Abstract
In the title compound, 5-(4-chlorophenyl)-1-(4-methoxyphenyl)-3-(trifluoromethyl)-1H-pyrazole (SC-560), C17H12ClF3N2O, a COX-1-selective inhibitor, the dihedral angles between the heterocycle and the chlorobenzene and methoxybenzene rings are 41.66 (6) and 43.08 (7)°, respectively. The dihedral angle between the two phenyl rings is 59.94 (6)°. No classic hydrogen bonds are possible in the crystal, and intermolecular interactions must be mainly of the dispersion type. This information may aid the identification of dosage formulations with improved oral bioavailability.
Related literature
For background literature, see: Choi et al. (2008 ▶); Cusimano et al. (2007 ▶); Kundu & Fulton (2002 ▶); Penning et al. (1997 ▶); Smith et al. (2000 ▶); Teng et al. (2003 ▶); Tiano et al. (2002 ▶); For related structures, see: Allen (2002 ▶); Charlier et al. (2004 ▶); Norris et al. (2005 ▶); Sonar et al. (2004 ▶); Zhu et al. (2004 ▶).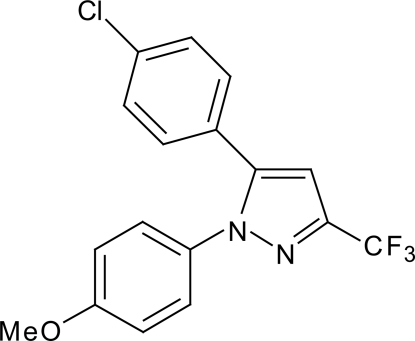
Experimental
Crystal data
C17H12ClF3N2O
M r = 352.74
Monoclinic,
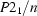
a = 15.585 (3) Å
b = 7.1671 (14) Å
c = 15.789 (3) Å
β = 116.81 (3)°
V = 1574.1 (5) Å3
Z = 4
Mo Kα radiation
μ = 0.28 mm−1
T = 90 (2) K
0.20 × 0.10 × 0.10 mm
Data collection
Nonius KappaCCD diffractometer
Absorption correction: multi-scan (SCALEPACK; Otwinowski & Minor, 1997 ▶) T min = 0.946, T max = 0.972
6895 measured reflections
3608 independent reflections
3030 reflections with I > 2σ(I)
R int = 0.020
Refinement
R[F 2 > 2σ(F 2)] = 0.035
wR(F 2) = 0.094
S = 1.04
3608 reflections
218 parameters
H-atom parameters constrained
Δρmax = 0.34 e Å−3
Δρmin = −0.29 e Å−3
Data collection: COLLECT (Hooft, 1998 ▶); cell refinement: DENZO-SMN (Otwinowski & Minor, 1997 ▶); data reduction: DENZO-SMN; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL and local procedures.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809001779/pv2130sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809001779/pv2130Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Acknowledgments
SL and TL are grateful for financial support by the NSF (DMR– 0449633). The authors also thank Dr Sean Parkin for providing laboratory facilities and for helpful discussions.
supplementary crystallographic information
Comment
Inhibition of the two cyclooxygenase (COX) isoforms is considered the primary mechanism responsible for both the therapeutic and toxic effects of nonsteroidal anti-inflammatory drugs (NSAIDS) (Smith, et al., 2000). Both of the COX isoforms have been shown to contribute to inflammation and tumor genesis, and therapeutic benefits may result from either COX-1 or COX-2 selective inhibition (Tiano, et al. 2002; Choi, et al. 2008; Kundu & Fulton, 2002; Cusimano, et al. 2007). The development of the COX-2-selective inhibitor celecoxib led to identification of a variety of structurally related compounds with varying selectivity for the COX isoforms, SC-560 was one of them (Penning, et al. 1997; Choi, et al. 2008). However, SC-560's poor bioavailability may limit its effects (Teng, et al. 2003). The information from crystal structure may provide direction in suitable dosage formulation. Herein, we describe the first crystal structure of SC-560, (I), a selective and potent inhibitor of the cyclooxygenase-1 isoform (Penning, et al. 1997).
