Abstract
A novel N-heterocyclic carbene derivative, C28H33N2O3 +·Br−·H2O, was synthesized and characterized by elemental analysis, 1H and 13C-NMR and IR spectroscopy and a single-crystal X-ray diffraction study. Ions of the title compound are linked by π⋯π stacking interactions (face–face separation 3.441 Å) and C—H⋯Br and O—H⋯Br interactions. Intra- and intermolecular C—H⋯O interactions are also present. The C—N bond lengths for the compound [1.329 (3), 1.325 (3), 1.389 (3) and 1.391 (3) Å] are all shorter than the average single C—N bond length of 1.48 Å, thus showing varying degrees of double-bond character.
Related literature
For the synthesis, see: Yaşar et al. (2008 ▶). For general background, see: Herrmann (2002 ▶); Arduengo & Krafczyc (1998 ▶); Herrmann et al. (1995 ▶, 1998 ▶); Navarro et al. (2006 ▶). For related compounds, see: Yaşar et al. (2008 ▶); Arslan et al. (2009 ▶ and references therein). For bond-length data, see: Allen et al. (1987 ▶).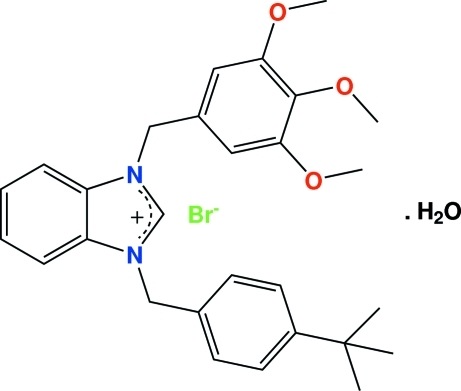
Experimental
Crystal data
C28H33N2O3 +·Br−·H2O
M r = 543.49
Triclinic,
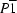
a = 10.389 (2) Å
b = 10.436 (2) Å
c = 14.038 (3) Å
α = 109.79 (3)°
β = 90.70 (3)°
γ = 103.57 (3)°
V = 1385.1 (6) Å3
Z = 2
Mo Kα radiation
μ = 1.52 mm−1
T = 298 (2) K
0.48 × 0.29 × 0.26 mm
Data collection
Mercury CCD diffractometer
Absorption correction: multi-scan (REQAB; Jacobson, 1998 ▶) T min = 0.514, T max = 0.673
11938 measured reflections
4860 independent reflections
3921 reflections with I > 2σ(I)
R int = 0.022
Refinement
R[F 2 > 2σ(F 2)] = 0.042
wR(F 2) = 0.112
S = 1.08
4860 reflections
328 parameters
2 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.31 e Å−3
Δρmin = −0.55 e Å−3
Data collection: CrystalClear (Rigaku/MSC, 2001 ▶); cell refinement: CrystalClear; data reduction: CrystalClear; program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: SHELXTL and Mercury (Macrae et al., 2006 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks I. DOI: 10.1107/S1600536808043250/hg2459sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808043250/hg2459Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
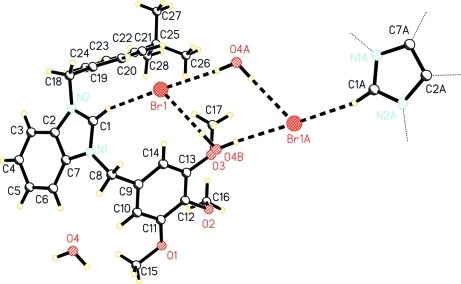
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O4—H4A⋯Br1i | 0.87 (3) | 2.54 (3) | 3.393 (3) | 169 (5) |
| O4—H4B⋯Br1ii | 0.88 (5) | 2.52 (5) | 3.399 (3) | 176 (5) |
| C1—H1⋯Br1 | 0.96 | 2.65 | 3.587 (3) | 165 |
| C3—H3⋯O2iii | 0.96 | 2.57 | 3.294 (4) | 132 |
| C6—H6⋯O4 | 0.96 | 2.38 | 3.305 (5) | 161 |
| C10—H10⋯O4 | 0.96 | 2.59 | 3.463 (5) | 152 |
| C14—H14⋯Br1 | 0.96 | 2.88 | 3.823 (3) | 167 |
| C18—H18A⋯Br1iv | 0.96 | 2.82 | 3.718 (3) | 155 |
Symmetry codes: (i) 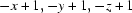 ; (ii)
; (ii) 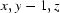 ; (iii)
; (iii) 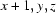 ; (iv)
; (iv) 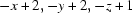 .
.
Acknowledgments
We thank the Technological and Scientific Research Council of Turkey TÜBİTAK-CNRS [TBAG-U/181 (106 T716)] and İnönü University Research Fund (İÜ BAP: 2008/Güdümlü 3) for financial support.
supplementary crystallographic information
Comment
N-heterocyclic carbenes have attracted much interest as a new class of compound in organometallic chemistry. The applications of N-heterocyclic carbenes were first reported by Herrmann in 1998 (Herrmann et al., 1998). Recently, Herrmann et al. and other researchers have designed new N-heterocyclic carbene compounds and have used them to prepare new catalysts for Suzuki-Miyura, Sonogashira, Stille and Heck reactions (Herrmann, 2002; Herrmann et al., 1995; Navarro et al., 2006; Arduengo & Krafczyc, 1998).
