Abstract
In the title compound, C12H14N2O2, the benzene ring and imidazole ring are almost coplanar, making a dihedral angle of 2.47 (14)°. Intermolecular O—H⋯N, N—H⋯O and C—H⋯O hydrogen bonds stabilize the crystal structure.
Related literature
For the use of the title compound as an intermediate in the preparation of telmisartan, see: Ries et al. (1993 ▶). For the biological activity of telmisartan, see: Engeli et al. (2000 ▶); Goossens et al. (2003 ▶). For reference structural data, see: Allen et al. (1987 ▶). For related literature, see: Kintscher et al. (2004 ▶).
Experimental
Crystal data
C12H14N2O2
M r = 218.25
Tetragonal,

a = 12.0548 (17) Å
c = 31.899 (6) Å
V = 4635.5 (13) Å3
Z = 16
Mo Kα radiation
μ = 0.09 mm−1
T = 293 (2) K
0.30 × 0.30 × 0.10 mm
Data collection
Enraf–Nonuis CAD-4 diffractometer
Absorption correction: ψ scan (North et al., 1968 ▶) T min = 0.975, T max = 0.991
4426 measured reflections
2105 independent reflections
1304 reflections with I > 2σ(I)
R int = 0.042
3 standard reflections every 200 reflections intensity decay: none
Refinement
R[F 2 > 2σ(F 2)] = 0.072
wR(F 2) = 0.195
S = 1.00
2105 reflections
133 parameters
2 restraints
H-atom parameters constrained
Δρmax = 0.62 e Å−3
Δρmin = −0.39 e Å−3
Data collection: CAD-4 Software (Enraf–Nonius,1989 ▶); cell refinement: CAD-4 Software; data reduction: XCAD4 (Harms & Wocadlo,1995 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808043420/sj2567sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808043420/sj2567Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C3—H3B⋯O2i | 0.97 | 2.56 | 3.399 (4) | 144 |
| C11—H11A⋯O2ii | 0.96 | 2.66 | 3.587 (4) | 163 |
| O2—H2C⋯N1iii | 0.82 | 1.80 | 2.562 (3) | 153 |
| N2—H2D⋯O1iv | 0.86 | 1.83 | 2.673 (3) | 165 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv) 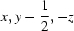 .
.
Acknowledgments
The authors thank the Center of Testing and Analysis, Nanjing University, for support.
supplementary crystallographic information
Comment
The title compound (I), Fig. 1, 4-methyl-2-n-propyl-1H-benzimidazole-6-carboxylic acid is an important intermediate in the preparation of the angiotensin II receptor blocker telmisartan (Ries et al.,1993). Telmisartan is used as a therapeutic tool for metabolic problems, including visceral obesity (Engeli et al., 2000; Goossens et al., 2003). Bond lengths in the compound are within normal ranges (Allen et al., 1987) and the benzene and imidazole rings are coplanar with a dihedral angle of 2.47 (14)° between them.
In the crystal structure, intermolecular O—H···N, N—H···O and C—H···O hydrogen bonds, Table 1, link the molecules, Fig. 2, and stabilise the crystal packing.
Experimental
The title compound was prepared from methyl 4-(butyrylamino)-5-methyl -3-aminobenzoate (13 g 52 mmol) in xylene (60 mL) and hydrochloric acid (130 mL). The mixture was refluxed for 3 h at 423 K, concentrated under reduced pressure then 80mL of methanol and 110 mL 15% sodium hydroxide were added. This solution was further , refluxed for 5 h at 383 K. The pH was adjusted to below 7 and the product filtered. Recrystallization of product from a mixture of ethanol/water (5:1) afforded the white powder. Crystals were obtained by dissolving the product (1.0 g) in acetone (10 ml) and evaporating slowly at room temperature for about 30 d.
Refinement
H All atoms were positioned geometrically, with N—H = 0.86 Å (for NH) and C—H = 0.93, 0.97 and 0.96 Å for aromatic, methine and methyl H, and constrained to ride on their parent atoms, with Uiso(H) = xUeq(C,N), where x = 1.5 for methyl H, and x = 1.2 for all other H atoms.
Figures
Fig. 1.
The molecular structure of the title molecule, with the atom-numbering scheme. Displacement ellipsoids are drawn at the 30% probability level.
Fig. 2.
Crystal packing of the title compound with hydrogen bonds drawn as dashed lines.
