Abstract
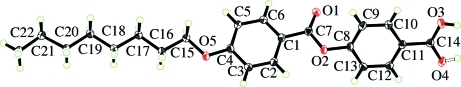
The title compound, C22H26O5, is an important intermediate for the synthesis of side-chain ligands for polymeric liquid crystals. The octyl group is coplanar with the central C6O moiety, where the maximum deviation of a C atom in the octyl group from the C6O plane is 0.161 (5) Å. The crystal structure is stabilized by intermolecular O—H⋯O hydrogen bonds.
Related literature
For studies of aromatic carboxylic acids and their applications, see: Naoum et al. (2008 ▶); Nazir et al. (2008a
▶,b
▶); Gabert et al. (2006 ▶); Aranzazu et al. (2006 ▶); Hussain et al. (2005 ▶); Shafiq et al. (2005 ▶); Ahmad et al. (2003 ▶); Ribeiro et al. (2008 ▶); Hameed & Rama (2004 ▶); For related structures, see: Muhammad et al. (2008 ▶); Hartung et al. (1997 ▶)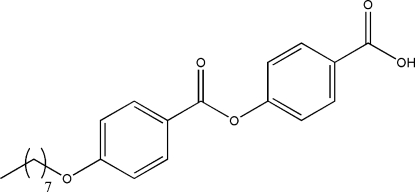
Experimental
Crystal data
C22H26O5
M r = 370.43
Monoclinic,
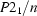
a = 13.528 (8) Å
b = 7.245 (4) Å
c = 20.903 (12) Å
β = 111.407 (8)°
V = 1907.5 (18) Å3
Z = 4
Mo Kα radiation
μ = 0.09 mm−1
T = 123 K
0.40 × 0.30 × 0.15 mm
Data collection
Rigaku/MSC Mercury CCD diffractometer
Absorption correction: none
14669 measured reflections
4358 independent reflections
3870 reflections with I > 2σ(I)
R int = 0.048
Refinement
R[F 2 > 2σ(F 2)] = 0.081
wR(F 2) = 0.138
S = 1.31
4358 reflections
247 parameters
H-atom parameters constrained
Δρmax = 0.28 e Å−3
Δρmin = −0.19 e Å−3
Data collection: CrystalClear (Molecular Structure Corporation & Rigaku, 2001 ▶); cell refinement: CrystalClear; data reduction: TEXSAN (Molecular Structure Corporation & Rigaku, 2004 ▶); program(s) used to solve structure: SIR97 (Altomare et al., 1999 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEPII (Johnson, 1976 ▶); software used to prepare material for publication: SHELXL97 and TEXSAN.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809003298/hg2473sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809003298/hg2473Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O3—H3O⋯O4i | 0.84 | 1.85 | 2.659 (3) | 161 |
| O4—H4O⋯O3i | 0.84 | 1.83 | 2.659 (3) | 171 |
Symmetry code: (i) 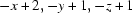 .
.
Acknowledgments
MKR is grateful to the Higher Education Commission of Pakistan for financial support under the International Support Initiative Programme for a doctoral fellowship at Gifu University, Japan.
supplementary crystallographic information
Comment
Aromatic carboxylic acids bearing different substituents have been investigated for their liquid crystalline properties (Naoum et al., 2008; Nazir et al., 2008a,b). Such acids have been utilized in the synthesis of both side-chain (Gabert et al., 2006) and main-chain (Aranzazu et al., 2006) liquid crystal polymers. In addition to their use as intermediates in the synthesis of a large number of organic compounds (Hussain et al., 2005; Shafiq et al., 2005; Ahmad et al., 2003), carboxylic acids has also been used in the pharmaceutical industry (Ribeiro et al., 2008; Hameed & Rama 2004). The title compound (I) was synthesized as an intermediate in the synthesis of side-chain liquid crystal polymers. In the present report, the crystal structure of (I) is being presented. Bond lengths and angles are within the normal ranges as given for benzoyloxybenzoic acids (Muhammad et al., 2008; Hartung et al., 1997). The C7—O1 and C7—O2 bond lengths are 1.205 (3) and 1.369 (3) Å, respectively, that reflect their double and single bond character. The very similar bond lengths of C14—O4 and C14—O3, 1.287 (3) and 1.261 (3) Å, are due to disorder of CO2H moiety. The octyl group is coplanar with the central C6O moiety where the max dviation of C atom in octyl group from the C6O moiety is 0.161 (5) Å. Two molecules related by an inversion center form a dimer via two hydrogen bonds composed of two carboxyl groups as shown in Fig. 2.
