6—C5 | 65.10 (12) | C6—C5—C10—C1 | 175.55 (10) |
| C15—C4—C3—O1 | −20.6 (2) | C4—C5—C10—C1 | −56.42 (12) |
| C5—C4—C3—O1 | 161.68 (12) | C6—C5—C10—C9 | 55.39 (12) |
| C15—C4—C3—C2 | 158.27 (13) | C4—C5—C10—C9 | −176.57 (10) |
| C5—C4—C3—C2 | −19.48 (16) | C6—C5—C10—C14 | −68.06 (12) |
| C8—C7—C11—C13 | −19.4 (2) | C4—C5—C10—C14 | 59.98 (12) |
| C6—C7—C11—C13 | −142.26 (15) | C6—O2—C12—O3 | 170.70 (12) |
| C8—C7—C11—C12 | 155.99 (11) | C6—O2—C12—C11 | −10.45 (13) |
| C6—C7—C11—C12 | 33.14 (12) | C13—C11—C12—O3 | −21.0 (2) |
| O2—C6—C5—C4 | 58.69 (14) | C7—C11—C12—O3 | 163.19 (14) |
| C7—C6—C5—C4 | 175.49 (10) | C13—C11—C12—O2 | 160.33 (12) |
| O2—C6—C5—C10 | −177.59 (9) | C7—C11—C12—O2 | −15.53 (13) |
| C7—C6—C5—C10 | −60.79 (12) | C10—C1—C2—C3 | 2.2 (2) |
| C15—C4—C5—C6 | −2.05 (18) | O1—C3—C2—C1 | 169.28 (13) |
| C3—C4—C5—C6 | 175.53 (10) | C4—C3—C2—C1 | −9.6 (2) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C6—H6···O1i | 0.98 | 2.39 | 3.360 (2) | 171 |
| C14—H14A···O1i | 0.96 | 2.57 | 3.393 (2) | 143 |
Symmetry codes: (i) −x+3/2, −y, z−1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: BI2330).
References
- Blessing, R. H. (1995). Acta Cryst. A51, 33–38. [DOI] [PubMed]
- Bohlmann, F. & Zdero, C. (1982). Phytochemistry, 21, 647–651.
- Farrugia, L. J. (1999). J. Appl. Cryst.32, 837–838.
- Grass, S., Zidorn, C., Ellmerer, E. P. & Stuppner, H. (2004). Chem. Biodivers.1, 353–360. [DOI] [PubMed]
- Higuchi, Y., Shimota, F., Koyanagi, R., Suda, K., Mitsui, T., Kataoka, T., Nagai, K. & Ando, M. (2003). J. Nat. Prod.66, 588–594. [DOI] [PubMed]
- Macrae, C. F., Edgington, P. R., McCabe, P., Pidcock, E., Shields, G. P., Taylor, R., Towler, M. & van de Streek, J. (2006). J. Appl. Cryst.39, 453–457.
- Oxford Diffraction (2006). CrysAlis CCD and CrysAlis RED Oxford Diffraction Ltd, Abingdon, England.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2003). J. Appl. Cryst.36, 7–13.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808042402/bi2330sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808042402/bi2330Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report