Crystal data
| C12H14N2O2 | Dx = 1.251 Mg m−3 |
| Mr = 218.25 | Mo Kα radiation, λ = 0.71073 Å |
| Tetragonal, I41/a | Cell parameters from 25 reflections |
| Hall symbol: -I 4ad | θ = 10–13° |
| a = 12.0548 (17) Å | µ = 0.09 mm−1 |
| c = 31.899 (6) Å | T = 293 K |
| V = 4635.5 (13) Å3 | Block, colorless |
| Z = 16 | 0.30 × 0.30 × 0.10 mm |
| F(000) = 1856 |
Data collection
| Enraf–Nonuis CAD-4 diffractometer | 1304 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.042 |
| graphite | θmax = 25.2°, θmin = 1.8° |
| ω/2θ scans | h = −9→10 |
| Absorption correction: ψ scan (North et al., 1968) | k = 0→14 |
| Tmin = 0.975, Tmax = 0.991 | l = 0→38 |
| 4426 measured reflections | 3 standard reflections every 200 reflections |
| 2105 independent reflections | intensity decay: none |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.072 | H-atom parameters constrained |
| wR(F2) = 0.195 | w = 1/[σ2(Fo2) + (0.1P)2 + 5P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.00 | (Δ/σ)max < 0.001 |
| 2105 reflections | Δρmax = 0.62 e Å−3 |
| 133 parameters | Δρmin = −0.39 e Å−3 |
| 2 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008) |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.039 (3) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.7815 (2) | 0.4947 (2) | 0.07388 (7) | 0.0591 (8) | |
| O2 | 0.7256 (2) | 0.32125 (18) | 0.07006 (6) | 0.0471 (7) | |
| H2C | 0.7077 | 0.3341 | 0.0944 | 0.071* | |
| N1 | 0.8807 (2) | 0.3584 (2) | −0.11544 (7) | 0.0376 (7) | |
| N2 | 0.8170 (2) | 0.2123 (2) | −0.08282 (7) | 0.0420 (7) | |
| H2D | 0.7940 | 0.1458 | −0.0785 | 0.050* | |
| C1 | 0.6545 (5) | 0.2486 (5) | −0.18175 (17) | 0.104 | |
| H1B | 0.5891 | 0.2171 | −0.1941 | 0.155* | |
| H1C | 0.6843 | 0.3042 | −0.2001 | 0.155* | |
| H1D | 0.6358 | 0.2818 | −0.1553 | 0.155* | |
| C2 | 0.7407 (4) | 0.1577 (5) | −0.17491 (18) | 0.100 | |
| H2A | 0.7097 | 0.1043 | −0.1554 | 0.120* | |
| H2B | 0.7513 | 0.1195 | −0.2014 | 0.120* | |
| C3 | 0.8501 (3) | 0.1904 (3) | −0.15925 (10) | 0.0561 (10) | |
| H3A | 0.8860 | 0.2355 | −0.1805 | 0.067* | |
| H3B | 0.8945 | 0.1239 | −0.1557 | 0.067* | |
| C4 | 0.8509 (3) | 0.2525 (3) | −0.11926 (9) | 0.0402 (8) | |
| C5 | 0.8656 (2) | 0.3874 (3) | −0.07355 (9) | 0.0324 (7) | |
| C6 | 0.8820 (2) | 0.4885 (2) | −0.05283 (9) | 0.0341 (7) | |
| C7 | 0.8539 (2) | 0.4896 (3) | −0.01107 (9) | 0.0351 (7) | |
| H7A | 0.8653 | 0.5543 | 0.0042 | 0.042* | |
| C8 | 0.8085 (2) | 0.3966 (2) | 0.00960 (9) | 0.0315 (7) | |
| C9 | 0.7938 (3) | 0.2969 (2) | −0.01128 (9) | 0.0353 (8) | |
| H9A | 0.7645 | 0.2350 | 0.0021 | 0.042* | |
| C10 | 0.8246 (3) | 0.2941 (2) | −0.05289 (9) | 0.0325 (7) | |
| C11 | 0.9269 (3) | 0.5876 (3) | −0.07572 (11) | 0.0523 (10) | |
| H11A | 0.9410 | 0.5681 | −0.1044 | 0.078* | |
| H11B | 0.9947 | 0.6112 | −0.0627 | 0.078* | |