Experimental
To a solution of 4-hydroxybenzaldehyde (0.032 moles) in 50 ml of triethylamine (TEA), was added an equivalent amount of 4-octyloxybenzoylchloride with stirring and the mixture heated at 60°C for 1 h. The excess TEA was removed in vacuo and the product, after recrystallization from hot ethanol, was subjected to KMnO4 oxidation.The 4-(4-octyloxybenzoyloxy)benzaldehyde (0.025 moles) was dissolved in acetone (100 ml) and aqueous KMnO4 (0.025 moles) was added dropwise at room temperature with stirring. The stirring was continued for three hours when the reaction mixture was filtered and the filtrate acidified using 6M HCl. The precipitated product was purified by recrystallization from acetone.
Refinement
The O-bound H atom was refined isotropically. All the other H atoms were placed in idealized positions and treated as riding atoms, with C—H distance in the range 0.95–0.99 Å and Uiso(H) = 1.2Ueq(C) or 1.5Ueq(C).
Figures
Fig. 1.
Molecular structure of (I) showing atom-labelling scheme and displacement ellipsoids at the 30% probability level.
Fig. 2.
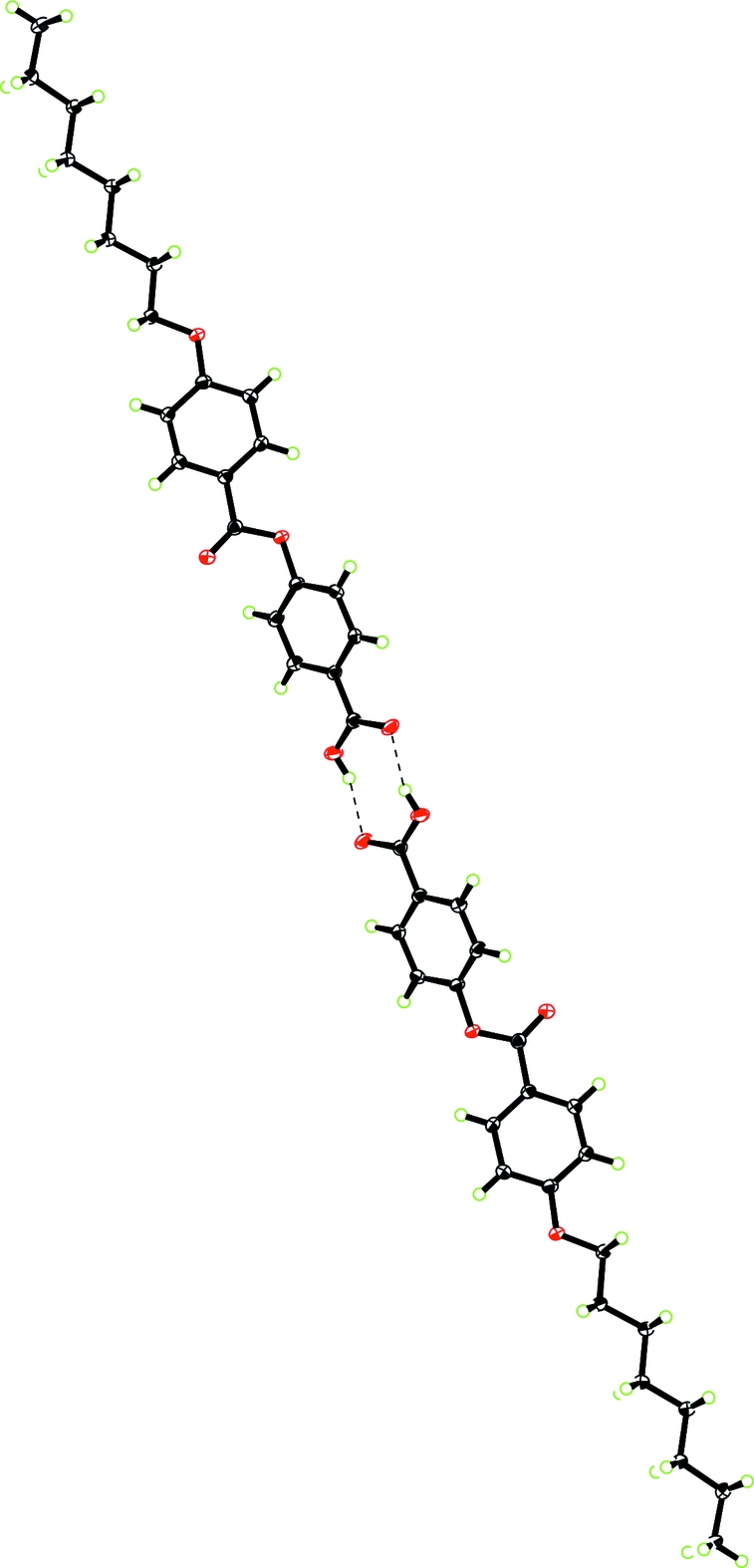
Showing hydrogen bonded molecules through O—H···O.
