Abstract
The title compound, C17H13N3OS, was obtained by the reaction of benzoyl chloride, ammonium thiocyanate and 5-aminoquinoline in the presence of polyethyleneglycol-400 (PEG-400) as a phase-transfer catalyst. The compound crystallized as discrete molecules linked by N—H⋯N and C—H⋯N hydrogen bonds involving all the potential donors, generating sheets parallel to (100). An intramolecular N—H⋯O bond is also present.
Related literature
For the biological activity of acyl thioureas, see: Hackmann (1960 ▶); Sarkis & Faisal (1985 ▶). For their application in the synthesis of supramolecular complexes, see: Pluta & Sadlej (2001 ▶); Kaminsky et al. (2002 ▶). For a related structure, see: Xue et al. (2004 ▶).
Experimental
Crystal data
C17H13N3OS
M r = 307.36
Monoclinic,

a = 5.0875 (1) Å
b = 16.1718 (4) Å
c = 18.2847 (4) Å
β = 95.892 (2)°
V = 1496.41 (6) Å3
Z = 4
Mo Kα radiation
μ = 0.22 mm−1
T = 296 (2) K
0.40 × 0.30 × 0.20 mm
Data collection
Enraf–Nonius CAD-4 diffractometer
Absorption correction: ψ scan (North et al., 1968 ▶) T min = 0.939, T max = 0.969
13322 measured reflections
3411 independent reflections
2184 reflections with I > 2σ(I)
R int = 0.032
3 standard reflections every 97 reflections intensity decay: 2.1%
Refinement
R[F 2 > 2σ(F 2)] = 0.048
wR(F 2) = 0.129
S = 1.04
3411 reflections
207 parameters
2 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.30 e Å−3
Δρmin = −0.22 e Å−3
Data collection: CAD-4 Software (Enraf–Nonius, 1989 ▶); cell refinement: CAD-4 Software; data reduction: XCAD4 (Harms & Wocadlo, 1995 ▶); program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: SHELXTL; software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809000932/hg2453sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809000932/hg2453Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H6⋯N3i | 0.825 (17) | 2.283 (17) | 3.100 (3) | 170.5 (18) |
| N2—H7⋯O1 | 0.91 (3) | 1.84 (3) | 2.619 (3) | 143 (3) |
| C6—H5⋯N3i | 0.93 | 2.46 | 3.252 (3) | 143 |
Symmetry code: (i)  .
.
Acknowledgments
We are grateful to the Science Foundation of Jiangsu Education Bureau (05KJD 150039), the Professor Foundation of Huaiyin Teachers College (05 HSJS018) and the Science Foundation of Jangsu Key Laboratory for the Chemistry of Low-Dimensional Materials (JSKC 06028) for financial support.
supplementary crystallographic information
Comment
Acyl thioureas have extensive biological activities such as bacteriostasis, weeding (Hackmann, 1960) and plant growth regulating (Sarkis & Faisal, 1985). In addition, acyl thioureas are excellent ligands, and have been widely applied in synthesis of supramolecular complexes (Pluta & Sadlej, 2001; Kaminsky et al., 2002). The title compound, (I) crystallizes as discrete molecules (Fig. 1). The full molecule is a big conjugated system because the bond lengths of C1—C7, C7—N1, C8—N1, C8—N2 and C9—N2 become shorter than standard values, and the bond lengths of C7—O1 and C8—S1 become longer than standard values. In (I) the torsion angle for C17—C9—N2—C8 of -78.2 (3)° indicates the quinoline ring is approximately orthogonal to the rest of the molecule. The molecules in (I) are linked by N1—H6···N3, N2—H7···O1 and C6—H5···N3 hydrogen bonds involving all the potential donors, generating sheets parallel to (100), as shown in Fig. 2. In addition, the bond lengths of S—C (1.655 (2)Å) and O—C(1.223 (2)Å) in (I) are longer than the bond lengths of S—C(1.6503Å) and O—C(1.201Å) in N-(4,6-dimethylpyrimidin-2-ylcarbamothioyl)benzamide (Xue et al., 2004)
Experimental
The title compound was synthesized as following. A mixture of benzoyl chloride (1400 mg, 10 mmol), ammonium thiocyanate (1140 mg, 15 mmol), 5-aminoquinoline (1300 mg, 9 mmol) and dichloromethane (50 ml) in the presence of PEG-400 (1200 mg, 3 mmol) as phase transfer catalyst at room temperature for 8h with stirring. The reaction mixture was evaporated to give a residue. Singles crystals suitable for X-ray analysis were obtained by slow evaporation of a mixture solution of dichloromethane and ethanol.
