Abstract
In the structure of the title compound, C17H13BrN4O2S, the dihedral angle between the two benzene rings is 38.5 (1)°; the angle between the 4-bromobenzene and thiadiazole rings is 1.3 (1)°. The conformations of the N—H and C=O bonds are anti with respect to each other. The structure displays intermolecular N—H⋯O and C—H⋯O hydrogen bonding, with both interactions leading to inversion dimers.
Related literature
For 1,3,4-thiadiazole scaffold compounds and their biological activity, see: Tu et al. (2008 ▶). For the synthesis, see: Foroumadi et al. (1999 ▶); Levy & Palmer (1942 ▶); Song et al. (1992 ▶). For related structures, see: Gowda et al. (2008 ▶); Li, Huang et al. (2008 ▶); Li, Li et al. (2008 ▶).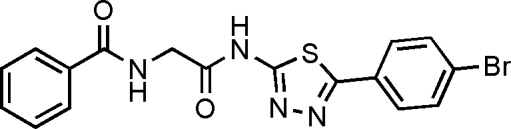
Experimental
Crystal data
C17H13BrN4O2S
M r = 417.28
Triclinic,
a = 4.020 (4) Å
b = 13.706 (9) Å
c = 16.210 (5) Å
α = 113.334 (17)°
β = 94.018 (19)°
γ = 92.78 (2)°
V = 815.2 (10) Å3
Z = 2
Mo Kα radiation
μ = 2.67 mm−1
T = 298 (2) K
0.54 × 0.17 × 0.04 mm
Data collection
Bruker X8 APEXII diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2008 ▶) T min = 0.591, T max = 0.914
4633 measured reflections
2592 independent reflections
1097 reflections with I > 2σ(I)
R int = 0.082
Refinement
R[F 2 > 2σ(F 2)] = 0.109
wR(F 2) = 0.281
S = 0.82
2592 reflections
227 parameters
6 restraints
H-atom parameters constrained
Δρmax = 1.67 e Å−3
Δρmin = −1.40 e Å−3
Data collection: APEX2 (Bruker, 2004 ▶); cell refinement: SAINT (Bruker, 2004 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: APEX2 (Bruker, 2004 ▶); software used to prepare material for publication: APEX2 (Bruker, 2004 ▶) and publCIF (Westrip, 2009 ▶).
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S160053680900124X/wn2304sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053680900124X/wn2304Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C16—H16A⋯O2i | 0.93 | 2.49 | 3.400 (5) | 168 |
| N2—H2A⋯O1ii | 0.86 | 1.99 | 2.835 (5) | 167 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Acknowledgments
The work was supported by the Science and Technology Research Project of JiangXi Provincial Educational Department (No. GJJ09076), the Science and Technology Planning Project of JiangXi Provincial Health Department (No. 20082015) and the Natural Science Foundation of JiangXi Province, China.
supplementary crystallographic information
Comment
In our previous work, 1,3,4-thiadiazole scaffold compounds and their biological activity have been studied (Tu et al., 2008). In view of the importance of these organic materials, the title compound (Fig. 1) was synthesized (Foroumadi et al., 1999; Levy & Palmer 1942; Song et al., 1992) and its crystal structure is reported here.
In the structure of the title compound, C17H13BrN4O2S, the dihedral angle between the p-bromobenzene and thiadiazole rings is 1.3 (1)°; the angle between the two benzene rings is 38.5 (1)°. The conformations of the N—H and C═O bonds are anti with respect to each other. Bond lengths and angles are in normal ranges and comparable to those in related structures (Gowda et al., 2008; Li, Huang et al., 2008; Li, Li et al., 2008). In the crystal structure, molecules are linked through intermolecular C—H···O and N—H···O hydrogen bonds, forming a three-dimensional network (Table 1, Figure 2).