Crystal data
| C22H26O5 | F(000) = 792 |
| Mr = 370.43 | Dx = 1.290 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71070 Å |
| Hall symbol: -P 2yn | Cell parameters from 4339 reflections |
| a = 13.528 (8) Å | θ = 3.0–27.5° |
| b = 7.245 (4) Å | µ = 0.09 mm−1 |
| c = 20.903 (12) Å | T = 123 K |
| β = 111.407 (8)° | Chip, colourless |
| V = 1907.5 (18) Å3 | 0.40 × 0.30 × 0.15 mm |
| Z = 4 |
Data collection
| Rigaku/MSC Mercury CCD diffractometer | 3870 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.048 |
| graphite | θmax = 27.5°, θmin = 3.2° |
| Detector resolution: 14.62 pixels mm-1 | h = −17→15 |
| ω scans | k = −7→9 |
| 14669 measured reflections | l = −25→27 |
| 4358 independent reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.081 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.138 | H-atom parameters constrained |
| S = 1.31 | w = 1/[σ2(Fo2) + (0.0263P)2 + 1.5055P] where P = (Fo2 + 2Fc2)/3 |
| 4358 reflections | (Δ/σ)max < 0.001 |
| 247 parameters | Δρmax = 0.28 e Å−3 |
| 0 restraints | Δρmin = −0.19 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| C1 | 0.22298 (16) | 0.5733 (3) | 0.30577 (10) | 0.0181 (4) | |
| C2 | 0.19154 (16) | 0.5148 (3) | 0.23723 (10) | 0.0218 (5) | |
| H2 | 0.2435 | 0.4795 | 0.2189 | 0.026* | |
| C3 | 0.08502 (17) | 0.5086 (3) | 0.19644 (11) | 0.0239 (5) | |
| H3 | 0.0638 | 0.4695 | 0.1500 | 0.029* | |
| C4 | 0.00858 (16) | 0.5596 (3) | 0.22337 (10) | 0.0202 (5) | |
| C5 | 0.03873 (16) | 0.6186 (3) | 0.29139 (10) | 0.0204 (5) | |
| H5 | −0.0132 | 0.6530 | 0.3099 | 0.024* | |
| C6 | 0.14626 (17) | 0.6261 (3) | 0.33158 (10) | 0.0211 (5) | |
| H6 | 0.1676 | 0.6683 | 0.3777 | 0.025* | |
| C7 | 0.33624 (16) | 0.5860 (3) | 0.35119 (10) | 0.0189 (4) | |
| O1 | 0.36934 (12) | 0.6558 (2) | 0.40736 (7) | 0.0245 (4) | |
| O2 | 0.40082 (11) | 0.5073 (2) | 0.32137 (7) | 0.0231 (4) | |
| C8 | 0.51107 (16) | 0.5074 (3) | 0.35729 (10) | 0.0198 (5) | |
| C9 | 0.55644 (17) | 0.4315 (3) | 0.42229 (11) | 0.0218 (5) | |
| H9 | 0.5133 | 0.3826 | 0.4453 | 0.026* | |
| C10 | 0.66633 (16) | 0.4284 (3) | 0.45304 (11) | 0.0201 (4) | |
| H10 | 0.6987 | 0.3783 | 0.4979 | 0.024* | |
| C11 | 0.72990 (16) | 0.4980 (3) | 0.41892 (10) | 0.0174 (4) | |
| C12 | 0.68191 (16) | 0.5711 (3) | 0.35287 (10) | 0.0191 (4) | |