Refinement
The atom H6 attached to N1 and the atom H7 attached to N2 was located in a difference Fourier map and refined with N—H distance restrained to 0.87 (2)Å, and with Uiso(H) = 0.85Ueq(N) and Uiso(H) = 1.91Ueq(N) All H atoms bound to carbon were refined using riding models with d(C—H) = 0.93Å and Uiso(H) = 1.2Ueq(C).
Figures
Fig. 1.
The molecular structure of (I), with atom labels and 50% probability displacement ellipsoids for non-H atoms.
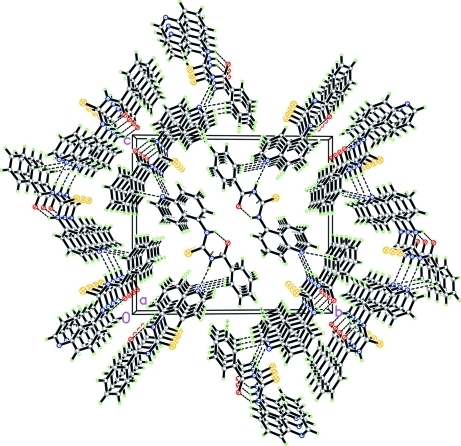
Fig. 2.
The packing of (I), viewed down the a axis, showing two layers of molecules connected by van der waals.
Crystal data
| C17H13N3OS | F(000) = 640 |
| Mr = 307.36 | Dx = 1.364 Mg m−3 |
| Monoclinic, P21/n | Melting point = 446.2–446.7 K |
| Hall symbol: -P 2yn | Mo Kα radiation, λ = 0.71073 Å |
| a = 5.0875 (1) Å | Cell parameters from 3090 reflections |
| b = 16.1718 (4) Å | θ = 2.2–22.4° |
| c = 18.2847 (4) Å | µ = 0.22 mm−1 |
| β = 95.892 (2)° | T = 296 K |
| V = 1496.41 (6) Å3 | Rod, yellow |
| Z = 4 | 0.40 × 0.30 × 0.20 mm |
Data collection
| Enraf–Nonius CAD-4 diffractometer | 2184 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.032 |
| graphite | θmax = 27.4°, θmin = 1.7° |
| ω/2θ scans | h = −6→6 |
| Absorption correction: ψ scan (North et al., 1968) | k = −20→18 |
| Tmin = 0.939, Tmax = 0.969 | l = −23→23 |
| 13322 measured reflections | 3 standard reflections every 97 reflections |
| 3411 independent reflections | intensity decay: 2.1% |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.048 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.129 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.04 | w = 1/[σ2(Fo2) + (0.0482P)2 + 0.4696P] where P = (Fo2 + 2Fc2)/3 |
| 3411 reflections | (Δ/σ)max < 0.001 |
| 207 parameters | Δρmax = 0.30 e Å−3 |
| 2 restraints | Δρmin = −0.22 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | −0.00333 (14) | 0.14855 (4) | 0.20925 (4) | 0.0704 (2) | |
| O1 | 0.5088 (4) | 0.09876 (10) | 0.02568 (9) | 0.0724 (5) | |
| N1 | 0.2883 (4) | 0.17950 (11) | 0.10117 (10) | 0.0503 (4) | |
| N2 | 0.2175 (4) | 0.04129 (12) | 0.12428 (10) | 0.0602 (5) | |
| C17 | 0.2711 (4) | −0.05169 (12) | 0.23117 (11) | 0.0442 (5) | |
| C13 | 0.1919 (4) | −0.12237 (12) | 0.26802 (11) | 0.0482 (5) | |
| C7 | 0.4431 (5) | 0.16753 (13) | 0.04478 (11) | 0.0524 (5) | |
| N3 | 0.3173 (4) | −0.14846 (11) | 0.33306 (10) | 0.0579 (5) | |
| C1 | 0.5204 (4) | 0.24240 (13) | 0.00465 (10) | 0.0470 (5) | |
| C16 | 0.4940 (4) | −0.00801 (14) | 0.26304 (13) | 0.0558 (6) | |
| H13 | 0.5538 | 0.0389 | 0.2404 | 0.067* | |
| C8 | 0.1748 (4) | 0.11985 (13) | 0.14277 (11) | 0.0511 (5) | |