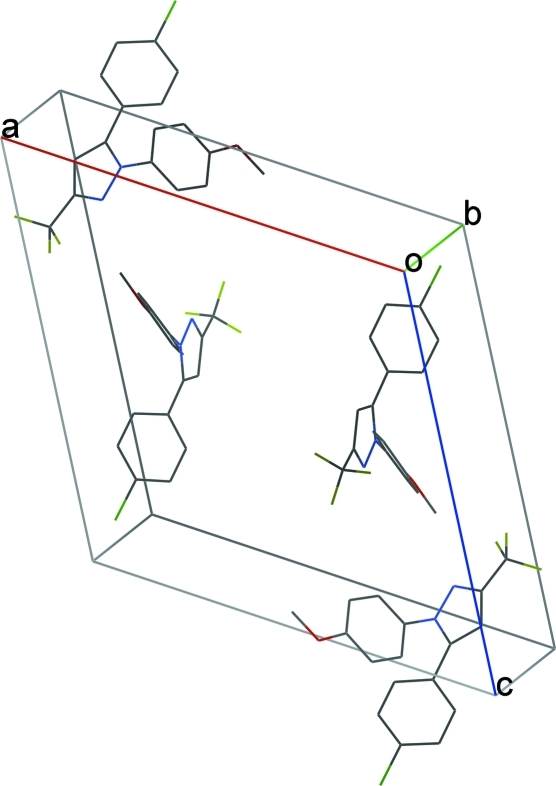
The crystal structure of (I) is presented in Fig. 1. The structure lacks the moieties necessary for hydrogen-bonding to occur between molecules (Fig. 2). Thus, the lattice energy mainly consists of dispersion energies, which typically result in a low melting point because of the weak intermolecular interactions (as confirmed by melting point measurement). Despite the entire chemical structure being fused together by three aromatic rings, a large conjugate system between the rings is not seen due to steric repulsion between the two phenyl rings. Similar structures are abundant in the Cambridge Structural Database (CSD - Version 5.29; Allen, 2002), EYISAG (Sonar, et al. 2004), IZAYUD (Charlier, et al. 2004), JAQBIN (Norris, et al. 2005), and MAJGUA (Zhu, et al. 2004) are few of them. To conclude, the single-crystal structure of SC-560 was solved. Because there is no hydrogen bonding in the structure, the major contribution for the lattice energy stems from weak dispersion energies leading to its low melting point at 335.5 K.
Experimental
Commercial SC-560 was dissolved in HPLC grade methanol in a glass vial at room temperature. The vial was sealed with Parafilm with numerous pin-size holes introduced to allow for evaporation of the solvent. Colorless block crystals were obtained following approximately one week of slow evaporation.
Refinement
H atoms were found in difference Fourier maps and those on the aromatic ring subsequently placed in idealized positions with C—H distances of 0.95 Å and isotropic displacement parameters equal to 1.2Ueq of the attached carbon atom. Hydrogen atom coordinates in the methyl group were placed in idealized positions with C—H distances of 0.98 Å and isotropic displacement parameters of 1.5Ueq of the carbon atom.
Figures
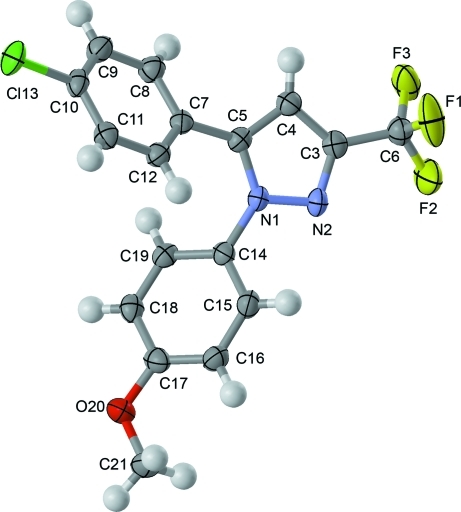
Fig. 1.
