Abstract
A new cadmium(II) coordination polymer, {[Cd(C8H4O4)(C12H8N2)(H2O)]·0.5C8H6O4}n, has been synthesized under hydrothermal conditions. The asymmetric unit contains one CdII atom, one benzene-1,4-dicarboxylate anion, one 1,10-phenanthroline ligand, one coordinated water molecule and half of an uncoordinated benzene-1,4-dicaboxylic acid solvent molecule. The CdII atom is in the centre of a monocapped distorted octahedron made up of four O atoms of two chelating benzene-1,4-dicarboxylate anions, one water O atom and two 1,10-phenanthroline N atoms. The metal centres are connected via bis-chelating benzene-1,4-dicarboxylate anions into a zigzag chain structure along [001]. These chains are further connected by O—H⋯O hydrogen bonds between the water molecules and adjacent carboxylate O atoms. Additional O—H⋯O hydrogen bonding between the uncoordinated benzene-1,4-dicaboxylic acid molecules along [010] consolidates the structure.
Related literature
For background to coordination polymers, see: Liang et al. (2002 ▶); McGarrah et al. (2001 ▶); Moulton et al. (2002 ▶); Wu et al. (2007 ▶). Zheng et al. (2004 ▶). For related structures, see: Shi et al. (2004 ▶); Wang et al. (2004 ▶).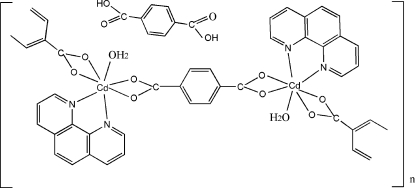
Experimental
Crystal data
[Cd(C8H4O4)(C12H8N2)(H2O)]·0.5C8H6O4
M r = 557.80
Monoclinic,

a = 26.108 (2) Å
b = 9.6928 (10) Å
c = 21.161 (2) Å
β = 126.494 (2)°
V = 4304.9 (7) Å3
Z = 8
Mo Kα radiation
μ = 1.07 mm−1
T = 298 (2) K
0.42 × 0.18 × 0.02 mm
Data collection
Bruker SMART CCD diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 2004 ▶) T min = 0.663, T max = 0.984
10895 measured reflections
3793 independent reflections
3061 reflections with I > 2σ(I)
R int = 0.021
Refinement
R[F 2 > 2σ(F 2)] = 0.025
wR(F 2) = 0.062
S = 1.07
3793 reflections
309 parameters
H-atom parameters constrained
Δρmax = 0.57 e Å−3
Δρmin = −0.28 e Å−3
Data collection: SMART (Bruker, 2001 ▶); cell refinement: SAINT (Bruker, 2001 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809000889/wm2211sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809000889/wm2211Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Selected bond lengths (Å).
| Cd1—O1 | 2.284 (3) |
| Cd1—O3 | 2.357 (2) |
| Cd1—N2 | 2.358 (2) |
| Cd1—N1 | 2.362 (2) |
| Cd1—O7 | 2.375 (2) |
| Cd1—O4 | 2.377 (2) |
| Cd1—O2 | 2.601 (3) |
Table 2. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O5—H5⋯O6i | 0.82 | 1.92 | 2.728 (3) | 167 |
| O7—H7A⋯O2ii | 0.85 | 1.88 | 2.665 (3) | 154 |
| O7—H7B⋯O4ii | 0.85 | 2.28 | 3.019 (3) | 146 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
supplementary crystallographic information
Comment
Carboxylic acid coordination polymers have an important position in coordination chemistry due to their interesting topologies and their potential applications in functional materials (Wu et al., 2007). The coordination ability of the 1,10-phenanthroline ligand stems from its chelate effect which frequently results in the formation of low-dimensional coordination polymers because of its terminal-group effect (Moulton et al., 2002). In this work, we report a new cadmium coordination polymer with a one-dimensional zigzag chain, {[Cd(C8H4O4)(C12H8N2)(H2O)]2(C8H6O4)}n, (I), that was synthesized from benzene-1,4-dicarboxylic acid, 1,10-phenanthroline and cadmium nitrate under hydrothermal conditions.
Each [Cd(C8H4O4)(C12H8N2)(H2O)]2(C8H6O4)] unit is composed of two Cd2+ ions, two 1,10-phenanthroline ligands, two benzene-1,4-dicarboxylate anions, two coordinated water molecules, and one uncoordinated neutral benzene-1,4-dicaboxylic acid solvent molecule. The Cd2+ ion in the complex is seven-coordinated with four oxygen atoms (O1, O2, O3 and O4) of two chelating bidentate carboxyl groups from two deprotonated benzene-1,4-dicaboxylic acid ligands, one O atom (O7) of a water molecule, and two nitrogen atoms (N1 and N2) from the chelating 1,10-phenanthroline ligand (Fig. 1). The Cd—O bond distances are between 2.284 (2) Å and 2.601 (2) Å, while the Cd—N bond distances are 2.358 (2) Å and 2.362 (2) Å, which all are in agreement with those of other reported Cd—O and Cd—N bond lengths (Wang et al., 2004; Shi et al., 2004). One Cd2+ ion connects two benzene-1,4-dicarboxylate ligands, and one benzene-1,4-dicarboxylic acid ligand also connects two Cd2+ ions, leading to a zigzag chain along [001], as shown in Fig. 2. These chains are further connected through O—H···O hydrogen bonding interactions between the water molecules and the oxygen atoms of the coordinated carboxyl groups of benzene-1,4-dicarboxylate ligands in neighboring chains to give an extended supramolecular two-dimensional layer structure, as illustrated in Fig. 3. Additional O—H···O hydrogen bonding between the uncoordinated benzene-1,4-dicaboxylic acid molecules along [010] consolidates the structure.