Experimental
N,N-Dicyclohexylcarbodiimide (5.7 mmol) was added to a cooled solution of N-benzoylglycine (5.6 mmol) and N-hydroxysuccinimide (5.6 mmol) in freshly distilled dioxane (30 ml). The reaction mixture was stirred overnight at room temperature. The insoluble material was filtered off and washed with cold dioxane. 2-Amino-5-(4-bromophenyl)-1,3,4-thiadiazole (5.5 mmol) was added to the filtrate and the reaction mixture was stirred for 48 h at room temperature. The solvent was removed under reduced pressure. The residue was dissolved in EtOAc and the insoluble material was filtered off. The filtrate was washed successively with saturated Na2CO3 solution (20 ml, x 3), water (20 ml, x 1), 0.1 M HCl (20 ml, x 3) and water (20 ml, x 1). The organic layer evaporated in vacuo, and the residue was recrystallized from methanol. Colorless block-shaped single crystals of the title compound suitable for X-ray diffraction analysis precipitated after several days.Yield: 37.0%; mp: 271–273°C.
Refinement
H atoms were positioned geometrically and refined using a riding model; Csp2—H = 0.93 Å, Csp3—H = 0.97 Å and N—H = 0.86 Å; Uĩso(H) = 1.2 Ueq(C,N). We made several attempts to obtain better quality data for this structure. However, due to poor crystal quality and possible disorder, the R and wR values are high. The maximum residual electron density occurs 1.23 Å from atom Br1, and the minimum residual electron density is located 1.29 Å from atom Br1.
Figures
Fig. 1.
Molecular structure of the title compound. Displacement ellipsoids are drawn at the 50% probability level. Hydrogen atoms are shown as spheres of arbitrary radius.
Fig. 2.
The crystal packing of the title compound, viewed along the a axis with hydrogen bonds drawn as dashed lines.
Crystal data
| C17H13BrN4O2S | Z = 2 |
| Mr = 417.28 | F(000) = 420.0 |
| Triclinic, P1 | Dx = 1.700 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 4.020 (4) Å | Cell parameters from 1179 reflections |
| b = 13.706 (9) Å | θ = 2.6–23.7° |
| c = 16.210 (5) Å | µ = 2.67 mm−1 |
| α = 113.334 (17)° | T = 298 K |
| β = 94.018 (19)° | Block, colourless |
| γ = 92.78 (2)° | 0.54 × 0.17 × 0.04 mm |
| V = 815.2 (10) Å3 |
Data collection
| Bruker X8 APEXII diffractometer | 2592 independent reflections |
| Radiation source: fine-focus sealed tube | 1097 reflections with I > 2σ(I) |
| graphite | Rint = 0.082 |
| φ and ω scans | θmax = 25.0°, θmin = 2.5° |
| Absorption correction: multi-scan (SADABS; Bruker, 2008) | h = −4→4 |
| Tmin = 0.591, Tmax = 0.914 | k = −16→16 |