The molecular structure of (I), showing atom displacement ellipsoids at the 50% probability level.
Fig. 2.
Crystal packing of SC-560.
Crystal data
| C17H12ClF3N2O | F(000) = 720 |
| Mr = 352.74 | Dx = 1.488 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| a = 15.585 (3) Å | Cell parameters from 3880 reflections |
| b = 7.1671 (14) Å | θ = 1.0–27.5° |
| c = 15.789 (3) Å | µ = 0.28 mm−1 |
| β = 116.81 (3)° | T = 90 K |
| V = 1574.1 (5) Å3 | Block, colorless |
| Z = 4 | 0.20 × 0.10 × 0.10 mm |
Data collection
| Nonius KappaCCD diffractometer | 3608 independent reflections |
| Radiation source: fine-focus sealed tube | 3030 reflections with I > 2σ(I) |
| graphite | Rint = 0.020 |
| Detector resolution: 18 pixels mm-1 | θmax = 27.5°, θmin = 1.5° |
| ω scans at fixed χ = 55° | h = −20→20 |
| Absorption correction: multi-scan (SCALEPACK; Otwinowski & Minor, 1997) | k = −9→9 |
| Tmin = 0.946, Tmax = 0.972 | l = −20→20 |
| 6895 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.035 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.094 | H-atom parameters constrained |
| S = 1.04 | w = 1/[σ2(Fo2) + (0.0421P)2 + 0.7994P] where P = (Fo2 + 2Fc2)/3 |
| 3608 reflections | (Δ/σ)max = 0.004 |
| 218 parameters | Δρmax = 0.34 e Å−3 |
| 0 restraints | Δρmin = −0.29 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl13 | 0.06492 (3) | 0.90044 (6) | 0.10611 (2) | 0.02831 (12) | |
| N2 | 0.38203 (8) | 0.87316 (17) | 0.67980 (8) | 0.0188 (2) | |
| O20 | 0.12419 (8) | 0.14611 (15) | 0.54981 (7) | 0.0287 (3) | |
| F3 | 0.56584 (7) | 1.19886 (15) | 0.74065 (7) | 0.0382 (3) | |
| F2 | 0.52162 (8) | 1.04490 (15) | 0.83041 (6) | 0.0443 (3) | |
| N1 | 0.32413 (8) | 0.81035 (16) | 0.59070 (7) | 0.0169 (2) | |
| C10 | 0.14398 (10) | 0.89725 (19) | 0.22720 (9) | 0.0199 (3) | |
| C5 | 0.33171 (9) | 0.91657 (19) | 0.52240 (9) | 0.0171 (3) | |
| C18 | 0.21726 (10) | 0.3440 (2) | 0.50702 (9) | 0.0196 (3) | |
| H18 | 0.2135 | 0.2529 | 0.4618 | 0.023* | |
| F1 | 0.44389 (7) | 1.28688 (15) | 0.75784 (7) | 0.0448 (3) | |
| C11 | 0.10734 (10) | 0.8619 (2) | 0.29082 (10) | 0.0199 (3) | |
| H11 | 0.0408 | 0.8367 | 0.2688 | 0.024* | |
| C19 | 0.26912 (9) | 0.50583 (19) | 0.51716 (9) | 0.0179 (3) | |
| H19 | 0.3033 | 0.5239 | 0.4809 | 0.021* | |
| C7 | 0.26748 (10) | 0.89925 (18) | 0.41982 (9) | 0.0170 (3) | |
| C14 | 0.27105 (9) | 0.64202 (19) | 0.58060 (9) | 0.0173 (3) | |