Recently, we have focused on the synthesis, characterization and use of palladium, platinum and ruthenium N-heterocyclic carbene complexes as catalysts for Suzuki-Miyura and Heck reactions (Yaşar et al., 2008; Arslan et al., 2009, and references therein).
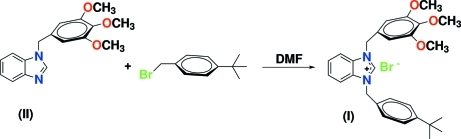
In the present work, we report the preparation and characterization of a novel N-heterocyclic carbene derivative, 1-(4-tert-butylbenzyl)-3-(3,4,5-trimethoxybenzyl)benzimidazolium bromide monohydrate, (I). The compound was purified by re-crystallization from ethanol:diethylether mixture (1:1) and characterized by elemental analysis, 1H and 13C-NMR and IR spectroscopy. The analytical and spectroscopic data are consistent with the proposed structure given in Scheme 1.
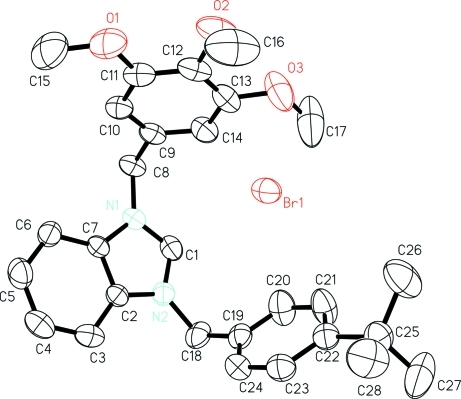
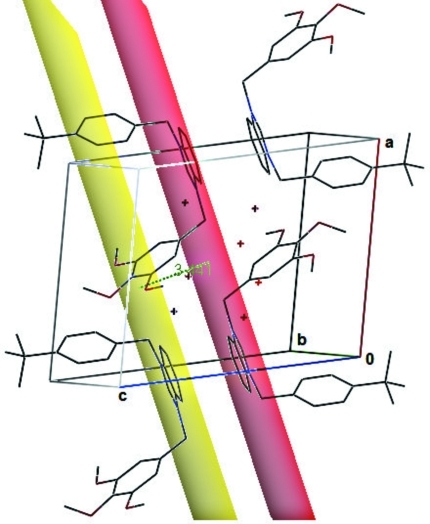
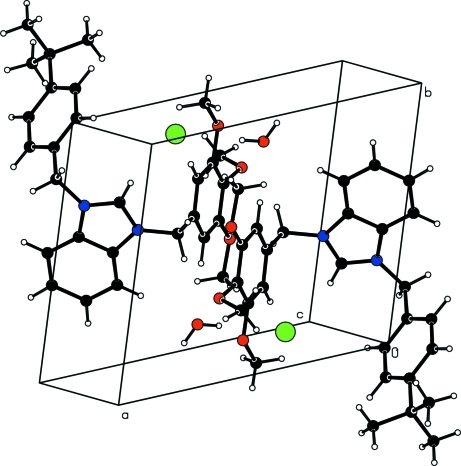
The molecular structure of the title compound is depicted in Figure 1. The crystal structure is composed of a 1-(4-tert-butylbenzyl)-3-(3,4,5-trimethoxybenzyl)benzimidazolium cation, a Br anion and solvent water molecules. All bond lengths in (I) are in normal ranges (Allen et al.,1987). The benzimidazole ring is almost coplanar with a maximum and a minimum deviation of 0.016 (2) Å for atom C2 and, 0.002 (2) Å for atom C6, respectively. In the crystal structure, π···π stacking interactions occurs between parallel benzimidazole rings, with a face-face separation of 3.441 Å (Figure 2) (Macrae et al., 2006). The dihedral angle between the benzimidazole ring and 4-tert-butylbenzyl and 3,4,5-trimethoxybenzyl groups are 70.23 (3)° and 73.48 (3) o, respectively.
The C—N bond lengths for the investigated compound are all shorter than the average single C—N bond length of 1.48 Å, being N1—C1 = 1.329 (3) Å, N2—C1 =1.325 (3) Å, N1—C7 = 1.389 (3) Å, and N2—C2 = 1.391 (3) Å thus showing varying degrees of double bond character in these C—N bonds. The other CN bond lengths are in agreement with the expected 1.48 Å C—N single bond lengths. This information indicates a partial electron delocalization within the C7—N1—C1—N2—C2 fragment. The N1—C1—N2 bond angle is also consistent with this hypothesis.
The crystal packing is shown in Figure 3. The intermolecular C—H···Br and O—H···Br hydrogen bonds (Figure 4, Table 1) and π···π stacking interactions link the molecules of the title compound.