Experimental
All regents used in the synthesis were of analytic grade and were used without further purification. A mixture of Cd(NO3)2.6H2O (0.3085 g), 1,10-phenanthroline (0.099 g), benzene-1,4-dicarboxylic acid (0.083 g) and water (20 mL) was sealed in a 40 mL stainless steel reactor with a Teflon inlay after adjustment of the pH to 7 by addition of a dilute NaOH solution. The autoclave was heated to 448 K for 6 d, and then cooled to room temperature. Colorless hexagonal-prismatic crystals were obtained. Elemental analysis, calculated: C, 51.68, H, 3.07, N 5.02; found: C, 50.87, H, 3.22, N 4.96.
The title compoud was further characterized by FT-IR spectroscopy (recorded over the 400 to 4000 cm-1 region on a Nicolet NEXUS 670 spectrometer with KBr pellets at room temperature); by thermogravimetric analysis (TGA) (performed on a Universal V4.1D TA-SDT Q600 thermal analyzer in N2 atmosphere with a heating rate of 10 °/min) and by its luminescent properties of the solid state under room temperature.
The FT-IR spectrum of the title compound exhibited the following absorption bands that were assigned referring to literature (Liang et al., 2002). A broad band at 3199 cm-1 corresponds to the O-H stretching vibration of the coordinated water molecule. The peak at 1682 cm-1 is attributed to the C=O stretching vibration of the carboxylate group, and the peak at 1380 cm-1 is due to the C-O stretching vibration of carboxylate group. A typical TG curve is shown in Figure 4. It shows that this compound has two steps of mass loss between 55 and 635 °C. The first step is completed at 376 °C, accompanied with 18.05 % mass loss, which is in good agreement with the theoretical mass loss of 18.12 %, corresponding to the evaporation of two water molecules and an uncoordinated neutral 1,4-benzenedecaboxylic acid molecule. In the second step, the mass loss is 70.31 % from 376 to 635 °C, which corresponds to the decomposition of two 1,10-phenanthroline and two deprotonated 1,4-benzenedecaboxylic acid ligands and can be compared with the calculated value of 70.37 %. After the two steps of mass loss, the mass fraction of the residue is 11.44 %, which is in agreement with the calculated mass summation of the remaining CdO (11.51 %). Previous studies have shown that coordination polymers containing zinc and cadmium ions exhibit photoluminescent properties (McGarrah et al., 2001). As shown in Figure 5, upon excitation of the solid sample at 370 nm, it exhibited strong cyan fluorescent emission bands at 504 nm. It is probably due to the (π*-π) transitions changing into the (π*-n) transitions after forming the coordination polymer (Zheng et al., 2004).
Refinement
H atoms were positioned in calculated positions (O–H = 0.82 Å and 0.82 Å, C–H = 0.93 Å) and were refined using the riding model approximation with Uiso(H) = 1.2Ueq of the parent atom.
Figures
Fig. 1.
The asymmetric unit in the structure of compound (I). Displacement ellipsoids are drawn at the 30% probability level.
Fig. 2.
A view of a one-dimensional zigzag chain in the structure of compound (I). The uncoordinated 1,4-benzenedecaboxylic acid solvent molecules were omitted for clarity.
Fig. 3.
The layered structure of compound (I), constructed by H-bonding.
Fig. 4.
The TG-curve of the thermal decomposition of the title compound (I).
Fig. 5.
Solid-state emission spectra of the title complex (I) at room temperature.