| H11C | 0.8737 | 0.6467 | −0.0747 | 0.078* | |
| C12 | 0.7704 (3) | 0.4069 (2) | 0.05433 (9) | 0.0352 (7) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.097 (2) | 0.0411 (14) | 0.0391 (14) | −0.0157 (14) | 0.0171 (13) | −0.0130 (11) |
| O2 | 0.0713 (17) | 0.0394 (13) | 0.0306 (12) | −0.0073 (12) | 0.0238 (11) | −0.0056 (10) |
| N1 | 0.0401 (16) | 0.0475 (17) | 0.0250 (13) | 0.0030 (13) | 0.0022 (11) | 0.0055 (12) |
| N2 | 0.0599 (19) | 0.0366 (15) | 0.0295 (13) | −0.0022 (13) | 0.0098 (13) | 0.0006 (12) |
| C1 | 0.104 | 0.104 | 0.104 | 0.000 | 0.000 | 0.000 |
| C2 | 0.100 | 0.100 | 0.100 | 0.000 | 0.000 | 0.000 |
| C3 | 0.062 (2) | 0.079 (3) | 0.0275 (18) | 0.006 (2) | 0.0032 (16) | −0.0106 (17) |
| C4 | 0.0392 (19) | 0.056 (2) | 0.0250 (16) | 0.0079 (16) | 0.0066 (14) | 0.0000 (15) |
| C5 | 0.0309 (16) | 0.0392 (18) | 0.0271 (15) | 0.0063 (13) | 0.0052 (13) | 0.0077 (13) |
| C6 | 0.0356 (17) | 0.0303 (17) | 0.0365 (17) | −0.0015 (13) | 0.0005 (14) | 0.0093 (13) |
| C7 | 0.0379 (18) | 0.0330 (17) | 0.0345 (16) | −0.0008 (13) | 0.0032 (13) | −0.0027 (13) |
| C8 | 0.0410 (18) | 0.0273 (16) | 0.0261 (15) | 0.0038 (13) | 0.0033 (13) | 0.0040 (12) |
| C9 | 0.0472 (19) | 0.0318 (17) | 0.0269 (16) | −0.0020 (14) | 0.0091 (13) | 0.0032 (13) |
| C10 | 0.0454 (19) | 0.0293 (16) | 0.0229 (14) | 0.0015 (13) | 0.0046 (13) | 0.0015 (12) |
| C11 | 0.064 (2) | 0.041 (2) | 0.052 (2) | −0.0089 (17) | 0.0104 (18) | 0.0146 (17) |
| C12 | 0.0430 (19) | 0.0321 (17) | 0.0305 (16) | 0.0009 (14) | 0.0040 (14) | −0.0042 (14) |
Geometric parameters (Å, °)
| O1—C12 | 1.237 (4) | C3—H3A | 0.9700 |
| O2—C12 | 1.268 (3) | C3—H3B | 0.9700 |
| O2—H2C | 0.8200 | C5—C10 | 1.394 (4) |
| N1—C4 | 1.332 (4) | C5—C6 | 1.400 (4) |
| N1—C5 | 1.393 (4) | C6—C7 | 1.374 (4) |
| N2—C4 | 1.324 (4) | C6—C11 | 1.501 (4) |
| N2—C10 | 1.376 (4) | C7—C8 | 1.411 (4) |
| N2—H2D | 0.8600 | C7—H7A | 0.9300 |
| C1—C2 | 1.526 (6) | C8—C9 | 1.385 (4) |
| C1—H1B | 0.9600 | C8—C12 | 1.504 (4) |
| C1—H1C | 0.9600 | C9—C10 | 1.379 (4) |
| C1—H1D | 0.9600 | C9—H9A | 0.9300 |
| C2—C3 | 1.465 (6) | C11—H11A | 0.9600 |
| C2—H2A | 0.9700 | C11—H11B | 0.9600 |
| C2—H2B | 0.9700 | C11—H11C | 0.9600 |
| C3—C4 | 1.479 (4) | ||
| C12—O2—H2C | 109.5 | N1—C5—C6 | 130.7 (3) |
| C4—N1—C5 | 107.0 (2) | C10—C5—C6 | 121.9 (3) |
| C4—N2—C10 | 109.1 (3) | C7—C6—C5 | 115.5 (3) |
| C4—N2—H2D | 125.5 | C7—C6—C11 | 123.5 (3) |
| C10—N2—H2D | 125.5 | C5—C6—C11 | 120.9 (3) |
| C2—C1—H1B | 109.5 | C6—C7—C8 | 122.7 (3) |
| C2—C1—H1C | 109.5 | C6—C7—H7A | 118.6 |
| H1B—C1—H1C | 109.5 | C8—C7—H7A | 118.6 |
| C2—C1—H1D | 109.5 | C9—C8—C7 | 120.9 (3) |
| H1B—C1—H1D | 109.5 | C9—C8—C12 | 119.2 (3) |
| H1C—C1—H1D | 109.5 | C7—C8—C12 | 119.8 (3) |
| C3—C2—C1 | 118.0 (5) | C10—C9—C8 | 116.7 (3) |