Experimental
4-tertbutylbenzyl bromide (2.27 g, 10.0 mmol) was slowly added to a solution of 1-(3,4,5-trimethoxylbenzyl) benzimidazole (II) (2.98 g, 10.0 mmol) in DMF (5 mL) and the resulting mixture was stirred at room temperature for 5 h (Scheme 2). Diethylether (10 ml) was added to obtain a white crystalline solid which was filtered off. The solid was washed with diethylether (3 x 10 ml) dried under vacuum and the crude product was re-crystallized from ethanole/diethylether. M.p. = 246–247°C; yield 4.47 g, 85%; ν(CN) = 1594 cm-1. 1H NMR (δ, 200.13 MHz, CDCl3): 1.25 (s, 9H, CH2C6H4C(CH3)3-p); 3.86 and 3.79 (s, 9H, CH2C6H2(OCH3)3-3,4,5); 5.81 (s,4H, CH2C6H2(OCH3)3-3,4,5 and CH2C6H4C(CH3)3-p); 6.90 (s, 2H, CH2C6H2(OCH3)3-3,4,5); 7.33 and 7.48 (m, 8H, NC6H4N and CH2C6H4C(CH3)3-p); 11.64 (s, 1H, NCHN). 13C NMR (δ, 50 MHz, CDCl3): 31.6 (CH2C6H4C(CH3)3-p); 35.1 (CH2C6H4C(CH3)3-p); 51.6 (CH2C6H4C(CH3)3-p); 52.1 (CH2C6H2(OCH3)3-3,4,5); 57.2 and 61.2 (CH2C6H2(OCH3)3-3,4,5); 106.5, 131.7, 138.9 and 152.9 (CH2C6H2(OCH3)3-3,4,5); 114.2, 127.5, 128.7, 130.1, 131.8 and 143.2 (NC6H4N); 114.1, 126.7, 128.5 and 130.9 (CH2C6H4C(CH3)3-p); 154.2 (NCHN). Anal. Found: C, 63.96; H, 6.28; N: 5.35. Calc. for C28H33N2O3Br: C, 64.00; H, 6.33; N, 5.33.
Refinement
All H atoms attached to carbons were geometrically fixed and allowed to ride on the corresponding non-H atom with C—H = 0.96 Å, and Uiso(H) = 1.5Ueq(C) of the attached C atom for methyl H atoms and 1.2Ueq(C) for other H atoms. The water H atoms were located from a Fourier map and their distances were constrained to 0.86 Å and the Uiso(H) = 1.5Ueq(O).
Figures
Fig. 1.
The molecular structure of the title compound, showing the atom-numbering scheme and displacement ellipsoids drawn at the 50% probability level.
Fig. 2.
A packing diagram for (I).
Fig. 3.
A packing diagram for (I).
Fig. 4.
Hydrogen bonding for (I). Symmetry: O4A = x, 1 + y, z; O4B =1 - x,1 - y,1 - z; Br1A, etc. = 1 - x, 2 - y, 1 - z.
Fig. 5.
The preparation of the title compound.
Crystal data
| C28H33N2O3+·Br−·H2O | Z = 2 |
| Mr = 543.49 | F(000) = 568 |
| Triclinic, P1 | Dx = 1.303 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 10.389 (2) Å | Cell parameters from 4659 reflections |
| b = 10.436 (2) Å | θ = 3.2–26.4° |
| c = 14.038 (3) Å | µ = 1.52 mm−1 |
| α = 109.79 (3)° | T = 298 K |
| β = 90.70 (3)° | Rod, colorless |
| γ = 103.57 (3)° | 0.48 × 0.29 × 0.26 mm |
| V = 1385.1 (6) Å3 |
Data collection
| Mercury CCD diffractometer | 4860 independent reflections |
| Radiation source: Sealed Tube | 3921 reflections with I > 2σ(I) |
| Graphite Monochromator | Rint = 0.022 |
| Detector resolution: 14.6306 pixels mm-1 | θmax = 25.0°, θmin = 3.2° |
| ω scans | h = −12→12 |
| Absorption correction: multi-scan (REQAB; Jacobson, 1998) | k = −12→12 |
| Tmin = 0.514, Tmax = 0.673 | l = −16→16 |
| 11938 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.042 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.112 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.08 | w = 1/[σ2(Fo2) + (0.0505P)2 + 0.6557P] where P = (Fo2 + 2Fc2)/3 |
| 4860 reflections | (Δ/σ)max = 0.001 |