| 4633 measured reflections | l = −19→19 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.109 | H-atom parameters constrained |
| wR(F2) = 0.281 | w = 1/[σ2(Fo2) + (0.1951P)2] where P = (Fo2 + 2Fc2)/3 |
| S = 0.82 | (Δ/σ)max = 0.019 |
| 2592 reflections | Δρmax = 1.67 e Å−3 |
| 227 parameters | Δρmin = −1.39 e Å−3 |
| 6 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.022 (2) |
Special details
| Experimental. 1H-NMR (DMSO-d6): δ 4.24–4.25(d, J=5.08 Hz, 2H), 7.49–7.57 (m, 3H),7.73–7.75 (d, J=8.04 Hz, 2H), 7.89–7.91(t, J=3.60 Hz, 4H), 9.01 (s, 1H),12.91 (s, 1H). ESI-MS: m/z [M+H]+ 417.3. |
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Br1 | 0.19570 (14) | 0.16142 (4) | 0.64242 (3) | 0.07526 (19) | |
| S1 | 0.3284 (3) | 0.41677 (8) | 0.33059 (7) | 0.0550 (4) | |
| O1 | 0.2257 (8) | 0.6333 (2) | −0.00717 (17) | 0.0636 (9) | |
| O2 | 0.2253 (9) | 0.5689 (2) | 0.25867 (18) | 0.0687 (8) | |
| N1 | 0.2711 (9) | 0.6635 (2) | 0.1388 (2) | 0.0551 (12) | |
| H1A | 0.2352 | 0.7034 | 0.1931 | 0.066* | |
| N3 | 0.5949 (10) | 0.2787 (3) | 0.1998 (2) | 0.0621 (13) | |
| N4 | 0.5645 (10) | 0.2351 (2) | 0.2632 (2) | 0.0597 (8) | |
| N2 | 0.4834 (9) | 0.4276 (2) | 0.1728 (2) | 0.0539 (8) | |
| H2A | 0.5726 | 0.3992 | 0.1229 | 0.065* | |
| C4 | −0.2078 (13) | 0.9136 (3) | 0.0368 (3) | 0.0711 (18) | |
| H4B | −0.3108 | 0.9254 | −0.0112 | 0.085* | |
| C5 | −0.0907 (12) | 0.8176 (3) | 0.0241 (3) | 0.0610 (16) | |
| H5A | −0.1231 | 0.7628 | −0.0332 | 0.073* | |
| C10 | 0.4756 (11) | 0.3703 (3) | 0.2263 (3) | 0.0512 (11) | |
| C16 | 0.1797 (12) | 0.3039 (3) | 0.5556 (3) | 0.0632 (17) | |
| H16A | 0.0838 | 0.3486 | 0.6061 | 0.076* | |
| C15 | 0.2691 (13) | 0.2067 (3) | 0.5481 (3) | 0.0603 (10) | |
| C12 | 0.3711 (10) | 0.2674 (3) | 0.4098 (2) | 0.0471 (9) | |
| C14 | 0.4058 (11) | 0.1370 (3) | 0.4729 (3) | 0.0598 (11) | |
| H14A | 0.4596 | 0.0701 | 0.4688 | 0.072* | |
| C17 | 0.2354 (12) | 0.3345 (3) | 0.4857 (3) | 0.0587 (11) | |
| H17A | 0.1807 | 0.4014 | 0.4900 | 0.070* | |
| C8 | 0.4114 (12) | 0.5638 (3) | 0.1200 (3) | 0.0560 (15) | |
| H8A | 0.6486 | 0.5723 | 0.1144 | 0.067* | |
| H8B | 0.3068 | 0.5116 | 0.0627 | 0.067* | |
| C9 | 0.3647 (11) | 0.5237 (3) | 0.1913 (2) | 0.0475 (10) | |
| C11 | 0.4297 (11) | 0.2963 (3) | 0.3341 (2) | 0.0483 (10) | |
| C13 | 0.4598 (12) | 0.1698 (3) | 0.4041 (3) | 0.0601 (16) | |
| H13A | 0.5571 | 0.1253 | 0.3537 | 0.072* | |
| C7 | 0.1970 (11) | 0.6938 (3) | 0.0722 (3) | 0.0521 (13) | |
| C6 | 0.0730 (11) | 0.7991 (3) | 0.0926 (2) | 0.0489 (14) | |