| H12 | 0.7246 | 0.6174 | 0.3291 | 0.023* | |
| C13 | 0.57211 (17) | 0.5762 (3) | 0.32203 (10) | 0.0210 (5) | |
| H13 | 0.5392 | 0.6262 | 0.2772 | 0.025* | |
| C14 | 0.84738 (16) | 0.4969 (3) | 0.45311 (10) | 0.0182 (4) | |
| O3 | 0.88859 (12) | 0.4343 (2) | 0.51339 (7) | 0.0257 (4) | |
| H3O | 0.9550 | 0.4364 | 0.5257 | 0.038* | 0.50 |
| O4 | 0.90241 (12) | 0.5614 (2) | 0.41929 (8) | 0.0258 (4) | |
| H4O | 0.9671 | 0.5599 | 0.4443 | 0.039* | 0.50 |
| O5 | −0.09388 (11) | 0.5459 (2) | 0.17868 (7) | 0.0249 (4) | |
| C15 | −0.17837 (16) | 0.5852 (3) | 0.20264 (10) | 0.0205 (5) | |
| H15A | −0.1738 | 0.7146 | 0.2188 | 0.025* | |
| H15B | −0.1743 | 0.5023 | 0.2412 | 0.025* | |
| C16 | −0.28096 (16) | 0.5541 (3) | 0.14232 (10) | 0.0196 (4) | |
| H16A | −0.2839 | 0.4239 | 0.1273 | 0.024* | |
| H16B | −0.2814 | 0.6332 | 0.1036 | 0.024* | |
| C17 | −0.37919 (16) | 0.5958 (3) | 0.15880 (10) | 0.0214 (5) | |
| H17A | −0.3764 | 0.7258 | 0.1740 | 0.026* | |
| H17B | −0.3792 | 0.5161 | 0.1973 | 0.026* | |
| C18 | −0.48182 (16) | 0.5646 (3) | 0.09742 (10) | 0.0212 (5) | |
| H18A | −0.4790 | 0.6383 | 0.0582 | 0.025* | |
| H18B | −0.4857 | 0.4329 | 0.0841 | 0.025* | |
| C19 | −0.58294 (16) | 0.6147 (3) | 0.10966 (10) | 0.0219 (5) | |
| H19A | −0.5865 | 0.5417 | 0.1488 | 0.026* | |
| H19B | −0.5806 | 0.7470 | 0.1221 | 0.026* | |
| C20 | −0.68233 (16) | 0.5782 (3) | 0.04655 (10) | 0.0205 (5) | |
| H20A | −0.6752 | 0.6432 | 0.0068 | 0.025* | |
| H20B | −0.6864 | 0.4443 | 0.0364 | 0.025* | |
| C21 | −0.78576 (17) | 0.6380 (4) | 0.05333 (11) | 0.0264 (5) | |
| H21A | −0.7825 | 0.7718 | 0.0635 | 0.032* | |
| H21B | −0.7944 | 0.5718 | 0.0924 | 0.032* | |
| C22 | −0.88171 (17) | 0.5991 (3) | −0.01178 (12) | 0.0261 (5) | |
| H22A | −0.8708 | 0.6557 | −0.0513 | 0.039* | |
| H22B | −0.9456 | 0.6510 | −0.0070 | 0.039* | |
| H22C | −0.8904 | 0.4654 | −0.0189 | 0.039* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0150 (10) | 0.0197 (11) | 0.0193 (9) | −0.0018 (8) | 0.0060 (8) | −0.0005 (8) |
| C2 | 0.0153 (10) | 0.0294 (13) | 0.0218 (10) | 0.0000 (9) | 0.0080 (8) | −0.0036 (9) |
| C3 | 0.0195 (11) | 0.0332 (13) | 0.0178 (10) | −0.0005 (10) | 0.0055 (8) | −0.0049 (9) |
| C4 | 0.0153 (10) | 0.0229 (12) | 0.0202 (10) | −0.0004 (9) | 0.0039 (8) | −0.0010 (9) |