Crystal data
| [Cd(C8H4O4)(C12H8N2)(H2O)]·0.5C8H6O4 | F(000) = 2232 |
| Mr = 557.80 | Dx = 1.721 Mg m−3 |
| Monoclinic, C2/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -C 2yc | Cell parameters from 5356 reflections |
| a = 26.108 (2) Å | θ = 2.4–27.4° |
| b = 9.6928 (10) Å | µ = 1.07 mm−1 |
| c = 21.161 (2) Å | T = 298 K |
| β = 126.494 (2)° | Prism, colourless |
| V = 4304.9 (7) Å3 | 0.42 × 0.18 × 0.02 mm |
| Z = 8 |
Data collection
| Bruker SMART CCD diffractometer | 3793 independent reflections |
| Radiation source: fine-focus sealed tube | 3061 reflections with I > 2σ(I) |
| graphite | Rint = 0.021 |
| φ and ω scans | θmax = 25.0°, θmin = 2.0° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 2004) | h = −30→30 |
| Tmin = 0.663, Tmax = 0.984 | k = −11→10 |
| 10895 measured reflections | l = −18→25 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.025 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.062 | H-atom parameters constrained |
| S = 1.07 | w = 1/[σ2(Fo2) + (0.0233P)2 + 5.6765P] where P = (Fo2 + 2Fc2)/3 |
| 3793 reflections | (Δ/σ)max = 0.001 |
| 309 parameters | Δρmax = 0.57 e Å−3 |
| 0 restraints | Δρmin = −0.28 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| Cd1 | 0.163966 (9) | 0.80224 (2) | 0.186508 (12) | 0.03638 (8) | |
| N1 | 0.05395 (11) | 0.8470 (2) | 0.11727 (14) | 0.0355 (5) | |
| N2 | 0.10891 (11) | 0.5924 (2) | 0.15983 (14) | 0.0415 (6) | |
| O1 | 0.18828 (13) | 0.8727 (3) | 0.30445 (14) | 0.0762 (8) | |
| O2 | 0.23664 (13) | 0.6773 (4) | 0.32126 (16) | 0.0928 (10) | |
| O3 | 0.14899 (10) | 0.7998 (2) | 0.06513 (12) | 0.0509 (6) | |
| O4 | 0.24360 (10) | 0.7435 (2) | 0.16977 (12) | 0.0502 (6) | |
| O5 | 0.05219 (11) | 1.1134 (2) | 0.28343 (16) | 0.0729 (8) | |
| H5 | 0.0460 | 1.1969 | 0.2794 | 0.109* | 0.50 |
| O6 | 0.05277 (11) | 0.3946 (2) | 0.28845 (15) | 0.0701 (7) | |
| H6 | 0.0467 | 0.3111 | 0.2840 | 0.105* | 0.50 |
| O7 | 0.18243 (10) | 1.0437 (2) | 0.19469 (12) | 0.0504 (6) | |
| H7A | 0.2076 | 1.0625 | 0.1829 | 0.061* | |
| H7B | 0.1988 | 1.0725 | 0.2410 | 0.061* | |
| C1 | 0.22048 (16) | 0.7708 (4) | 0.3456 (2) | 0.0569 (10) | |
| C2 | 0.23658 (13) | 0.7594 (3) | 0.42642 (17) | 0.0382 (7) | |
| C3 | 0.23231 (15) | 0.8742 (3) | 0.46128 (19) | 0.0478 (8) | |
| H3 | 0.2202 | 0.9585 | 0.4351 | 0.057* | |
| C4 | 0.25404 (15) | 0.6350 (3) | 0.46504 (19) | 0.0477 (8) | |
| H4 | 0.2567 | 0.5570 | 0.4415 | 0.057* | |
| C5 | 0.20622 (14) | 0.7676 (3) | 0.09733 (18) | 0.0373 (7) | |
| C6 | 0.22902 (13) | 0.7568 (3) | 0.04736 (17) | 0.0332 (6) | |
| C7 | 0.18645 (13) | 0.7628 (3) | −0.03373 (18) | 0.0425 (7) | |
| H7 | 0.1432 | 0.7718 | −0.0571 | 0.051* | |
| C8 | 0.29305 (14) | 0.7441 (3) | 0.08069 (18) | 0.0428 (7) | |
| H8 | 0.3226 | 0.7403 | 0.1351 | 0.051* | |
| C9 | 0.0000 | 1.0513 (4) | 0.2500 | 0.0443 (11) | |