| C2 | −0.0154 (12) | 0.9776 (3) | 0.1919 (3) | 0.0601 (16) | |
| H2B | 0.0074 | 1.0320 | 0.2494 | 0.072* | |
| C3 | −0.1698 (12) | 0.9930 (3) | 0.1230 (3) | 0.0643 (17) | |
| H3B | −0.2533 | 1.0585 | 0.1332 | 0.077* | |
| C1 | 0.1113 (12) | 0.8805 (3) | 0.1780 (3) | 0.0555 (15) | |
| H1B | 0.2216 | 0.8701 | 0.2259 | 0.067* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Br1 | 0.0941 (4) | 0.0888 (3) | 0.0604 (2) | 0.0226 (3) | 0.0361 (2) | 0.04206 (19) |
| S1 | 0.0804 (8) | 0.0441 (5) | 0.0406 (5) | 0.0193 (5) | 0.0286 (5) | 0.0118 (4) |
| O1 | 0.106 (2) | 0.0501 (14) | 0.0414 (6) | 0.0279 (14) | 0.0433 (10) | 0.0174 (7) |
| O2 | 0.1043 (12) | 0.0616 (16) | 0.0469 (7) | 0.0256 (13) | 0.0439 (6) | 0.0207 (9) |
| N1 | 0.082 (3) | 0.0452 (17) | 0.0435 (16) | 0.0255 (16) | 0.0332 (18) | 0.0165 (13) |
| N3 | 0.099 (3) | 0.0488 (17) | 0.0448 (17) | 0.0288 (18) | 0.0270 (19) | 0.0193 (13) |
| N4 | 0.1059 (15) | 0.0386 (15) | 0.0399 (8) | 0.0252 (12) | 0.0297 (7) | 0.0154 (8) |
| N2 | 0.075 (2) | 0.0474 (5) | 0.0474 (5) | 0.0157 (15) | 0.0353 (16) | 0.0212 (3) |
| C4 | 0.097 (4) | 0.049 (2) | 0.070 (3) | 0.025 (2) | 0.022 (3) | 0.022 (2) |
| C5 | 0.080 (3) | 0.042 (2) | 0.053 (2) | 0.012 (2) | 0.009 (2) | 0.0102 (18) |
| C10 | 0.073 (3) | 0.0390 (7) | 0.0442 (7) | 0.0091 (19) | 0.024 (2) | 0.0156 (4) |
| C16 | 0.080 (3) | 0.054 (2) | 0.051 (2) | 0.012 (2) | 0.034 (2) | 0.0102 (19) |
| C15 | 0.0825 (16) | 0.0564 (19) | 0.0442 (9) | −0.0018 (12) | 0.0194 (8) | 0.0211 (8) |
| C12 | 0.0546 (15) | 0.0498 (18) | 0.0351 (9) | 0.0036 (12) | 0.0172 (7) | 0.0131 (8) |
| C14 | 0.066 (2) | 0.066 (2) | 0.0600 (12) | 0.0136 (14) | 0.0282 (10) | 0.0337 (10) |
| C17 | 0.078 (2) | 0.050 (2) | 0.0448 (11) | 0.0131 (15) | 0.0261 (10) | 0.0109 (10) |
| C8 | 0.064 (3) | 0.061 (2) | 0.053 (2) | 0.026 (2) | 0.035 (2) | 0.0269 (16) |
| C9 | 0.0644 (16) | 0.041 (2) | 0.0353 (8) | 0.0082 (16) | 0.0210 (7) | 0.0104 (12) |
| C11 | 0.0753 (18) | 0.0407 (18) | 0.0336 (10) | 0.0106 (14) | 0.0223 (9) | 0.0165 (9) |
| C13 | 0.094 (3) | 0.042 (2) | 0.046 (2) | 0.010 (2) | 0.030 (2) | 0.0159 (16) |
| C7 | 0.071 (3) | 0.047 (2) | 0.0344 (7) | 0.0104 (19) | 0.0185 (14) | 0.0105 (9) |
| C6 | 0.061 (3) | 0.047 (2) | 0.0339 (18) | 0.0118 (19) | 0.030 (2) | 0.0067 (16) |
| C2 | 0.078 (3) | 0.043 (2) | 0.050 (2) | 0.009 (2) | 0.019 (2) | 0.0062 (18) |
| C3 | 0.080 (3) | 0.052 (2) | 0.065 (3) | 0.031 (2) | 0.025 (3) | 0.0211 (19) |
| C1 | 0.071 (3) | 0.047 (2) | 0.046 (2) | 0.016 (2) | 0.023 (2) | 0.0120 (17) |
Geometric parameters (Å, °)