| 328 parameters | Δρmax = 0.31 e Å−3 |
| 2 restraints | Δρmin = −0.55 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Br1 | 0.75295 (3) | 0.93707 (4) | 0.51515 (3) | 0.06917 (15) | |
| N1 | 0.8315 (2) | 0.5830 (2) | 0.57685 (15) | 0.0384 (5) | |
| N2 | 1.0326 (2) | 0.7189 (2) | 0.62364 (16) | 0.0407 (5) | |
| C1 | 0.9118 (3) | 0.7064 (3) | 0.5843 (2) | 0.0439 (6) | |
| H1 | 0.8858 | 0.7766 | 0.5640 | 0.053* | |
| C2 | 1.0322 (2) | 0.5974 (2) | 0.64403 (18) | 0.0363 (5) | |
| C3 | 1.1313 (3) | 0.5558 (3) | 0.6834 (2) | 0.0497 (7) | |
| H3 | 1.2196 | 0.6159 | 0.7050 | 0.060* | |
| C4 | 1.0966 (3) | 0.4233 (3) | 0.6901 (2) | 0.0609 (8) | |
| H4 | 1.1626 | 0.3902 | 0.7168 | 0.073* | |
| C5 | 0.9681 (3) | 0.3364 (3) | 0.6591 (2) | 0.0573 (7) | |
| H5 | 0.9483 | 0.2449 | 0.6650 | 0.069* | |
| C6 | 0.8695 (3) | 0.3772 (3) | 0.6207 (2) | 0.0449 (6) | |
| H6 | 0.7811 | 0.3170 | 0.5997 | 0.054* | |
| C7 | 0.9039 (2) | 0.5104 (2) | 0.61354 (18) | 0.0351 (5) | |
| C8 | 0.6876 (2) | 0.5379 (3) | 0.5428 (2) | 0.0454 (6) | |
| H8A | 0.6617 | 0.4377 | 0.5084 | 0.054* | |
| H8B | 0.6699 | 0.5814 | 0.4955 | 0.054* | |
| C9 | 0.6061 (2) | 0.5776 (3) | 0.6319 (2) | 0.0438 (6) | |
| C10 | 0.5443 (3) | 0.4785 (3) | 0.6729 (2) | 0.0456 (6) | |
| H10 | 0.5523 | 0.3835 | 0.6449 | 0.055* | |
| C11 | 0.4705 (3) | 0.5178 (3) | 0.7550 (2) | 0.0521 (7) | |
| C12 | 0.4583 (3) | 0.6547 (3) | 0.7952 (2) | 0.0537 (7) | |
| C13 | 0.5222 (3) | 0.7534 (3) | 0.7543 (2) | 0.0553 (7) | |
| C14 | 0.5963 (3) | 0.7155 (3) | 0.6720 (2) | 0.0508 (7) | |
| H14 | 0.6399 | 0.7839 | 0.6435 | 0.061* | |
| C15 | 0.4234 (4) | 0.2906 (4) | 0.7695 (3) | 0.0873 (12) | |
| H15A | 0.3873 | 0.2414 | 0.6998 | 0.131* | |
| H15B | 0.3783 | 0.2416 | 0.8112 | 0.131* | |
| H15C | 0.5166 | 0.2951 | 0.7756 | 0.131* | |
| C16 | 0.4304 (4) | 0.7401 (6) | 0.9699 (3) | 0.1083 (17) | |
| H16A | 0.4738 | 0.6735 | 0.9805 | 0.162* | |
| H16B | 0.3620 | 0.7526 | 1.0150 | 0.162* | |
| H16C | 0.4944 | 0.8286 | 0.9835 | 0.162* | |
| C17 | 0.5773 (7) | 0.9936 (4) | 0.7672 (4) | 0.119 (2) | |
| H17A | 0.6705 | 0.9984 | 0.7736 | 0.179* | |
| H17B | 0.5622 | 1.0823 | 0.8082 | 0.179* | |
| H17C | 0.5478 | 0.9731 | 0.6973 | 0.179* | |
| C18 | 1.1457 (3) | 0.8450 (3) | 0.6461 (2) | 0.0536 (7) | |
| H18A | 1.1451 | 0.8826 | 0.5925 | 0.064* | |
| H18B | 1.2277 | 0.8182 | 0.6483 | 0.064* | |
| C19 | 1.1387 (3) | 0.9570 (3) | 0.7460 (2) | 0.0450 (6) | |
| C20 | 1.0657 (4) | 1.0516 (3) | 0.7490 (2) | 0.0671 (9) | |
| H20 | 1.0205 | 1.0466 | 0.6873 | 0.081* | |
| C21 | 1.0563 (4) | 1.1546 (3) | 0.8403 (2) | 0.0677 (9) | |
| H21 | 1.0048 | 1.2198 | 0.8403 | 0.081* | |
| C22 | 1.1192 (3) | 1.1659 (3) | 0.9317 (2) | 0.0452 (6) | |
| C23 | 1.1931 (3) | 1.0704 (3) | 0.9268 (2) | 0.0532 (7) | |
| H23 | 1.2390 | 1.0752 | 0.9883 | 0.064* | |
| C24 | 1.2033 (3) | 0.9667 (3) | 0.8350 (2) | 0.0533 (7) | |
| H24 | 1.2558 | 0.9019 | 0.8342 | 0.064* | |
| C25 | 1.1111 (3) | 1.2836 (3) | 1.0308 (2) | 0.0591 (8) | |
| C26 | 0.9702 (4) | 1.3046 (5) | 1.0365 (3) | 0.0903 (13) | |
| H26A | 0.9665 | 1.3779 | 1.0993 | 0.135* | |