| C16 | 0.17333 (11) | 0.4491 (2) | 0.62713 (10) | 0.0244 (3) | |
| H16 | 0.1419 | 0.4285 | 0.6657 | 0.029* | |
| C6 | 0.48912 (10) | 1.1368 (2) | 0.74825 (10) | 0.0222 (3) | |
| C12 | 0.16930 (10) | 0.86393 (19) | 0.38715 (9) | 0.0190 (3) | |
| H12 | 0.1449 | 0.8411 | 0.4315 | 0.023* | |
| C4 | 0.39888 (9) | 1.0532 (2) | 0.56958 (9) | 0.0189 (3) | |
| H4 | 0.4216 | 1.1482 | 0.5428 | 0.023* | |
| C8 | 0.30233 (10) | 0.9337 (2) | 0.35415 (10) | 0.0211 (3) | |
| H8 | 0.3689 | 0.9576 | 0.3757 | 0.025* | |
| C3 | 0.42585 (9) | 1.0204 (2) | 0.66545 (9) | 0.0190 (3) | |
| C15 | 0.22267 (10) | 0.6153 (2) | 0.63461 (10) | 0.0222 (3) | |
| H15 | 0.2231 | 0.7102 | 0.6768 | 0.027* | |
| C17 | 0.17048 (10) | 0.3143 (2) | 0.56306 (10) | 0.0217 (3) | |
| C9 | 0.24066 (11) | 0.9335 (2) | 0.25715 (10) | 0.0238 (3) | |
| H9 | 0.2645 | 0.9578 | 0.2124 | 0.029* | |
| C21 | 0.07544 (14) | 0.1110 (3) | 0.60633 (11) | 0.0403 (5) | |
| H21A | 0.1214 | 0.1185 | 0.6737 | 0.060* | |
| H21B | 0.0468 | −0.0138 | 0.5921 | 0.060* | |
| H21C | 0.0248 | 0.2043 | 0.5919 | 0.060* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl13 | 0.0288 (2) | 0.0370 (2) | 0.01330 (17) | 0.00223 (16) | 0.00434 (14) | 0.00022 (14) |
| N2 | 0.0178 (6) | 0.0216 (6) | 0.0138 (5) | −0.0009 (5) | 0.0044 (4) | −0.0028 (4) |
| O20 | 0.0349 (6) | 0.0259 (6) | 0.0258 (5) | −0.0125 (5) | 0.0143 (5) | −0.0024 (4) |
| F3 | 0.0326 (5) | 0.0472 (6) | 0.0402 (5) | −0.0209 (5) | 0.0211 (4) | −0.0206 (5) |
| F2 | 0.0533 (6) | 0.0441 (6) | 0.0177 (4) | −0.0183 (5) | 0.0001 (4) | −0.0012 (4) |
| N1 | 0.0180 (5) | 0.0186 (6) | 0.0126 (5) | −0.0015 (5) | 0.0057 (4) | −0.0014 (4) |
| C10 | 0.0238 (7) | 0.0184 (7) | 0.0134 (6) | 0.0030 (5) | 0.0046 (5) | 0.0000 (5) |
| C5 | 0.0177 (6) | 0.0180 (7) | 0.0159 (6) | 0.0022 (5) | 0.0080 (5) | 0.0011 (5) |
| C18 | 0.0207 (7) | 0.0180 (7) | 0.0178 (6) | 0.0029 (5) | 0.0067 (5) | −0.0008 (5) |
| F1 | 0.0342 (5) | 0.0427 (6) | 0.0472 (6) | 0.0054 (5) | 0.0092 (5) | −0.0268 (5) |
| C11 | 0.0187 (6) | 0.0189 (7) | 0.0189 (6) | 0.0009 (5) | 0.0059 (5) | 0.0001 (5) |
| C19 | 0.0188 (6) | 0.0189 (7) | 0.0170 (6) | 0.0031 (5) | 0.0089 (5) | 0.0019 (5) |
| C7 | 0.0200 (6) | 0.0141 (6) | 0.0150 (6) | 0.0016 (5) | 0.0063 (5) | 0.0008 (5) |
| C14 | 0.0171 (6) | 0.0177 (7) | 0.0142 (6) | −0.0010 (5) | 0.0045 (5) | 0.0013 (5) |