| C12 | −0.0297 (5) | −0.16872 (13) | 0.23615 (14) | 0.0582 (6) | |
| H10 | −0.0850 | −0.2156 | 0.2597 | 0.070* | |
| C9 | 0.1265 (4) | −0.02866 (13) | 0.16352 (12) | 0.0520 (5) | |
| C11 | −0.1587 (5) | −0.14385 (14) | 0.17142 (14) | 0.0607 (6) | |
| H9 | −0.3023 | −0.1745 | 0.1508 | 0.073* | |
| C2 | 0.7141 (5) | 0.23490 (16) | −0.04248 (13) | 0.0650 (6) | |
| H1 | 0.8019 | 0.1848 | −0.0458 | 0.078* | |
| C15 | 0.6203 (5) | −0.03616 (15) | 0.32799 (13) | 0.0621 (6) | |
| H12 | 0.7686 | −0.0090 | 0.3502 | 0.075* | |
| C14 | 0.5228 (5) | −0.10613 (16) | 0.36003 (13) | 0.0637 (6) | |
| H11 | 0.6111 | −0.1242 | 0.4042 | 0.076* | |
| C6 | 0.3941 (5) | 0.31702 (15) | 0.00781 (13) | 0.0658 (7) | |
| H5 | 0.2610 | 0.3232 | 0.0386 | 0.079* | |
| C5 | 0.4626 (6) | 0.38332 (17) | −0.03438 (14) | 0.0784 (8) | |
| H4 | 0.3787 | 0.4340 | −0.0308 | 0.094* | |
| C4 | 0.6519 (6) | 0.37434 (18) | −0.08094 (14) | 0.0747 (7) | |
| H3 | 0.6947 | 0.4184 | −0.1102 | 0.090* | |
| C10 | −0.0822 (5) | −0.07367 (14) | 0.13504 (13) | 0.0599 (6) | |
| H8 | −0.1754 | −0.0578 | 0.0909 | 0.072* | |
| C3 | 0.7785 (5) | 0.30086 (19) | −0.08470 (15) | 0.0764 (8) | |
| H2 | 0.9100 | 0.2950 | −0.1161 | 0.092* | |
| H6 | 0.268 (4) | 0.2277 (10) | 0.1143 (10) | 0.043 (6)* | |
| H7 | 0.324 (5) | 0.0371 (19) | 0.0876 (14) | 0.115 (11)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0932 (5) | 0.0458 (4) | 0.0790 (4) | 0.0004 (3) | 0.0421 (4) | 0.0001 (3) |
| O1 | 0.1085 (14) | 0.0473 (10) | 0.0671 (10) | 0.0049 (9) | 0.0359 (10) | −0.0030 (8) |
| N1 | 0.0701 (12) | 0.0363 (10) | 0.0461 (10) | 0.0012 (9) | 0.0131 (9) | 0.0013 (8) |
| N2 | 0.0852 (15) | 0.0426 (11) | 0.0558 (12) | 0.0015 (10) | 0.0220 (11) | 0.0056 (9) |
| C17 | 0.0501 (11) | 0.0343 (10) | 0.0503 (11) | 0.0001 (9) | 0.0159 (9) | −0.0048 (9) |
| C13 | 0.0589 (13) | 0.0362 (11) | 0.0522 (12) | 0.0016 (9) | 0.0189 (10) | −0.0032 (9) |
| C7 | 0.0653 (14) | 0.0480 (13) | 0.0443 (11) | 0.0010 (10) | 0.0080 (10) | −0.0033 (9) |
| N3 | 0.0709 (13) | 0.0490 (11) | 0.0550 (11) | 0.0006 (10) | 0.0123 (10) | 0.0033 (9) |
| C1 | 0.0554 (12) | 0.0489 (12) | 0.0368 (10) | −0.0039 (10) | 0.0048 (9) | 0.0001 (9) |
| C16 | 0.0594 (14) | 0.0449 (13) | 0.0664 (14) | −0.0077 (10) | 0.0227 (12) | −0.0067 (11) |
| C8 | 0.0664 (14) | 0.0387 (12) | 0.0493 (11) | 0.0025 (10) | 0.0108 (10) | 0.0039 (9) |
| C12 | 0.0687 (15) | 0.0393 (12) | 0.0692 (15) | −0.0076 (10) | 0.0199 (12) | −0.0077 (10) |
| C9 | 0.0642 (14) | 0.0401 (12) | 0.0536 (12) | 0.0027 (10) | 0.0154 (11) | −0.0035 (10) |
| C11 | 0.0597 (14) | 0.0515 (14) | 0.0711 (15) | −0.0089 (11) | 0.0068 (12) | −0.0150 (12) |