| H26B | 0.9085 | 1.2186 | 1.0334 | 0.135* | |
| H26C | 0.9472 | 1.3306 | 0.9804 | 0.135* | |
| C27 | 1.2089 (5) | 1.4190 (4) | 1.0328 (4) | 0.1013 (15) | |
| H27A | 1.2976 | 1.4066 | 1.0315 | 0.152* | |
| H27B | 1.2037 | 1.4947 | 1.0937 | 0.152* | |
| H27C | 1.1868 | 1.4408 | 0.9745 | 0.152* | |
| C28 | 1.1443 (4) | 1.2485 (5) | 1.1241 (3) | 0.0840 (11) | |
| H28A | 1.2353 | 1.2435 | 1.1266 | 0.126* | |
| H28B | 1.0870 | 1.1594 | 1.1196 | 0.126* | |
| H28C | 1.1315 | 1.3204 | 1.1846 | 0.126* | |
| O1 | 0.4055 (2) | 0.4294 (3) | 0.80195 (18) | 0.0717 (6) | |
| O2 | 0.3740 (2) | 0.6898 (3) | 0.86948 (17) | 0.0741 (7) | |
| O3 | 0.5058 (3) | 0.8868 (3) | 0.8002 (2) | 0.0870 (8) | |
| O4 | 0.5832 (3) | 0.1696 (3) | 0.4946 (3) | 0.0969 (10) | |
| H4A | 0.499 (2) | 0.144 (6) | 0.501 (4) | 0.145* | |
| H4B | 0.623 (5) | 0.107 (5) | 0.501 (4) | 0.145* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Br1 | 0.0556 (2) | 0.0784 (2) | 0.0980 (3) | 0.02072 (16) | 0.01227 (17) | 0.0592 (2) |
| N1 | 0.0361 (11) | 0.0451 (11) | 0.0394 (12) | 0.0158 (9) | 0.0067 (9) | 0.0176 (9) |
| N2 | 0.0412 (12) | 0.0386 (11) | 0.0437 (12) | 0.0106 (9) | 0.0121 (9) | 0.0156 (9) |
| C1 | 0.0482 (15) | 0.0452 (14) | 0.0470 (15) | 0.0210 (12) | 0.0139 (12) | 0.0208 (12) |
| C2 | 0.0364 (12) | 0.0393 (12) | 0.0342 (13) | 0.0132 (10) | 0.0078 (10) | 0.0117 (10) |
| C3 | 0.0378 (14) | 0.0634 (17) | 0.0469 (16) | 0.0149 (13) | 0.0004 (12) | 0.0169 (13) |
| C4 | 0.0558 (18) | 0.073 (2) | 0.067 (2) | 0.0308 (16) | −0.0007 (15) | 0.0315 (16) |
| C5 | 0.067 (2) | 0.0494 (16) | 0.0648 (19) | 0.0205 (15) | 0.0037 (15) | 0.0284 (14) |
| C6 | 0.0463 (15) | 0.0391 (13) | 0.0477 (15) | 0.0082 (11) | 0.0045 (12) | 0.0148 (11) |
| C7 | 0.0348 (12) | 0.0396 (12) | 0.0330 (12) | 0.0147 (10) | 0.0060 (10) | 0.0119 (10) |
| C8 | 0.0360 (13) | 0.0599 (16) | 0.0433 (15) | 0.0190 (12) | 0.0027 (11) | 0.0173 (12) |
| C9 | 0.0329 (13) | 0.0567 (15) | 0.0432 (14) | 0.0176 (12) | 0.0004 (11) | 0.0155 (12) |
| C10 | 0.0366 (13) | 0.0535 (15) | 0.0483 (16) | 0.0120 (12) | 0.0020 (11) | 0.0195 (12) |
| C11 | 0.0363 (14) | 0.0673 (18) | 0.0546 (17) | 0.0098 (13) | 0.0039 (12) | 0.0260 (14) |
| C12 | 0.0355 (14) | 0.079 (2) | 0.0471 (16) | 0.0209 (14) | 0.0077 (12) | 0.0187 (14) |
| C13 | 0.0541 (17) | 0.0633 (18) | 0.0550 (17) | 0.0314 (15) | 0.0114 (14) | 0.0175 (14) |
| C14 | 0.0493 (16) | 0.0589 (17) | 0.0537 (17) | 0.0222 (13) | 0.0107 (13) | 0.0260 (13) |
| C15 | 0.091 (3) | 0.081 (3) | 0.099 (3) | 0.006 (2) | 0.019 (2) | 0.054 (2) |
| C16 | 0.078 (3) | 0.170 (5) | 0.054 (2) | 0.022 (3) | 0.013 (2) | 0.016 (3) |
| C17 | 0.195 (6) | 0.063 (2) | 0.112 (4) | 0.055 (3) | 0.059 (4) | 0.030 (2) |
| C18 | 0.0514 (16) | 0.0446 (15) | 0.0592 (18) | 0.0025 (12) | 0.0187 (14) | 0.0175 (13) |
| C19 | 0.0452 (14) | 0.0382 (13) | 0.0515 (16) | 0.0060 (11) | 0.0132 (12) | 0.0184 (11) |
| C20 | 0.097 (3) | 0.0610 (19) | 0.0488 (18) | 0.0340 (18) | −0.0027 (17) | 0.0174 (14) |
| C21 | 0.097 (3) | 0.0613 (19) | 0.0550 (19) | 0.0438 (19) | 0.0010 (18) | 0.0179 (15) |
| C22 | 0.0462 (15) | 0.0448 (14) | 0.0458 (15) | 0.0110 (12) | 0.0085 (12) | 0.0176 (12) |