| C10 | 0.0000 | 0.8973 (4) | 0.2500 | 0.0390 (10) | |
| C11 | 0.05674 (14) | 0.8252 (3) | 0.28751 (19) | 0.0479 (8) | |
| H11 | 0.0951 | 0.8730 | 0.3127 | 0.058* | |
| C12 | 0.05684 (15) | 0.6829 (3) | 0.2878 (2) | 0.0500 (8) | |
| H12 | 0.0952 | 0.6352 | 0.3134 | 0.060* | |
| C13 | 0.0000 | 0.6107 (4) | 0.2500 | 0.0400 (10) | |
| C14 | 0.0000 | 0.4565 (5) | 0.2500 | 0.0452 (11) | |
| C15 | 0.02751 (15) | 0.9707 (3) | 0.09881 (19) | 0.0470 (8) | |
| H15 | 0.0539 | 1.0478 | 0.1175 | 0.056* | |
| C16 | −0.03804 (15) | 0.9903 (4) | 0.0527 (2) | 0.0538 (9) | |
| H16 | −0.0549 | 1.0790 | 0.0409 | 0.065* | |
| C17 | −0.07692 (15) | 0.8796 (4) | 0.0251 (2) | 0.0547 (9) | |
| H17 | −0.1209 | 0.8918 | −0.0067 | 0.066* | |
| C18 | −0.05130 (14) | 0.7468 (3) | 0.04402 (19) | 0.0452 (8) | |
| C19 | 0.01571 (13) | 0.7355 (3) | 0.09130 (16) | 0.0344 (6) | |
| C20 | 0.04452 (14) | 0.6005 (3) | 0.11358 (17) | 0.0390 (7) | |
| C21 | 0.00562 (16) | 0.4833 (3) | 0.0881 (2) | 0.0520 (8) | |
| C22 | 0.0362 (2) | 0.3557 (4) | 0.1137 (3) | 0.0774 (12) | |
| H22 | 0.0120 | 0.2754 | 0.0981 | 0.093* | |
| C23 | 0.1001 (2) | 0.3473 (4) | 0.1607 (3) | 0.0796 (13) | |
| H23 | 0.1204 | 0.2622 | 0.1781 | 0.096* | |
| C24 | 0.13532 (18) | 0.4689 (4) | 0.1827 (2) | 0.0626 (10) | |
| H24 | 0.1795 | 0.4628 | 0.2152 | 0.075* | |
| C25 | −0.08894 (16) | 0.6240 (4) | 0.0181 (2) | 0.0647 (10) | |
| H25 | −0.1331 | 0.6313 | −0.0142 | 0.078* | |
| C26 | −0.06213 (18) | 0.4987 (4) | 0.0391 (2) | 0.0692 (11) | |
| H26 | −0.0879 | 0.4207 | 0.0217 | 0.083* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cd1 | 0.03152 (12) | 0.04161 (13) | 0.03571 (13) | 0.00181 (10) | 0.01983 (10) | 0.00116 (11) |
| N1 | 0.0354 (13) | 0.0350 (13) | 0.0368 (14) | 0.0012 (11) | 0.0219 (12) | 0.0022 (11) |
| N2 | 0.0383 (14) | 0.0368 (14) | 0.0448 (15) | 0.0046 (11) | 0.0223 (13) | −0.0005 (12) |
| O1 | 0.085 (2) | 0.091 (2) | 0.0430 (15) | −0.0133 (17) | 0.0329 (15) | 0.0057 (15) |
| O2 | 0.0747 (19) | 0.157 (3) | 0.0568 (17) | 0.0122 (19) | 0.0443 (16) | −0.0235 (19) |
| O3 | 0.0394 (12) | 0.0742 (15) | 0.0446 (12) | 0.0144 (11) | 0.0280 (11) | 0.0073 (12) |
| O4 | 0.0433 (12) | 0.0731 (15) | 0.0366 (13) | 0.0063 (11) | 0.0251 (11) | 0.0046 (11) |
| O5 | 0.0487 (14) | 0.0436 (15) | 0.101 (2) | −0.0060 (12) | 0.0307 (15) | −0.0069 (14) |
| O6 | 0.0583 (16) | 0.0435 (14) | 0.0852 (19) | 0.0088 (12) | 0.0300 (15) | −0.0008 (14) |
| O7 | 0.0564 (13) | 0.0473 (13) | 0.0485 (13) | −0.0114 (11) | 0.0317 (12) | −0.0060 (11) |
| C1 | 0.0359 (18) | 0.091 (3) | 0.044 (2) | −0.0160 (19) | 0.0237 (17) | −0.012 (2) |
| C2 | 0.0297 (15) | 0.0521 (18) | 0.0326 (16) | −0.0034 (13) | 0.0185 (14) | −0.0059 (15) |
| C3 | 0.0518 (19) | 0.0381 (18) | 0.0470 (19) | 0.0049 (15) | 0.0260 (17) | 0.0044 (15) |
| C4 | 0.0513 (19) | 0.0443 (19) | 0.049 (2) | 0.0033 (15) | 0.0302 (17) | −0.0118 (17) |