| Br1—C15 | 1.898 (5) | C16—C17 | 1.382 (7) |
| S1—C10 | 1.714 (4) | C16—H16A | 0.9300 |
| S1—C11 | 1.741 (5) | C15—C14 | 1.387 (6) |
| O1—C7 | 1.242 (4) | C12—C13 | 1.370 (6) |
| O2—C9 | 1.214 (5) | C12—C17 | 1.380 (5) |
| N1—C7 | 1.323 (6) | C12—C11 | 1.461 (6) |
| N1—C8 | 1.430 (5) | C14—C13 | 1.383 (7) |
| N1—H1A | 0.8600 | C14—H14A | 0.9300 |
| N3—C10 | 1.285 (5) | C17—H17A | 0.9300 |
| N3—N4 | 1.387 (6) | C8—C9 | 1.483 (7) |
| N4—C11 | 1.299 (5) | C8—H8A | 0.9700 |
| N2—C9 | 1.349 (5) | C8—H8B | 0.9700 |
| N2—C10 | 1.381 (6) | C13—H13A | 0.9300 |
| N2—H2A | 0.8600 | C7—C6 | 1.469 (6) |
| C4—C5 | 1.361 (7) | C6—C1 | 1.383 (5) |
| C4—C3 | 1.382 (6) | C2—C3 | 1.335 (7) |
| C4—H4B | 0.9300 | C2—C1 | 1.388 (6) |
| C5—C6 | 1.367 (7) | C2—H2B | 0.9300 |
| C5—H5A | 0.9300 | C3—H3B | 0.9300 |
| C16—C15 | 1.359 (7) | C1—H1B | 0.9300 |
| C10—S1—C11 | 86.0 (2) | C16—C17—H17A | 119.5 |
| C7—N1—C8 | 119.6 (3) | N1—C8—C9 | 112.3 (3) |
| C7—N1—H1A | 120.2 | N1—C8—H8A | 109.1 |
| C8—N1—H1A | 120.2 | C9—C8—H8A | 109.1 |
| C10—N3—N4 | 110.7 (4) | N1—C8—H8B | 109.1 |
| C11—N4—N3 | 113.3 (3) | C9—C8—H8B | 109.1 |
| C9—N2—C10 | 126.3 (3) | H8A—C8—H8B | 107.9 |
| C9—N2—H2A | 116.8 | O2—C9—N2 | 121.9 (4) |
| C10—N2—H2A | 116.8 | O2—C9—C8 | 125.0 (4) |
| C5—C4—C3 | 118.3 (5) | N2—C9—C8 | 113.1 (3) |
| C5—C4—H4B | 120.9 | N4—C11—C12 | 123.3 (4) |
| C3—C4—H4B | 120.9 | N4—C11—S1 | 113.4 (3) |
| C4—C5—C6 | 122.2 (4) | C12—C11—S1 | 123.3 (3) |
| C4—C5—H5A | 118.9 | C12—C13—C14 | 120.4 (4) |
| C6—C5—H5A | 118.9 | C12—C13—H13A | 119.8 |
| N3—C10—N2 | 119.8 (4) | C14—C13—H13A | 119.8 |
| N3—C10—S1 | 116.5 (4) | O1—C7—N1 | 120.5 (4) |
| N2—C10—S1 | 123.6 (3) | O1—C7—C6 | 120.0 (4) |
| C15—C16—C17 | 118.0 (4) | N1—C7—C6 | 119.4 (3) |
| C15—C16—H16A | 121.0 | C5—C6—C1 | 118.3 (4) |
| C17—C16—H16A | 121.0 | C5—C6—C7 | 118.5 (3) |
| C16—C15—C14 | 122.6 (5) | C1—C6—C7 | 123.2 (4) |
| C16—C15—Br1 | 119.4 (3) | C3—C2—C1 | 120.2 (4) |
| C14—C15—Br1 | 118.0 (4) | C3—C2—H2B | 119.9 |
| C13—C12—C17 | 119.7 (4) | C1—C2—H2B | 119.9 |
| C13—C12—C11 | 117.6 (3) | C2—C3—C4 | 121.2 (4) |
| C17—C12—C11 | 122.6 (4) | C2—C3—H3B | 119.4 |
| C13—C14—C15 | 118.2 (4) | C4—C3—H3B | 119.4 |
| C13—C14—H14A | 120.9 | C6—C1—C2 | 119.8 (4) |
| C15—C14—H14A | 120.9 | C6—C1—H1B | 120.1 |
| C12—C17—C16 | 121.0 (4) | C2—C1—H1B | 120.1 |
| C12—C17—H17A | 119.5 | ||
| C10—N3—N4—C11 | −1.0 (5) | C13—C12—C11—N4 | −0.9 (6) |
| C3—C4—C5—C6 | −2.5 (8) | C17—C12—C11—N4 | 178.3 (4) |
| N4—N3—C10—N2 | 179.7 (4) | C13—C12—C11—S1 | 179.6 (3) |