| C16 | 0.0259 (7) | 0.0312 (8) | 0.0191 (6) | −0.0075 (6) | 0.0127 (6) | −0.0023 (6) |
| C6 | 0.0208 (7) | 0.0240 (8) | 0.0207 (7) | −0.0022 (6) | 0.0084 (6) | −0.0033 (6) |
| C12 | 0.0215 (7) | 0.0192 (7) | 0.0171 (6) | 0.0008 (5) | 0.0093 (5) | 0.0011 (5) |
| C4 | 0.0182 (6) | 0.0186 (7) | 0.0189 (6) | 0.0001 (5) | 0.0073 (5) | 0.0009 (5) |
| C8 | 0.0199 (7) | 0.0237 (7) | 0.0193 (7) | −0.0004 (6) | 0.0084 (5) | 0.0003 (5) |
| C3 | 0.0166 (6) | 0.0195 (7) | 0.0191 (6) | 0.0013 (5) | 0.0066 (5) | −0.0008 (5) |
| C15 | 0.0238 (7) | 0.0262 (8) | 0.0172 (6) | −0.0032 (6) | 0.0097 (6) | −0.0044 (6) |
| C17 | 0.0204 (7) | 0.0213 (7) | 0.0188 (6) | −0.0037 (6) | 0.0048 (5) | 0.0024 (5) |
| C9 | 0.0270 (7) | 0.0280 (8) | 0.0177 (6) | 0.0000 (6) | 0.0113 (6) | 0.0009 (6) |
| C21 | 0.0495 (11) | 0.0482 (11) | 0.0253 (8) | −0.0297 (9) | 0.0188 (8) | −0.0057 (7) |
Geometric parameters (Å, °)
| Cl13—C10 | 1.7461 (15) | C19—C14 | 1.3892 (19) |
| N2—C3 | 1.3307 (18) | C19—H19 | 0.9500 |
| N2—N1 | 1.3605 (15) | C7—C8 | 1.3924 (19) |
| O20—C17 | 1.3714 (18) | C7—C12 | 1.3998 (19) |
| O20—C21 | 1.4312 (19) | C14—C15 | 1.383 (2) |
| F3—C6 | 1.3308 (17) | C16—C17 | 1.385 (2) |
| F2—C6 | 1.3345 (17) | C16—C15 | 1.393 (2) |
| N1—C5 | 1.3680 (17) | C16—H16 | 0.9500 |
| N1—C14 | 1.4305 (17) | C6—C3 | 1.4884 (19) |
| C10—C11 | 1.384 (2) | C12—H12 | 0.9500 |
| C10—C9 | 1.385 (2) | C4—C3 | 1.3962 (19) |
| C5—C4 | 1.3807 (19) | C4—H4 | 0.9500 |
| C5—C7 | 1.4759 (18) | C8—C9 | 1.394 (2) |
| C18—C19 | 1.381 (2) | C8—H8 | 0.9500 |
| C18—C17 | 1.393 (2) | C15—H15 | 0.9500 |
| C18—H18 | 0.9500 | C9—H9 | 0.9500 |
| F1—C6 | 1.3313 (18) | C21—H21A | 0.9800 |
| C11—C12 | 1.3861 (19) | C21—H21B | 0.9800 |
| C11—H11 | 0.9500 | C21—H21C | 0.9800 |
| C3—N2—N1 | 103.79 (11) | F1—C6—F2 | 106.06 (12) |
| C17—O20—C21 | 116.75 (12) | F3—C6—C3 | 111.98 (12) |
| N2—N1—C5 | 112.22 (11) | F1—C6—C3 | 112.21 (12) |
| N2—N1—C14 | 118.35 (11) | F2—C6—C3 | 112.80 (13) |
| C5—N1—C14 | 129.25 (11) | C11—C12—C7 | 120.73 (13) |
| C11—C10—C9 | 121.85 (13) | C11—C12—H12 | 119.6 |
| C11—C10—Cl13 | 118.59 (11) | C7—C12—H12 | 119.6 |
| C9—C10—Cl13 | 119.55 (11) | C5—C4—C3 | 104.46 (12) |
| N1—C5—C4 | 106.47 (11) | C5—C4—H4 | 127.8 |
| N1—C5—C7 | 124.14 (12) | C3—C4—H4 | 127.8 |
| C4—C5—C7 | 128.73 (12) | C7—C8—C9 | 120.73 (13) |
| C19—C18—C17 | 120.10 (13) | C7—C8—H8 | 119.6 |
| C19—C18—H18 | 120.0 | C9—C8—H8 | 119.6 |