| C5 | 0.0387 (17) | 0.0345 (16) | 0.0414 (18) | 0.0001 (13) | 0.0253 (15) | 0.0002 (13) |
| C6 | 0.0339 (15) | 0.0303 (14) | 0.0367 (16) | −0.0003 (12) | 0.0217 (14) | −0.0008 (13) |
| C7 | 0.0277 (15) | 0.059 (2) | 0.0396 (18) | 0.0030 (14) | 0.0196 (14) | 0.0025 (15) |
| C8 | 0.0337 (16) | 0.0604 (19) | 0.0299 (16) | 0.0016 (14) | 0.0166 (14) | −0.0007 (15) |
| C9 | 0.046 (3) | 0.042 (3) | 0.040 (3) | 0.000 | 0.023 (2) | 0.000 |
| C10 | 0.042 (2) | 0.040 (2) | 0.034 (2) | 0.000 | 0.023 (2) | 0.000 |
| C11 | 0.0376 (17) | 0.044 (2) | 0.055 (2) | −0.0039 (14) | 0.0236 (16) | −0.0058 (16) |
| C12 | 0.0386 (18) | 0.046 (2) | 0.057 (2) | 0.0050 (15) | 0.0235 (17) | 0.0006 (16) |
| C13 | 0.043 (2) | 0.039 (3) | 0.038 (2) | 0.000 | 0.024 (2) | 0.000 |
| C14 | 0.048 (3) | 0.044 (3) | 0.041 (3) | 0.000 | 0.025 (2) | 0.000 |
| C15 | 0.0451 (19) | 0.0393 (18) | 0.058 (2) | 0.0037 (14) | 0.0311 (17) | 0.0069 (16) |
| C16 | 0.0446 (19) | 0.047 (2) | 0.066 (2) | 0.0145 (16) | 0.0312 (19) | 0.0146 (18) |
| C17 | 0.0318 (17) | 0.072 (3) | 0.053 (2) | 0.0116 (17) | 0.0216 (16) | 0.0107 (19) |
| C18 | 0.0329 (16) | 0.0537 (19) | 0.0446 (19) | −0.0028 (14) | 0.0207 (15) | −0.0013 (16) |
| C19 | 0.0344 (15) | 0.0394 (17) | 0.0301 (15) | −0.0020 (13) | 0.0195 (13) | −0.0014 (13) |
| C20 | 0.0405 (17) | 0.0398 (17) | 0.0377 (17) | −0.0029 (13) | 0.0238 (15) | −0.0045 (14) |
| C21 | 0.054 (2) | 0.044 (2) | 0.057 (2) | −0.0094 (16) | 0.0331 (19) | −0.0105 (17) |
| C22 | 0.084 (3) | 0.039 (2) | 0.105 (4) | −0.011 (2) | 0.054 (3) | −0.013 (2) |
| C23 | 0.085 (3) | 0.034 (2) | 0.110 (4) | 0.007 (2) | 0.052 (3) | 0.001 (2) |
| C24 | 0.058 (2) | 0.045 (2) | 0.072 (3) | 0.0138 (18) | 0.031 (2) | 0.0034 (19) |
| C25 | 0.0352 (19) | 0.077 (3) | 0.065 (2) | −0.0120 (19) | 0.0211 (18) | −0.009 (2) |
| C26 | 0.056 (2) | 0.062 (3) | 0.082 (3) | −0.027 (2) | 0.037 (2) | −0.020 (2) |
Geometric parameters (Å, °)
| Cd1—O1 | 2.284 (3) | C8—C7ii | 1.382 (4) |
| Cd1—O3 | 2.357 (2) | C8—H8 | 0.9300 |
| Cd1—N2 | 2.358 (2) | C9—O5iii | 1.254 (3) |
| Cd1—N1 | 2.362 (2) | C9—C10 | 1.493 (6) |
| Cd1—O7 | 2.375 (2) | C10—C11iii | 1.384 (4) |
| Cd1—O4 | 2.377 (2) | C10—C11 | 1.384 (4) |
| Cd1—O2 | 2.601 (3) | C11—C12 | 1.379 (4) |
| N1—C15 | 1.321 (4) | C11—H11 | 0.9300 |
| N1—C19 | 1.347 (4) | C12—C13 | 1.385 (4) |
| N2—C24 | 1.321 (4) | C12—H12 | 0.9300 |
| N2—C20 | 1.354 (4) | C13—C12iii | 1.385 (4) |
| O1—C1 | 1.253 (4) | C13—C14 | 1.494 (6) |
| O2—C1 | 1.235 (4) | C14—O6iii | 1.260 (3) |
| O3—C5 | 1.258 (3) | C15—C16 | 1.390 (4) |
| O4—C5 | 1.256 (3) | C15—H15 | 0.9300 |
| O5—C9 | 1.254 (3) | C16—C17 | 1.348 (5) |
| O5—H5 | 0.8200 | C16—H16 | 0.9300 |
| O6—C14 | 1.260 (3) | C17—C18 | 1.395 (5) |
| O6—H6 | 0.8200 | C17—H17 | 0.9300 |
| O7—H7A | 0.8501 | C18—C19 | 1.411 (4) |