| N4—N3—C10—S1 | 2.2 (5) | C17—C12—C11—S1 | −1.2 (6) |
| C9—N2—C10—N3 | 179.2 (4) | C10—S1—C11—N4 | 1.5 (3) |
| C9—N2—C10—S1 | −3.6 (6) | C10—S1—C11—C12 | −178.9 (4) |
| C11—S1—C10—N3 | −2.2 (4) | C17—C12—C13—C14 | 1.8 (6) |
| C11—S1—C10—N2 | −179.5 (4) | C11—C12—C13—C14 | −179.0 (4) |
| C17—C16—C15—C14 | −1.5 (7) | C15—C14—C13—C12 | −1.8 (7) |
| C17—C16—C15—Br1 | −179.8 (3) | C8—N1—C7—O1 | 5.2 (6) |
| C16—C15—C14—C13 | 1.7 (7) | C8—N1—C7—C6 | −176.1 (4) |
| Br1—C15—C14—C13 | −179.9 (3) | C4—C5—C6—C1 | 1.8 (7) |
| C13—C12—C17—C16 | −1.6 (6) | C4—C5—C6—C7 | −178.7 (5) |
| C11—C12—C17—C16 | 179.2 (4) | O1—C7—C6—C5 | 17.2 (7) |
| C15—C16—C17—C12 | 1.4 (7) | N1—C7—C6—C5 | −161.5 (4) |
| C7—N1—C8—C9 | −159.0 (4) | O1—C7—C6—C1 | −163.3 (4) |
| C10—N2—C9—O2 | −2.3 (6) | N1—C7—C6—C1 | 18.0 (7) |
| C10—N2—C9—C8 | −180.0 (4) | C1—C2—C3—C4 | 0.1 (8) |
| N1—C8—C9—O2 | −0.2 (6) | C5—C4—C3—C2 | 1.6 (8) |
| N1—C8—C9—N2 | 177.4 (3) | C5—C6—C1—C2 | 0.0 (7) |
| N3—N4—C11—C12 | 179.8 (4) | C7—C6—C1—C2 | −179.5 (4) |
| N3—N4—C11—S1 | −0.6 (5) | C3—C2—C1—C6 | −0.9 (8) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C16—H16A···O2i | 0.93 | 2.49 | 3.400 (5) | 168 |
| N2—H2A···O1ii | 0.86 | 1.99 | 2.835 (5) | 167 |
Symmetry codes: (i) −x, −y+1, −z+1; (ii) −x+1, −y+1, −z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: WN2304).
References
- Bruker (2004). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2008). SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Foroumadi, A., Daneshtalab, M. & Shafiee, A. (1999). Arzneim. Forsch.49, 1035–1038. [PubMed]
- Gowda, B. T., Tokarčík, M., Kožíšek, J., Sowmya, B. P. & Fuess, H. (2008). Acta Cryst. E64, o950. [DOI] [PMC free article] [PubMed]
- Levy, M. & Palmer, A. H. (1942). J. Biol. Chem.146, 493–495.
- Li, S.-H., Huang, H.-M., Kuang, B.-H., Tu, G.-G. & Liu, C.-M. (2008). Acta Cryst. E64, o2006. [DOI] [PMC free article] [PubMed]
- Li, S.-H., Li, G., Huang, H.-M., Tu, G.-G. & Liu, C.-M. (2008). Acta Cryst. E64, o1887. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Song, K. S., Ishikawa, Y., Kobayashi, S., Sankawa, U. & Ebizuka, Y. (1992). Phytochemistry, 31, 823–826.
- Tu, G. G., Li, S. H., Huang, H. M., Li, G., Xiong, F., Mai, X., Zhu, H. W., Kuang, B. H. & Xu, W. F. (2008). Bioorg. Med. Chem.16, 6663–6668. [DOI] [PubMed]
- Westrip, S. P. (2009). publCIF In preparation.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S160053680900124X/wn2304sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053680900124X/wn2304Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