| C17—C18—H18 | 120.0 | N2—C3—C4 | 113.05 (12) |
| C10—C11—C12 | 118.89 (13) | N2—C3—C6 | 118.86 (12) |
| C10—C11—H11 | 120.6 | C4—C3—C6 | 127.88 (13) |
| C12—C11—H11 | 120.6 | C14—C15—C16 | 119.99 (13) |
| C18—C19—C14 | 119.66 (12) | C14—C15—H15 | 120.0 |
| C18—C19—H19 | 120.2 | C16—C15—H15 | 120.0 |
| C14—C19—H19 | 120.2 | O20—C17—C16 | 124.47 (13) |
| C8—C7—C12 | 119.10 (12) | O20—C17—C18 | 115.32 (13) |
| C8—C7—C5 | 120.19 (12) | C16—C17—C18 | 120.21 (13) |
| C12—C7—C5 | 120.48 (12) | C10—C9—C8 | 118.70 (13) |
| C15—C14—C19 | 120.45 (13) | C10—C9—H9 | 120.6 |
| C15—C14—N1 | 119.79 (12) | C8—C9—H9 | 120.6 |
| C19—C14—N1 | 119.76 (12) | O20—C21—H21A | 109.5 |
| C17—C16—C15 | 119.53 (13) | O20—C21—H21B | 109.5 |
| C17—C16—H16 | 120.2 | H21A—C21—H21B | 109.5 |
| C15—C16—H16 | 120.2 | O20—C21—H21C | 109.5 |
| F3—C6—F1 | 106.49 (12) | H21A—C21—H21C | 109.5 |
| F3—C6—F2 | 106.86 (12) | H21B—C21—H21C | 109.5 |
| C3—N2—N1—C5 | −0.10 (14) | C12—C7—C8—C9 | 0.1 (2) |
| C3—N2—N1—C14 | 175.45 (11) | C5—C7—C8—C9 | −174.46 (13) |
| N2—N1—C5—C4 | 0.92 (15) | N1—N2—C3—C4 | −0.79 (15) |
| C14—N1—C5—C4 | −174.02 (12) | N1—N2—C3—C6 | 174.38 (12) |
| N2—N1—C5—C7 | −170.48 (12) | C5—C4—C3—N2 | 1.34 (16) |
| C14—N1—C5—C7 | 14.6 (2) | C5—C4—C3—C6 | −173.29 (13) |
| C9—C10—C11—C12 | 0.3 (2) | F3—C6—C3—N2 | 143.23 (13) |
| Cl13—C10—C11—C12 | −178.35 (11) | F1—C6—C3—N2 | −97.07 (16) |
| C17—C18—C19—C14 | −2.7 (2) | F2—C6—C3—N2 | 22.64 (18) |
| N1—C5—C7—C8 | −147.72 (14) | F3—C6—C3—C4 | −42.4 (2) |
| C4—C5—C7—C8 | 42.9 (2) | F1—C6—C3—C4 | 77.29 (18) |
| N1—C5—C7—C12 | 37.8 (2) | F2—C6—C3—C4 | −163.00 (14) |
| C4—C5—C7—C12 | −131.61 (15) | C19—C14—C15—C16 | 1.4 (2) |
| C18—C19—C14—C15 | 1.0 (2) | N1—C14—C15—C16 | −178.04 (12) |
| C18—C19—C14—N1 | −179.52 (12) | C17—C16—C15—C14 | −2.2 (2) |
| N2—N1—C14—C15 | 45.45 (17) | C21—O20—C17—C16 | 0.4 (2) |
| C5—N1—C14—C15 | −139.87 (14) | C21—O20—C17—C18 | 179.96 (14) |
| N2—N1—C14—C19 | −134.02 (13) | C15—C16—C17—O20 | 180.00 (13) |
| C5—N1—C14—C19 | 40.7 (2) | C15—C16—C17—C18 | 0.5 (2) |
| C10—C11—C12—C7 | −0.6 (2) | C19—C18—C17—O20 | −177.60 (12) |
| C8—C7—C12—C11 | 0.4 (2) | C19—C18—C17—C16 | 2.0 (2) |
| C5—C7—C12—C11 | 174.95 (13) | C11—C10—C9—C8 | 0.2 (2) |
| N1—C5—C4—C3 | −1.30 (15) | Cl13—C10—C9—C8 | 178.84 (11) |
| C7—C5—C4—C3 | 169.57 (13) | C7—C8—C9—C10 | −0.4 (2) |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: PV2130).