| C2 | 0.0682 (15) | 0.0633 (16) | 0.0668 (15) | 0.0042 (12) | 0.0220 (13) | 0.0018 (12) |
| C15 | 0.0542 (14) | 0.0669 (16) | 0.0655 (15) | −0.0073 (12) | 0.0077 (12) | −0.0149 (12) |
| C14 | 0.0670 (16) | 0.0664 (16) | 0.0581 (14) | 0.0028 (13) | 0.0084 (12) | 0.0030 (12) |
| C6 | 0.0821 (17) | 0.0613 (15) | 0.0584 (14) | 0.0085 (13) | 0.0276 (13) | 0.0110 (11) |
| C5 | 0.104 (2) | 0.0613 (16) | 0.0740 (16) | 0.0143 (15) | 0.0295 (16) | 0.0208 (13) |
| C4 | 0.0806 (18) | 0.0751 (19) | 0.0706 (16) | −0.0080 (15) | 0.0187 (14) | 0.0243 (14) |
| C10 | 0.0687 (15) | 0.0509 (14) | 0.0597 (14) | 0.0019 (12) | 0.0054 (12) | −0.0091 (11) |
| C3 | 0.0728 (17) | 0.086 (2) | 0.0761 (17) | −0.0040 (15) | 0.0362 (14) | 0.0121 (15) |
Geometric parameters (Å, °)
| S1—C8 | 1.655 (2) | C12—C11 | 1.354 (3) |
| O1—C7 | 1.223 (2) | C12—H10 | 0.9300 |
| N1—C7 | 1.374 (3) | C9—C10 | 1.347 (3) |
| N1—C8 | 1.390 (3) | C11—C10 | 1.391 (3) |
| N1—H6 | 0.826 (15) | C11—H9 | 0.9300 |
| N2—C8 | 1.338 (3) | C2—C3 | 1.376 (3) |
| N2—C9 | 1.441 (3) | C2—H1 | 0.9300 |
| N2—H7 | 0.909 (17) | C15—C14 | 1.389 (3) |
| C17—C13 | 1.407 (3) | C15—H12 | 0.9300 |
| C17—C16 | 1.410 (3) | C14—H11 | 0.9300 |
| C17—C9 | 1.422 (3) | C6—C5 | 1.386 (3) |
| C13—N3 | 1.358 (3) | C6—H5 | 0.9300 |
| C13—C12 | 1.427 (3) | C5—C4 | 1.357 (4) |
| C7—C1 | 1.490 (3) | C5—H4 | 0.9300 |
| N3—C14 | 1.304 (3) | C4—C3 | 1.357 (4) |
| C1—C6 | 1.371 (3) | C4—H3 | 0.9300 |
| C1—C2 | 1.379 (3) | C10—H8 | 0.9300 |
| C16—C15 | 1.369 (3) | C3—H2 | 0.9300 |
| C16—H13 | 0.9300 | ||
| C7—N1—C8 | 127.96 (19) | C10—C9—N2 | 120.8 (2) |
| C7—N1—H6 | 116.7 (14) | C17—C9—N2 | 118.35 (19) |
| C8—N1—H6 | 115.2 (14) | C12—C11—C10 | 121.7 (2) |
| C8—N2—C9 | 123.42 (19) | C12—C11—H9 | 119.1 |
| C8—N2—H7 | 112 (2) | C10—C11—H9 | 119.1 |
| C9—N2—H7 | 124 (2) | C3—C2—C1 | 120.6 (2) |
| C13—C17—C16 | 117.8 (2) | C3—C2—H1 | 119.7 |
| C13—C17—C9 | 118.79 (19) | C1—C2—H1 | 119.7 |
| C16—C17—C9 | 123.39 (19) | C16—C15—C14 | 118.7 (2) |
| N3—C13—C17 | 122.7 (2) | C16—C15—H12 | 120.6 |
| N3—C13—C12 | 118.3 (2) | C14—C15—H12 | 120.6 |
| C17—C13—C12 | 118.9 (2) | N3—C14—C15 | 125.1 (2) |
| O1—C7—N1 | 122.5 (2) | N3—C14—H11 | 117.4 |
| O1—C7—C1 | 120.3 (2) | C15—C14—H11 | 117.4 |
| N1—C7—C1 | 117.14 (19) | C1—C6—C5 | 120.8 (2) |
| C14—N3—C13 | 117.0 (2) | C1—C6—H5 | 119.6 |
| C6—C1—C2 | 118.1 (2) | C5—C6—H5 | 119.6 |
| C6—C1—C7 | 123.1 (2) | C4—C5—C6 | 120.1 (3) |
| C2—C1—C7 | 118.6 (2) | C4—C5—H4 | 119.9 |
| C15—C16—C17 | 118.6 (2) | C6—C5—H4 | 119.9 |
| C15—C16—H13 | 120.7 | C3—C4—C5 | 119.8 (2) |
| C17—C16—H13 | 120.7 | C3—C4—H3 | 120.1 |