| C3—C2—H2A | 107.8 | C10—C9—H9A | 121.6 |
| C1—C2—H2A | 107.8 | C8—C9—H9A | 121.6 |
| C3—C2—H2B | 107.8 | N2—C10—C9 | 131.9 (3) |
| C1—C2—H2B | 107.8 | N2—C10—C5 | 105.9 (2) |
| H2A—C2—H2B | 107.2 | C9—C10—C5 | 122.1 (3) |
| C2—C3—C4 | 115.8 (4) | C6—C11—H11A | 109.5 |
| C2—C3—H3A | 108.3 | C6—C11—H11B | 109.5 |
| C4—C3—H3A | 108.3 | H11A—C11—H11B | 109.5 |
| C2—C3—H3B | 108.3 | C6—C11—H11C | 109.5 |
| C4—C3—H3B | 108.3 | H11A—C11—H11C | 109.5 |
| H3A—C3—H3B | 107.4 | H11B—C11—H11C | 109.5 |
| N2—C4—N1 | 110.7 (3) | O1—C12—O2 | 122.9 (3) |
| N2—C4—C3 | 124.8 (3) | O1—C12—C8 | 121.1 (3) |
| N1—C4—C3 | 124.5 (3) | O2—C12—C8 | 116.0 (3) |
| N1—C5—C10 | 107.3 (3) | ||
| C1—C2—C3—C4 | 56.3 (6) | C6—C7—C8—C12 | −174.6 (3) |
| C10—N2—C4—N1 | −0.2 (4) | C7—C8—C9—C10 | −0.6 (5) |
| C10—N2—C4—C3 | −177.3 (3) | C12—C8—C9—C10 | 176.5 (3) |
| C5—N1—C4—N2 | 0.5 (4) | C4—N2—C10—C9 | 175.5 (3) |
| C5—N1—C4—C3 | 177.5 (3) | C4—N2—C10—C5 | −0.1 (4) |
| C2—C3—C4—N2 | 63.9 (5) | C8—C9—C10—N2 | −176.8 (3) |
| C2—C3—C4—N1 | −112.7 (4) | C8—C9—C10—C5 | −1.8 (5) |
| C4—N1—C5—C10 | −0.5 (3) | N1—C5—C10—N2 | 0.4 (3) |
| C4—N1—C5—C6 | −178.6 (3) | C6—C5—C10—N2 | 178.6 (3) |
| N1—C5—C6—C7 | 177.2 (3) | N1—C5—C10—C9 | −175.7 (3) |
| C10—C5—C6—C7 | −0.6 (4) | C6—C5—C10—C9 | 2.5 (5) |
| N1—C5—C6—C11 | −2.5 (5) | C9—C8—C12—O1 | −179.5 (3) |
| C10—C5—C6—C11 | 179.7 (3) | C7—C8—C12—O1 | −2.4 (5) |
| C5—C6—C7—C8 | −1.8 (4) | C9—C8—C12—O2 | −0.4 (4) |
| C11—C6—C7—C8 | 177.9 (3) | C7—C8—C12—O2 | 176.7 (3) |
| C6—C7—C8—C9 | 2.4 (5) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C3—H3B···O2i | 0.97 | 2.56 | 3.399 (4) | 144 |
| C11—H11A···O2ii | 0.96 | 2.66 | 3.587 (4) | 163 |
| O2—H2C···N1iii | 0.82 | 1.80 | 2.562 (3) | 153 |
| N2—H2D···O1iv | 0.86 | 1.83 | 2.673 (3) | 165 |
Symmetry codes: (i) y+3/4, −x+3/4, z−1/4; (ii) −y+5/4, x−1/4, z−1/4; (iii) y+1/4, −x+5/4, z+1/4; (iv) x, y−1/2, −z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: SJ2567).
References
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
- Engeli, S., Negrel, R. & Sharma, A. M. (2000). Hypertension, 35, 1270–1277. [DOI] [PubMed]
- Enraf–Nonius (1989). CAD-4 Software Enraf–Nonius, Delft, The Netherlands.
- Goossens, G. H., Blaak, E. E. & van Baak, M. A. (2003). Obes. Rev.4, 43–55. [DOI] [PubMed]
- Harms, K. & Wocadlo, S. (1995). XCAD4 University of Marburg, Germany.
- Kintscher, U., Lyon, C. J. & Law, R. E. (2004). Front. Biosci 9, 359–369. [DOI] [PubMed]
- North, A. C. T., Phillips, D. C. & Mathews, F. S. (1968). Acta Cryst. A24, 351–359.
- Ries, U. J., Mihm, G. & Narr, B. (1993). J. Med. Chem.36, 4040–4051. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808043420/sj2567sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808043420/sj2567Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