| C5 | 0.0156 (10) | 0.0266 (12) | 0.0201 (10) | 0.0003 (9) | 0.0078 (8) | 0.0002 (8) |
| C6 | 0.0190 (11) | 0.0273 (12) | 0.0170 (10) | 0.0002 (9) | 0.0066 (8) | −0.0008 (8) |
| C7 | 0.0176 (10) | 0.0195 (11) | 0.0204 (10) | −0.0012 (9) | 0.0080 (8) | 0.0012 (8) |
| O1 | 0.0180 (8) | 0.0345 (10) | 0.0195 (7) | −0.0007 (7) | 0.0049 (6) | −0.0042 (7) |
| O2 | 0.0122 (7) | 0.0339 (10) | 0.0212 (7) | 0.0011 (7) | 0.0037 (6) | −0.0064 (7) |
| C8 | 0.0124 (10) | 0.0240 (12) | 0.0208 (10) | 0.0011 (9) | 0.0035 (8) | −0.0050 (9) |
| C9 | 0.0169 (10) | 0.0244 (12) | 0.0255 (10) | −0.0020 (9) | 0.0095 (8) | −0.0002 (9) |
| C10 | 0.0174 (10) | 0.0210 (11) | 0.0210 (10) | 0.0011 (9) | 0.0059 (8) | 0.0006 (9) |
| C11 | 0.0153 (10) | 0.0159 (11) | 0.0206 (9) | 0.0005 (8) | 0.0060 (8) | −0.0026 (8) |
| C12 | 0.0184 (10) | 0.0219 (11) | 0.0179 (9) | 0.0005 (9) | 0.0077 (8) | −0.0030 (8) |
| C13 | 0.0191 (10) | 0.0266 (12) | 0.0151 (9) | 0.0021 (9) | 0.0036 (8) | −0.0026 (8) |
| C14 | 0.0180 (10) | 0.0172 (10) | 0.0200 (9) | 0.0004 (9) | 0.0075 (8) | −0.0022 (8) |
| O3 | 0.0160 (7) | 0.0338 (10) | 0.0232 (8) | 0.0005 (7) | 0.0024 (6) | 0.0036 (7) |
| O4 | 0.0140 (7) | 0.0347 (10) | 0.0291 (8) | −0.0015 (7) | 0.0084 (6) | 0.0029 (7) |
| O5 | 0.0130 (7) | 0.0395 (10) | 0.0206 (7) | 0.0009 (7) | 0.0042 (6) | −0.0046 (7) |
| C15 | 0.0158 (10) | 0.0259 (12) | 0.0203 (10) | 0.0000 (9) | 0.0071 (8) | 0.0012 (9) |
| C16 | 0.0154 (10) | 0.0211 (11) | 0.0205 (10) | 0.0005 (9) | 0.0045 (8) | −0.0001 (8) |
| C17 | 0.0162 (10) | 0.0266 (12) | 0.0194 (10) | 0.0008 (9) | 0.0042 (8) | −0.0015 (9) |
| C18 | 0.0170 (10) | 0.0253 (12) | 0.0189 (10) | −0.0003 (9) | 0.0038 (8) | −0.0007 (9) |
| C19 | 0.0173 (10) | 0.0267 (13) | 0.0201 (10) | −0.0004 (9) | 0.0050 (8) | −0.0019 (9) |
| C20 | 0.0158 (10) | 0.0248 (12) | 0.0198 (10) | −0.0002 (9) | 0.0050 (8) | 0.0003 (9) |
| C21 | 0.0191 (11) | 0.0376 (14) | 0.0231 (11) | 0.0044 (10) | 0.0084 (9) | 0.0004 (10) |
| C22 | 0.0152 (10) | 0.0282 (13) | 0.0333 (12) | 0.0005 (9) | 0.0068 (9) | −0.0001 (10) |
Geometric parameters (Å, °)
| C1—C6 | 1.387 (3) | O3—H3O | 0.8400 |
| C1—C2 | 1.403 (3) | O4—H4O | 0.8400 |
| C1—C7 | 1.482 (3) | O5—C15 | 1.434 (3) |
| C2—C3 | 1.382 (3) | C15—C16 | 1.513 (3) |