| O7—H7B | 0.8499 | C18—C25 | 1.429 (5) |
| C1—C2 | 1.503 (5) | C19—C20 | 1.442 (4) |
| C2—C4 | 1.374 (4) | C20—C21 | 1.400 (4) |
| C2—C3 | 1.376 (4) | C21—C22 | 1.395 (5) |
| C3—C4i | 1.380 (5) | C21—C26 | 1.430 (5) |
| C3—H3 | 0.9300 | C22—C23 | 1.345 (5) |
| C4—C3i | 1.380 (5) | C22—H22 | 0.9300 |
| C4—H4 | 0.9300 | C23—C24 | 1.393 (5) |
| C5—C6 | 1.494 (4) | C23—H23 | 0.9300 |
| C6—C8 | 1.380 (4) | C24—H24 | 0.9300 |
| C6—C7 | 1.384 (4) | C25—C26 | 1.339 (5) |
| C7—C8ii | 1.382 (4) | C25—H25 | 0.9300 |
| C7—H7 | 0.9300 | C26—H26 | 0.9300 |
| O1—Cd1—O3 | 162.20 (9) | C6—C8—C7ii | 120.4 (3) |
| O1—Cd1—N2 | 104.69 (9) | C6—C8—H8 | 119.8 |
| O3—Cd1—N2 | 92.75 (8) | C7ii—C8—H8 | 119.8 |
| O1—Cd1—N1 | 93.90 (9) | O5—C9—O5iii | 122.6 (4) |
| O3—Cd1—N1 | 88.48 (7) | O5—C9—C10 | 118.7 (2) |
| N2—Cd1—N1 | 70.53 (8) | O5iii—C9—C10 | 118.7 (2) |
| O1—Cd1—O7 | 73.32 (9) | C11iii—C10—C11 | 119.3 (4) |
| O3—Cd1—O7 | 89.10 (8) | C11iii—C10—C9 | 120.3 (2) |
| N2—Cd1—O7 | 159.42 (8) | C11—C10—C9 | 120.3 (2) |
| N1—Cd1—O7 | 89.04 (8) | C12—C11—C10 | 120.4 (3) |
| O1—Cd1—O4 | 122.37 (9) | C12—C11—H11 | 119.8 |
| O3—Cd1—O4 | 55.12 (7) | C10—C11—H11 | 119.8 |
| N2—Cd1—O4 | 102.73 (8) | C11—C12—C13 | 120.3 (3) |
| N1—Cd1—O4 | 143.17 (8) | C11—C12—H12 | 119.9 |
| O7—Cd1—O4 | 95.13 (8) | C13—C12—H12 | 119.9 |
| O1—Cd1—O2 | 52.68 (10) | C12—C13—C12iii | 119.3 (4) |
| O3—Cd1—O2 | 136.90 (8) | C12—C13—C14 | 120.3 (2) |
| N2—Cd1—O2 | 78.69 (10) | C12iii—C13—C14 | 120.3 (2) |
| N1—Cd1—O2 | 126.21 (9) | O6iii—C14—O6 | 123.1 (4) |
| O7—Cd1—O2 | 113.27 (9) | O6iii—C14—C13 | 118.5 (2) |
| O4—Cd1—O2 | 85.37 (8) | O6—C14—C13 | 118.5 (2) |
| C15—N1—C19 | 118.5 (2) | N1—C15—C16 | 122.7 (3) |
| C15—N1—Cd1 | 125.38 (19) | N1—C15—H15 | 118.6 |
| C19—N1—Cd1 | 115.99 (18) | C16—C15—H15 | 118.6 |
| C24—N2—C20 | 118.1 (3) | C17—C16—C15 | 119.4 (3) |
| C24—N2—Cd1 | 125.7 (2) | C17—C16—H16 | 120.3 |
| C20—N2—Cd1 | 116.18 (19) | C15—C16—H16 | 120.3 |
| C1—O1—Cd1 | 99.2 (2) | C16—C17—C18 | 120.1 (3) |
| C1—O2—Cd1 | 84.7 (2) | C16—C17—H17 | 120.0 |
| C5—O3—Cd1 | 92.19 (18) | C18—C17—H17 | 120.0 |
| C5—O4—Cd1 | 91.35 (17) | C17—C18—C19 | 117.1 (3) |
| C9—O5—H5 | 109.5 | C17—C18—C25 | 123.7 (3) |
| C14—O6—H6 | 109.5 | C19—C18—C25 | 119.1 (3) |
| Cd1—O7—H7A | 110.4 | N1—C19—C18 | 122.2 (3) |
| Cd1—O7—H7B | 110.4 | N1—C19—C20 | 118.6 (2) |
| H7A—O7—H7B | 108.7 | C18—C19—C20 | 119.2 (3) |
| O2—C1—O1 | 122.9 (4) | N2—C20—C21 | 122.4 (3) |
| O2—C1—C2 | 119.2 (4) | N2—C20—C19 | 118.1 (3) |
| O1—C1—C2 | 117.8 (3) | C21—C20—C19 | 119.5 (3) |
| C4—C2—C3 | 119.7 (3) | C22—C21—C20 | 117.0 (3) |
| C4—C2—C1 | 120.7 (3) | C22—C21—C26 | 123.3 (3) |
| C3—C2—C1 | 119.6 (3) | C20—C21—C26 | 119.7 (3) |
| C2—C3—C4i | 120.3 (3) | C23—C22—C21 | 120.8 (4) |
| C2—C3—H3 | 119.9 | C23—C22—H22 | 119.6 |
| C4i—C3—H3 | 119.9 | C21—C22—H22 | 119.6 |