| C23 | 0.0498 (16) | 0.0567 (17) | 0.0518 (17) | 0.0147 (13) | −0.0032 (13) | 0.0170 (13) |
| C24 | 0.0456 (15) | 0.0498 (15) | 0.0653 (19) | 0.0185 (13) | 0.0051 (14) | 0.0170 (14) |
| C25 | 0.0622 (19) | 0.0573 (17) | 0.0523 (18) | 0.0180 (15) | 0.0122 (15) | 0.0103 (14) |
| C26 | 0.091 (3) | 0.113 (3) | 0.072 (3) | 0.056 (3) | 0.024 (2) | 0.018 (2) |
| C27 | 0.127 (4) | 0.053 (2) | 0.093 (3) | 0.002 (2) | 0.024 (3) | 0.000 (2) |
| C28 | 0.087 (3) | 0.112 (3) | 0.0454 (19) | 0.032 (2) | 0.0076 (18) | 0.0146 (19) |
| O1 | 0.0604 (14) | 0.0848 (16) | 0.0758 (16) | 0.0089 (12) | 0.0203 (12) | 0.0416 (13) |
| O2 | 0.0492 (12) | 0.1158 (19) | 0.0554 (14) | 0.0316 (13) | 0.0177 (10) | 0.0202 (13) |
| O3 | 0.111 (2) | 0.0785 (16) | 0.0900 (19) | 0.0592 (16) | 0.0421 (16) | 0.0286 (14) |
| O4 | 0.0533 (14) | 0.0568 (14) | 0.174 (3) | 0.0082 (12) | −0.0014 (18) | 0.0367 (17) |
Geometric parameters (Å, °)
| N1—C1 | 1.329 (3) | C16—H16A | 0.9599 |
| N1—C7 | 1.389 (3) | C16—H16B | 0.9599 |
| N1—C8 | 1.478 (3) | C16—H16C | 0.9599 |
| N2—C1 | 1.325 (3) | C17—O3 | 1.407 (5) |
| N2—C2 | 1.391 (3) | C17—H17A | 0.9599 |
| N2—C18 | 1.481 (3) | C17—H17B | 0.9599 |
| C1—H1 | 0.9600 | C17—H17C | 0.9599 |
| C2—C3 | 1.382 (4) | C18—C19 | 1.507 (4) |
| C2—C7 | 1.393 (3) | C18—H18A | 0.9600 |
| C3—C4 | 1.380 (4) | C18—H18B | 0.9600 |
| C3—H3 | 0.9600 | C19—C20 | 1.370 (4) |
| C4—C5 | 1.394 (5) | C19—C24 | 1.372 (4) |
| C4—H4 | 0.9600 | C20—C21 | 1.389 (4) |
| C5—C6 | 1.368 (4) | C20—H20 | 0.9600 |
| C5—H5 | 0.9600 | C21—C22 | 1.388 (4) |
| C6—C7 | 1.390 (3) | C21—H21 | 0.9600 |
| C6—H6 | 0.9600 | C22—C23 | 1.380 (4) |
| C8—C9 | 1.513 (4) | C22—C25 | 1.532 (4) |
| C8—H8A | 0.9600 | C23—C24 | 1.399 (4) |
| C8—H8B | 0.9600 | C23—H23 | 0.9600 |
| C9—C10 | 1.383 (4) | C24—H24 | 0.9600 |
| C9—C14 | 1.386 (4) | C25—C27 | 1.526 (5) |
| C10—C11 | 1.390 (4) | C25—C26 | 1.529 (5) |
| C10—H10 | 0.9600 | C25—C28 | 1.530 (5) |
| C11—O1 | 1.369 (3) | C26—H26A | 0.9599 |
| C11—C12 | 1.385 (4) | C26—H26B | 0.9599 |
| C12—O2 | 1.380 (3) | C26—H26C | 0.9599 |
| C12—C13 | 1.385 (4) | C27—H27A | 0.9599 |
| C13—O3 | 1.374 (4) | C27—H27B | 0.9599 |
| C13—C14 | 1.392 (4) | C27—H27C | 0.9599 |
| C14—H14 | 0.9600 | C28—H28A | 0.9599 |
| C15—O1 | 1.422 (5) | C28—H28B | 0.9599 |
| C15—H15A | 0.9599 | C28—H28C | 0.9599 |
| C15—H15B | 0.9599 | O4—H4A | 0.87 (2) |
| C15—H15C | 0.9599 | O4—H4B | 0.88 (5) |
| C16—O2 | 1.394 (5) | ||
| C1—N1—C7 | 108.2 (2) | H16A—C16—H16C | 109.5 |
| C1—N1—C8 | 125.0 (2) | H16B—C16—H16C | 109.5 |
| C7—N1—C8 | 126.6 (2) | O3—C17—H17A | 109.5 |
| C1—N2—C2 | 108.4 (2) | O3—C17—H17B | 109.5 |
| C1—N2—C18 | 124.9 (2) | H17A—C17—H17B | 109.5 |
| C2—N2—C18 | 126.6 (2) | O3—C17—H17C | 109.5 |
| N2—C1—N1 | 110.5 (2) | H17A—C17—H17C | 109.5 |
| N2—C1—H1 | 124.8 | H17B—C17—H17C | 109.5 |
| N1—C1—H1 | 124.8 | N2—C18—C19 | 111.7 (2) |
| C3—C2—N2 | 132.0 (2) | N2—C18—H18A | 109.3 |
| C3—C2—C7 | 121.6 (2) | C19—C18—H18A | 109.3 |
| N2—C2—C7 | 106.3 (2) | N2—C18—H18B | 109.3 |
| C4—C3—C2 | 116.5 (3) | C19—C18—H18B | 109.3 |
| C4—C3—H3 | 121.7 | H18A—C18—H18B | 107.9 |