| C2—H2 | 0.9500 | C15—H15A | 0.9900 |
| C3—C4 | 1.396 (3) | C15—H15B | 0.9900 |
| C3—H3 | 0.9500 | C16—C17 | 1.520 (3) |
| C4—O5 | 1.363 (2) | C16—H16A | 0.9900 |
| C4—C5 | 1.396 (3) | C16—H16B | 0.9900 |
| C5—C6 | 1.390 (3) | C17—C18 | 1.525 (3) |
| C5—H5 | 0.9500 | C17—H17A | 0.9900 |
| C6—H6 | 0.9500 | C17—H17B | 0.9900 |
| C7—O1 | 1.205 (3) | C18—C19 | 1.525 (3) |
| C7—O2 | 1.369 (3) | C18—H18A | 0.9900 |
| O2—C8 | 1.404 (2) | C18—H18B | 0.9900 |
| C8—C9 | 1.384 (3) | C19—C20 | 1.524 (3) |
| C8—C13 | 1.385 (3) | C19—H19A | 0.9900 |
| C9—C10 | 1.388 (3) | C19—H19B | 0.9900 |
| C9—H9 | 0.9500 | C20—C21 | 1.520 (3) |
| C10—C11 | 1.397 (3) | C20—H20A | 0.9900 |
| C10—H10 | 0.9500 | C20—H20B | 0.9900 |
| C11—C12 | 1.398 (3) | C21—C22 | 1.526 (3) |
| C11—C14 | 1.486 (3) | C21—H21A | 0.9900 |
| C12—C13 | 1.387 (3) | C21—H21B | 0.9900 |
| C12—H12 | 0.9500 | C22—H22A | 0.9800 |
| C13—H13 | 0.9500 | C22—H22B | 0.9800 |
| C14—O3 | 1.261 (3) | C22—H22C | 0.9800 |
| C14—O4 | 1.287 (3) | ||
| C6—C1—C2 | 119.32 (19) | O5—C15—H15A | 110.4 |
| C6—C1—C7 | 118.64 (19) | C16—C15—H15A | 110.4 |
| C2—C1—C7 | 122.02 (19) | O5—C15—H15B | 110.4 |
| C3—C2—C1 | 119.9 (2) | C16—C15—H15B | 110.4 |
| C3—C2—H2 | 120.0 | H15A—C15—H15B | 108.6 |
| C1—C2—H2 | 120.0 | C15—C16—C17 | 113.15 (18) |
| C2—C3—C4 | 120.1 (2) | C15—C16—H16A | 108.9 |
| C2—C3—H3 | 119.9 | C17—C16—H16A | 108.9 |
| C4—C3—H3 | 119.9 | C15—C16—H16B | 108.9 |
| O5—C4—C5 | 124.50 (19) | C17—C16—H16B | 108.9 |
| O5—C4—C3 | 114.96 (19) | H16A—C16—H16B | 107.8 |
| C5—C4—C3 | 120.54 (19) | C16—C17—C18 | 112.50 (18) |
| C6—C5—C4 | 118.7 (2) | C16—C17—H17A | 109.1 |
| C6—C5—H5 | 120.7 | C18—C17—H17A | 109.1 |
| C4—C5—H5 | 120.7 | C16—C17—H17B | 109.1 |
| C1—C6—C5 | 121.39 (19) | C18—C17—H17B | 109.1 |
| C1—C6—H6 | 119.3 | H17A—C17—H17B | 107.8 |
| C5—C6—H6 | 119.3 | C19—C18—C17 | 114.88 (18) |
| O1—C7—O2 | 123.14 (19) | C19—C18—H18A | 108.5 |
| O1—C7—C1 | 125.5 (2) | C17—C18—H18A | 108.5 |
| O2—C7—C1 | 111.40 (17) | C19—C18—H18B | 108.5 |
| C7—O2—C8 | 119.08 (16) | C17—C18—H18B | 108.5 |
| C9—C8—C13 | 121.90 (19) | H18A—C18—H18B | 107.5 |
| C9—C8—O2 | 121.9 (2) | C20—C19—C18 | 112.06 (18) |
| C13—C8—O2 | 116.03 (18) | C20—C19—H19A | 109.2 |
| C8—C9—C10 | 118.5 (2) | C18—C19—H19A | 109.2 |