| C2—C4—C3i | 120.0 (3) | C22—C23—C24 | 118.6 (4) |
| C2—C4—H4 | 120.0 | C22—C23—H23 | 120.7 |
| C3i—C4—H4 | 120.0 | C24—C23—H23 | 120.7 |
| O4—C5—O3 | 121.2 (3) | N2—C24—C23 | 123.2 (3) |
| O4—C5—C6 | 120.2 (3) | N2—C24—H24 | 118.4 |
| O3—C5—C6 | 118.6 (3) | C23—C24—H24 | 118.4 |
| C8—C6—C7 | 118.2 (3) | C26—C25—C18 | 121.6 (3) |
| C8—C6—C5 | 121.1 (3) | C26—C25—H25 | 119.2 |
| C7—C6—C5 | 120.7 (3) | C18—C25—H25 | 119.2 |
| C8ii—C7—C6 | 121.4 (3) | C25—C26—C21 | 120.8 (3) |
| C8ii—C7—H7 | 119.3 | C25—C26—H26 | 119.6 |
| C6—C7—H7 | 119.3 | C21—C26—H26 | 119.6 |
| O1—Cd1—N1—C15 | 73.5 (3) | Cd1—O4—C5—C6 | 176.8 (2) |
| O3—Cd1—N1—C15 | −88.8 (3) | Cd1—O3—C5—O4 | 4.1 (3) |
| N2—Cd1—N1—C15 | 177.7 (3) | Cd1—O3—C5—C6 | −176.7 (2) |
| O7—Cd1—N1—C15 | 0.3 (3) | O4—C5—C6—C8 | −11.4 (4) |
| O4—Cd1—N1—C15 | −97.0 (3) | O3—C5—C6—C8 | 169.4 (3) |
| O2—Cd1—N1—C15 | 118.8 (2) | O4—C5—C6—C7 | 170.3 (3) |
| O1—Cd1—N1—C19 | −110.4 (2) | O3—C5—C6—C7 | −8.9 (4) |
| O3—Cd1—N1—C19 | 87.2 (2) | C8—C6—C7—C8ii | 0.3 (5) |
| N2—Cd1—N1—C19 | −6.22 (19) | C5—C6—C7—C8ii | 178.6 (3) |
| O7—Cd1—N1—C19 | 176.4 (2) | C7—C6—C8—C7ii | −0.3 (5) |
| O4—Cd1—N1—C19 | 79.1 (2) | C5—C6—C8—C7ii | −178.6 (3) |
| O2—Cd1—N1—C19 | −65.1 (2) | O5—C9—C10—C11iii | −178.8 (2) |
| O1—Cd1—N2—C24 | −88.3 (3) | O5iii—C9—C10—C11iii | 1.2 (2) |
| O3—Cd1—N2—C24 | 95.3 (3) | O5—C9—C10—C11 | 1.2 (2) |
| N1—Cd1—N2—C24 | −177.3 (3) | O5iii—C9—C10—C11 | −178.8 (2) |
| O7—Cd1—N2—C24 | −169.9 (3) | C11iii—C10—C11—C12 | −0.2 (2) |
| O4—Cd1—N2—C24 | 40.5 (3) | C9—C10—C11—C12 | 179.8 (2) |
| O2—Cd1—N2—C24 | −42.1 (3) | C10—C11—C12—C13 | 0.4 (5) |
| O1—Cd1—N2—C20 | 95.2 (2) | C11—C12—C13—C12iii | −0.2 (2) |
| O3—Cd1—N2—C20 | −81.1 (2) | C11—C12—C13—C14 | 179.8 (2) |
| N1—Cd1—N2—C20 | 6.3 (2) | C12—C13—C14—O6iii | −176.3 (2) |
| O7—Cd1—N2—C20 | 13.6 (4) | C12iii—C13—C14—O6iii | 3.7 (2) |
| O4—Cd1—N2—C20 | −136.0 (2) | C12—C13—C14—O6 | 3.7 (2) |
| O2—Cd1—N2—C20 | 141.5 (2) | C12iii—C13—C14—O6 | −176.3 (2) |
| O3—Cd1—O1—C1 | −133.0 (3) | C19—N1—C15—C16 | −1.1 (5) |
| N2—Cd1—O1—C1 | 58.9 (2) | Cd1—N1—C15—C16 | 174.8 (2) |
| N1—Cd1—O1—C1 | 129.8 (2) | N1—C15—C16—C17 | −0.1 (5) |
| O7—Cd1—O1—C1 | −142.3 (2) | C15—C16—C17—C18 | 1.1 (5) |
| O4—Cd1—O1—C1 | −56.9 (2) | C16—C17—C18—C19 | −1.0 (5) |
| O2—Cd1—O1—C1 | −4.0 (2) | C16—C17—C18—C25 | 179.3 (3) |
| O1—Cd1—O2—C1 | 4.0 (2) | C15—N1—C19—C18 | 1.3 (4) |
| O3—Cd1—O2—C1 | 163.68 (19) | Cd1—N1—C19—C18 | −175.1 (2) |
| N2—Cd1—O2—C1 | −114.5 (2) | C15—N1—C19—C20 | −177.9 (3) |
| N1—Cd1—O2—C1 | −59.1 (3) | Cd1—N1—C19—C20 | 5.7 (3) |
| O7—Cd1—O2—C1 | 47.9 (2) | C17—C18—C19—N1 | −0.2 (5) |
| O4—Cd1—O2—C1 | 141.5 (2) | C25—C18—C19—N1 | 179.5 (3) |
| O1—Cd1—O3—C5 | 85.7 (3) | C17—C18—C19—C20 | 179.0 (3) |
| N2—Cd1—O3—C5 | −105.85 (18) | C25—C18—C19—C20 | −1.3 (5) |
| N1—Cd1—O3—C5 | −176.28 (18) | C24—N2—C20—C21 | −1.6 (5) |