| C2—C3—H3 | 121.7 | C20—C19—C24 | 118.7 (3) |
| C3—C4—C5 | 121.6 (3) | C20—C19—C18 | 119.6 (3) |
| C3—C4—H4 | 119.2 | C24—C19—C18 | 121.7 (3) |
| C5—C4—H4 | 119.2 | C19—C20—C21 | 120.7 (3) |
| C6—C5—C4 | 122.1 (3) | C19—C20—H20 | 119.7 |
| C6—C5—H5 | 118.9 | C21—C20—H20 | 119.7 |
| C4—C5—H5 | 118.9 | C22—C21—C20 | 121.9 (3) |
| C5—C6—C7 | 116.5 (3) | C22—C21—H21 | 119.1 |
| C5—C6—H6 | 121.8 | C20—C21—H21 | 119.1 |
| C7—C6—H6 | 121.8 | C23—C22—C21 | 116.4 (3) |
| N1—C7—C6 | 131.8 (2) | C23—C22—C25 | 122.7 (3) |
| N1—C7—C2 | 106.6 (2) | C21—C22—C25 | 120.8 (3) |
| C6—C7—C2 | 121.6 (2) | C22—C23—C24 | 121.9 (3) |
| N1—C8—C9 | 111.2 (2) | C22—C23—H23 | 119.1 |
| N1—C8—H8A | 109.4 | C24—C23—H23 | 119.1 |
| C9—C8—H8A | 109.4 | C19—C24—C23 | 120.4 (3) |
| N1—C8—H8B | 109.4 | C19—C24—H24 | 119.8 |
| C9—C8—H8B | 109.4 | C23—C24—H24 | 119.8 |
| H8A—C8—H8B | 108.0 | C27—C25—C26 | 109.6 (3) |
| C10—C9—C14 | 120.8 (2) | C27—C25—C28 | 110.0 (3) |
| C10—C9—C8 | 120.6 (2) | C26—C25—C28 | 107.1 (3) |
| C14—C9—C8 | 118.6 (2) | C27—C25—C22 | 108.2 (3) |
| C9—C10—C11 | 119.5 (3) | C26—C25—C22 | 110.5 (3) |
| C9—C10—H10 | 120.3 | C28—C25—C22 | 111.5 (3) |
| C11—C10—H10 | 120.3 | C25—C26—H26A | 109.5 |
| O1—C11—C12 | 114.9 (3) | C25—C26—H26B | 109.5 |
| O1—C11—C10 | 124.7 (3) | H26A—C26—H26B | 109.5 |
| C12—C11—C10 | 120.4 (3) | C25—C26—H26C | 109.5 |
| O2—C12—C11 | 120.2 (3) | H26A—C26—H26C | 109.5 |
| O2—C12—C13 | 120.0 (3) | H26B—C26—H26C | 109.5 |
| C11—C12—C13 | 119.6 (3) | C25—C27—H27A | 109.5 |
| O3—C13—C12 | 115.3 (3) | C25—C27—H27B | 109.5 |
| O3—C13—C14 | 124.1 (3) | H27A—C27—H27B | 109.5 |
| C12—C13—C14 | 120.6 (3) | C25—C27—H27C | 109.5 |
| C9—C14—C13 | 119.1 (3) | H27A—C27—H27C | 109.5 |
| C9—C14—H14 | 120.5 | H27B—C27—H27C | 109.5 |
| C13—C14—H14 | 120.5 | C25—C28—H28A | 109.5 |
| O1—C15—H15A | 109.5 | C25—C28—H28B | 109.5 |
| O1—C15—H15B | 109.5 | H28A—C28—H28B | 109.5 |
| H15A—C15—H15B | 109.5 | C25—C28—H28C | 109.5 |
| O1—C15—H15C | 109.5 | H28A—C28—H28C | 109.5 |
| H15A—C15—H15C | 109.5 | H28B—C28—H28C | 109.5 |
| H15B—C15—H15C | 109.5 | C11—O1—C15 | 117.4 (3) |
| O2—C16—H16A | 109.5 | C12—O2—C16 | 116.4 (3) |
| O2—C16—H16B | 109.5 | C13—O3—C17 | 117.6 (3) |
| H16A—C16—H16B | 109.5 | H4A—O4—H4B | 110 (5) |
| O2—C16—H16C | 109.5 | ||
| C2—N2—C1—N1 | −0.4 (3) | O2—C12—C13—O3 | 6.4 (4) |
| C18—N2—C1—N1 | −177.7 (2) | C11—C12—C13—O3 | −179.1 (3) |
| C7—N1—C1—N2 | 0.1 (3) | O2—C12—C13—C14 | −173.2 (3) |
| C8—N1—C1—N2 | 175.6 (2) | C11—C12—C13—C14 | 1.4 (5) |
| C1—N2—C2—C3 | 179.0 (3) | C10—C9—C14—C13 | −0.4 (4) |
| C18—N2—C2—C3 | −3.8 (4) | C8—C9—C14—C13 | −179.7 (3) |
| C1—N2—C2—C7 | 0.6 (3) | O3—C13—C14—C9 | 179.9 (3) |
| C18—N2—C2—C7 | 177.8 (2) | C12—C13—C14—C9 | −0.5 (4) |
| N2—C2—C3—C4 | −177.6 (3) | C1—N2—C18—C19 | 81.3 (3) |
| C7—C2—C3—C4 | 0.5 (4) | C2—N2—C18—C19 | −95.5 (3) |
| C2—C3—C4—C5 | −0.3 (4) | N2—C18—C19—C20 | −86.2 (3) |
| C3—C4—C5—C6 | −0.1 (5) | N2—C18—C19—C24 | 93.5 (3) |
| C4—C5—C6—C7 | 0.2 (4) | C24—C19—C20—C21 | −0.5 (5) |
| C1—N1—C7—C6 | −178.2 (3) | C18—C19—C20—C21 | 179.1 (3) |
| C8—N1—C7—C6 | 6.3 (4) | C19—C20—C21—C22 | −0.3 (6) |