| N2—C8—N1 | 115.68 (19) | C5—C4—H3 | 120.1 |
| N2—C8—S1 | 124.54 (17) | C9—C10—C11 | 120.3 (2) |
| N1—C8—S1 | 119.78 (16) | C9—C10—H8 | 119.9 |
| C11—C12—C13 | 119.5 (2) | C11—C10—H8 | 119.9 |
| C11—C12—H10 | 120.2 | C4—C3—C2 | 120.6 (2) |
| C13—C12—H10 | 120.2 | C4—C3—H2 | 119.7 |
| C10—C9—C17 | 120.8 (2) | C2—C3—H2 | 119.7 |
| C16—C17—C13—N3 | 1.5 (3) | C16—C17—C9—C10 | 178.8 (2) |
| C9—C17—C13—N3 | −179.51 (18) | C13—C17—C9—N2 | −176.79 (18) |
| C16—C17—C13—C12 | −178.68 (18) | C16—C17—C9—N2 | 2.1 (3) |
| C9—C17—C13—C12 | 0.3 (3) | C8—N2—C9—C10 | 105.1 (3) |
| C8—N1—C7—O1 | −2.7 (4) | C8—N2—C9—C17 | −78.2 (3) |
| C8—N1—C7—C1 | 174.7 (2) | C13—C12—C11—C10 | −0.4 (3) |
| C17—C13—N3—C14 | −1.8 (3) | C6—C1—C2—C3 | 0.3 (3) |
| C12—C13—N3—C14 | 178.4 (2) | C7—C1—C2—C3 | 175.3 (2) |
| O1—C7—C1—C6 | 160.1 (2) | C17—C16—C15—C14 | −0.4 (3) |
| N1—C7—C1—C6 | −17.3 (3) | C13—N3—C14—C15 | 1.0 (4) |
| O1—C7—C1—C2 | −14.6 (3) | C16—C15—C14—N3 | 0.1 (4) |
| N1—C7—C1—C2 | 168.0 (2) | C2—C1—C6—C5 | −1.0 (4) |
| C13—C17—C16—C15 | −0.4 (3) | C7—C1—C6—C5 | −175.7 (2) |
| C9—C17—C16—C15 | −179.3 (2) | C1—C6—C5—C4 | 1.7 (4) |
| C9—N2—C8—N1 | 176.8 (2) | C6—C5—C4—C3 | −1.6 (4) |
| C9—N2—C8—S1 | −4.2 (3) | C17—C9—C10—C11 | −0.3 (3) |
| C7—N1—C8—N2 | −1.6 (3) | N2—C9—C10—C11 | 176.3 (2) |
| C7—N1—C8—S1 | 179.31 (18) | C12—C11—C10—C9 | 0.6 (4) |
| N3—C13—C12—C11 | 179.8 (2) | C5—C4—C3—C2 | 0.9 (4) |
| C17—C13—C12—C11 | 0.0 (3) | C1—C2—C3—C4 | −0.3 (4) |
| C13—C17—C9—C10 | −0.1 (3) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H6···N3i | 0.83 (2) | 2.28 (2) | 3.100 (3) | 171 (2) |
| N2—H7···O1 | 0.91 (3) | 1.84 (3) | 2.619 (3) | 143 (3) |
| C6—H5···N3i | 0.93 | 2.46 | 3.252 (3) | 143 |
Symmetry codes: (i) −x+1/2, y+1/2, −z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HG2453).
References
- Enraf–Nonius (1989). CAD-4 Software Enraf–Nonius, Delft, The Netherlands.
- Hackmann, J. T. (1960). US Patent No. 2 923 656.
- Harms, K. & Wocadlo, S. (1995). XCAD4 University of Marburg, Germany.
- Kaminsky, W., Goldberg, K. I. & West, D. X. (2002). J. Mol. Struct.605, 9–15.
- North, A. C. T., Phillips, D. C. & Mathews, F. S. (1968). Acta Cryst. A24, 351–359.
- Pluta, T. & Sadlej, A. J. (2001). J. Chem. Phys.114, 136–146.
- Sarkis, G. Y. & Faisal, E. D. (1985). J. Heterocycl. Chem.22, 137–140.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Xue, S. J., Duan, L. P., Ke, S. Y. & Zhu, J. M. (2004). Chin. J. Struct. Chem.23, 441–444.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809000932/hg2453sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809000932/hg2453Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report