| O7—Cd1—O3—C5 | 94.65 (18) | Cd1—N2—C20—C21 | 175.1 (2) |
| O4—Cd1—O3—C5 | −2.24 (16) | C24—N2—C20—C19 | 177.4 (3) |
| O2—Cd1—O3—C5 | −29.5 (2) | Cd1—N2—C20—C19 | −5.9 (3) |
| O1—Cd1—O4—C5 | −156.55 (18) | N1—C19—C20—N2 | 0.1 (4) |
| O3—Cd1—O4—C5 | 2.24 (16) | C18—C19—C20—N2 | −179.1 (3) |
| N2—Cd1—O4—C5 | 86.65 (18) | N1—C19—C20—C21 | 179.2 (3) |
| N1—Cd1—O4—C5 | 12.2 (2) | C18—C19—C20—C21 | −0.1 (4) |
| O7—Cd1—O4—C5 | −83.06 (18) | N2—C20—C21—C22 | 1.2 (5) |
| O2—Cd1—O4—C5 | 163.94 (19) | C19—C20—C21—C22 | −177.8 (3) |
| Cd1—O2—C1—O1 | −7.0 (3) | N2—C20—C21—C26 | −179.9 (3) |
| Cd1—O2—C1—C2 | 169.4 (3) | C19—C20—C21—C26 | 1.1 (5) |
| Cd1—O1—C1—O2 | 8.0 (4) | C20—C21—C22—C23 | −0.1 (6) |
| Cd1—O1—C1—C2 | −168.4 (2) | C26—C21—C22—C23 | −179.0 (4) |
| O2—C1—C2—C4 | −16.4 (5) | C21—C22—C23—C24 | −0.5 (7) |
| O1—C1—C2—C4 | 160.2 (3) | C20—N2—C24—C23 | 0.9 (6) |
| O2—C1—C2—C3 | 165.2 (3) | Cd1—N2—C24—C23 | −175.4 (3) |
| O1—C1—C2—C3 | −18.3 (4) | C22—C23—C24—N2 | 0.1 (7) |
| C4—C2—C3—C4i | 0.5 (5) | C17—C18—C25—C26 | −178.6 (4) |
| C1—C2—C3—C4i | 179.0 (3) | C19—C18—C25—C26 | 1.7 (6) |
| C3—C2—C4—C3i | −0.5 (5) | C18—C25—C26—C21 | −0.6 (6) |
| C1—C2—C4—C3i | −179.0 (3) | C22—C21—C26—C25 | 178.0 (4) |
| Cd1—O4—C5—O3 | −4.0 (3) | C20—C21—C26—C25 | −0.8 (6) |
Symmetry codes: (i) −x+1/2, −y+3/2, −z+1; (ii) −x+1/2, −y+3/2, −z; (iii) −x, y, −z+1/2.
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O5—H5···O6iv | 0.82 | 1.92 | 2.728 (3) | 167 |
| O7—H7A···O2v | 0.85 | 1.88 | 2.665 (3) | 154 |
| O7—H7B···O4v | 0.85 | 2.28 | 3.019 (3) | 146 |
Symmetry codes: (iv) x, y+1, z; (v) −x+1/2, y+1/2, −z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: WM2211).
References
- Bruker (2001). SAINT and SMART Bruker AXS Inc., Madison, Wisconsin, USA.
- Liang, Y. C., Hong, M. C., Liu, J. C. & Cao, R. (2002). Inorg. Chim. Acta, 328, 152-158.
- McGarrah, J. E., Kim, Y.-J., Hissler, M. & Eisenberg, R. (2001). Inorg. Chem.40, 4510-4511. [DOI] [PubMed]
- Moulton, B., Lu, J., Hajndl, R., Hariharan, S. & Zaworotko, M. J. (2002). Angew. Chem. Int. Ed.41, 2821-2824. [DOI] [PubMed]
- Sheldrick, G. M. (2004). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Shi, X., Zhu, G. S., Fang, Q. R., Wu, G., Tian, G., Wang, R. W., Zhang, D. L., Xue, M. & Qiu, S. L. (2004). Eur. J. Inorg. Chem. pp. 185-191.
- Wang, X. L., Qin, C., Wang, E. B., Xu, L., Su, M. & Hu, C. W. (2004). Angew. Chem. Int. Ed.43, 5036-5040. [DOI] [PubMed]
- Wu, G., Wang, Y., Wang, X. F. Kawaguchi, H. & Sun, W. Y. (2007). J. Chem. Cryst.37, 199-205.
- Zheng, S. L., Yang, J. H., Yu, X. L., Chen, X. M. & Wong, W. T. (2004). Inorg. Chem.43, 830-836. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809000889/wm2211sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809000889/wm2211Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report