References
- Allen, F. H. (2002). Acta Cryst. B58, 380–388. [DOI] [PubMed]
- Charlier, C., Norberg, B., Goossens, L., Hénichart, J.-P. & Durant, F. (2004). Acta Cryst. C60, o648–o652. [DOI] [PubMed]
- Choi, S. H., Langenbach, R. & Bosetti, F. (2008). J. Fed. Am. Soc. Exp. Biol.22, 1491-1501. [DOI] [PMC free article] [PubMed]
- Cusimano, A., Fodera, D., D’Alessandro, N., Lampiasi, N., Azzolina, A., Montalto, G. & Cervello, M. (2007). Cancer Biol. Ther.6, 1461–1468. [DOI] [PubMed]
- Hooft, R. (1998). COLLECT Nonius BV, Delft, The Netherlands.
- Kundu, N. & Fulton, A. M. (2002). Cancer Res.62, 2343–2346. [PubMed]
- Norris, T., Colon-Cruz, R. & Ripin, D. H. (2005). Org. Biomol. Chem.3, 1844–1849. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods in Enzymology, Vol. 276, Macromolecular Crystallography, Part A, edited by C. W. Carter Jr & R. M. Sweet, pp. 307–326. New York: Academic Press.
- Penning, T. D., Talley, J. J., Bertenshaw, S. R., Carter, J. S., Collins, P. W., Docter, S., Graneto, M. J., Lee, L. F., Malecha, J. W., Miyashiro, J. M., Rogers, R. S., Rogier, D. J., Yu, S. S., Anderson, G. D., Burton, E. G., Cogburn, J. N., Gregory, S. A., Koboldt, C. M., Perkins, W. E., Seibert, K., Veenhuizen, A. W., Zhang, Y. Y. & Isakson, P. C. (1997). J. Med. Chem.40, 1347–1365. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Smith, W. L., DeWitt, D. L. & Garavito, R. M. (2000). Annu. Rev. Biochem.69, 145–182. [DOI] [PubMed]
- Sonar, V. N., Parkin, S. & Crooks, P. A. (2004). Acta Cryst. C60, o547–o549. [DOI] [PubMed]
- Teng, X. W., Abu-Mellal, A. K. & Davies, N. M. (2003). J. Pharm. Pharm. Sci.6, 205–210. [PubMed]
- Tiano, H. F., Loftin, C. D., Akunda, J., Lee, C. A., Spalding, J., Sessoms, A., Dunson, D. B., Rogan, E. G., Morham, S. G., Smart, R. C. & Langenbach, R. (2002). Cancer Res.62, 3395–3401. [PubMed]
- Zhu, H.-J., Wang, D.-D. & Ma, J. (2004). Acta Cryst. E60, o2144–o2146.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809001779/pv2130sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809001779/pv2130Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report