| C1—N1—C7—C2 | 0.3 (3) | C20—C21—C22—C23 | 0.8 (5) |
| C8—N1—C7—C2 | −175.2 (2) | C20—C21—C22—C25 | 177.9 (3) |
| C5—C6—C7—N1 | 178.4 (3) | C21—C22—C23—C24 | −0.6 (4) |
| C5—C6—C7—C2 | 0.0 (4) | C25—C22—C23—C24 | −177.7 (3) |
| C3—C2—C7—N1 | −179.1 (2) | C20—C19—C24—C23 | 0.7 (4) |
| N2—C2—C7—N1 | −0.5 (2) | C18—C19—C24—C23 | −178.9 (3) |
| C3—C2—C7—C6 | −0.4 (4) | C22—C23—C24—C19 | −0.1 (4) |
| N2—C2—C7—C6 | 178.2 (2) | C23—C22—C25—C27 | 98.5 (4) |
| C1—N1—C8—C9 | −90.8 (3) | C21—C22—C25—C27 | −78.4 (4) |
| C7—N1—C8—C9 | 84.0 (3) | C23—C22—C25—C26 | −141.5 (3) |
| N1—C8—C9—C10 | −100.3 (3) | C21—C22—C25—C26 | 41.6 (4) |
| N1—C8—C9—C14 | 78.9 (3) | C23—C22—C25—C28 | −22.5 (4) |
| C14—C9—C10—C11 | 0.5 (4) | C21—C22—C25—C28 | 160.6 (3) |
| C8—C9—C10—C11 | 179.7 (2) | C12—C11—O1—C15 | −175.0 (3) |
| C9—C10—C11—O1 | −179.8 (3) | C10—C11—O1—C15 | 5.2 (5) |
| C9—C10—C11—C12 | 0.3 (4) | C11—C12—O2—C16 | 92.8 (4) |
| O1—C11—C12—O2 | −6.6 (4) | C13—C12—O2—C16 | −92.7 (4) |
| C10—C11—C12—O2 | 173.3 (3) | C12—C13—O3—C17 | 175.0 (4) |
| O1—C11—C12—C13 | 178.9 (3) | C14—C13—O3—C17 | −5.5 (6) |
| C10—C11—C12—C13 | −1.2 (4) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O4—H4A···Br1i | 0.87 (3) | 2.54 (3) | 3.393 (3) | 169 (5) |
| O4—H4B···Br1ii | 0.88 (5) | 2.52 (5) | 3.399 (3) | 176 (5) |
| C1—H1···Br1 | 0.96 | 2.65 | 3.587 (3) | 165 |
| C3—H3···O2iii | 0.96 | 2.57 | 3.294 (4) | 132 |
| C6—H6···O4 | 0.96 | 2.38 | 3.305 (5) | 161 |
| C10—H10···O4 | 0.96 | 2.59 | 3.463 (5) | 152 |
| C14—H14···Br1 | 0.96 | 2.88 | 3.823 (3) | 167 |
| C18—H18A···Br1iv | 0.96 | 2.82 | 3.718 (3) | 155 |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) x, y−1, z; (iii) x+1, y, z; (iv) −x+2, −y+2, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HG2459).
References
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
- Arduengo, A. J. & Krafczyc, R. (1998). Chem. Ztg.32, 6–14.
- Arslan, H., VanDerveer, D., Yaşar, S., Özdemir, İ. & Çetinkaya, B. (2009). Acta Cryst. E65, o121–o122. [DOI] [PMC free article] [PubMed]
- Herrmann, W. A. (2002). Angew. Chem. Int. Ed.41, 1290–1309.
- Herrmann, W. A., Elison, M., Fischer, J., Köcher, C. & Artus, G. R. J. (1995). Angew. Chem. Int. Ed. Engl.34, 2371–2374.
- Herrmann, W. A., Reisinger, C. P. & Spiegler, M. (1998). J. Organomet. Chem.557, 93–96.
- Jacobson, R. (1998). REQAB. Molecular Structure Corporation, The Woodlands, Texas,USA.
- Macrae, C. F., Edgington, P. R., McCabe, P., Pidcock, E., Shields, G. P., Taylor, R., Towler, M. & van de Streek, J. (2006). J. Appl. Cryst.39, 453–457.
- Navarro, O., Marion, N., Oonishi, Y., Kelly, R. A. & Nolan, S. P. (2006). J. Org. Chem.71, 685–692. [DOI] [PubMed]
- Rigaku/MSC (2001). CrystalClear Rigaku/MSC, The Woodlands, Texas, USA.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Yaşar, S., Özdemir, I., Çetinkaya, B., Renaud, J. L. & Bruneau, L. (2008). Eur. J. Org. Chem.12, 2142–2149.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I. DOI: 10.1107/S1600536808043250/hg2459sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808043250/hg2459Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report