| C8—C9—H9 | 120.8 | C20—C19—H19B | 109.2 |
| C10—C9—H9 | 120.8 | C18—C19—H19B | 109.2 |
| C9—C10—C11 | 120.9 (2) | H19A—C19—H19B | 107.9 |
| C9—C10—H10 | 119.6 | C21—C20—C19 | 114.91 (18) |
| C11—C10—H10 | 119.6 | C21—C20—H20A | 108.5 |
| C10—C11—C12 | 119.38 (19) | C19—C20—H20A | 108.5 |
| C10—C11—C14 | 120.15 (19) | C21—C20—H20B | 108.5 |
| C12—C11—C14 | 120.46 (19) | C19—C20—H20B | 108.5 |
| C13—C12—C11 | 120.1 (2) | H20A—C20—H20B | 107.5 |
| C13—C12—H12 | 119.9 | C20—C21—C22 | 112.13 (19) |
| C11—C12—H12 | 119.9 | C20—C21—H21A | 109.2 |
| C8—C13—C12 | 119.24 (19) | C22—C21—H21A | 109.2 |
| C8—C13—H13 | 120.4 | C20—C21—H21B | 109.2 |
| C12—C13—H13 | 120.4 | C22—C21—H21B | 109.2 |
| O3—C14—O4 | 123.08 (19) | H21A—C21—H21B | 107.9 |
| O3—C14—C11 | 119.19 (19) | C21—C22—H22A | 109.5 |
| O4—C14—C11 | 117.73 (18) | C21—C22—H22B | 109.5 |
| C14—O3—H3O | 109.5 | H22A—C22—H22B | 109.5 |
| C14—O4—H4O | 109.5 | C21—C22—H22C | 109.5 |
| C4—O5—C15 | 119.13 (17) | H22A—C22—H22C | 109.5 |
| O5—C15—C16 | 106.55 (17) | H22B—C22—H22C | 109.5 |
| C6—C1—C2—C3 | −0.6 (3) | C9—C10—C11—C12 | −0.2 (3) |
| C7—C1—C2—C3 | −178.8 (2) | C9—C10—C11—C14 | 178.9 (2) |
| C1—C2—C3—C4 | −0.3 (4) | C10—C11—C12—C13 | 0.8 (3) |
| C2—C3—C4—O5 | −179.2 (2) | C14—C11—C12—C13 | −178.3 (2) |
| C2—C3—C4—C5 | 0.5 (4) | C9—C8—C13—C12 | −0.8 (3) |
| O5—C4—C5—C6 | 179.9 (2) | O2—C8—C13—C12 | −176.44 (19) |
| C3—C4—C5—C6 | 0.2 (3) | C11—C12—C13—C8 | −0.3 (3) |
| C2—C1—C6—C5 | 1.4 (3) | C10—C11—C14—O3 | −0.8 (3) |
| C7—C1—C6—C5 | 179.6 (2) | C12—C11—C14—O3 | 178.3 (2) |
| C4—C5—C6—C1 | −1.2 (3) | C10—C11—C14—O4 | 179.5 (2) |
| C6—C1—C7—O1 | −7.8 (4) | C12—C11—C14—O4 | −1.4 (3) |
| C2—C1—C7—O1 | 170.4 (2) | C5—C4—O5—C15 | −3.2 (3) |
| C6—C1—C7—O2 | 172.24 (19) | C3—C4—O5—C15 | 176.5 (2) |
| C2—C1—C7—O2 | −9.6 (3) | C4—O5—C15—C16 | −178.78 (19) |
| O1—C7—O2—C8 | −0.1 (3) | O5—C15—C16—C17 | −178.28 (18) |
| C1—C7—O2—C8 | 179.84 (18) | C15—C16—C17—C18 | 179.72 (19) |
| C7—O2—C8—C9 | 56.4 (3) | C16—C17—C18—C19 | −176.7 (2) |
| C7—O2—C8—C13 | −128.0 (2) | C17—C18—C19—C20 | −179.4 (2) |
| C13—C8—C9—C10 | 1.4 (3) | C18—C19—C20—C21 | −175.8 (2) |
| O2—C8—C9—C10 | 176.7 (2) | C19—C20—C21—C22 | 179.5 (2) |
| C8—C9—C10—C11 | −0.8 (3) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O3—H3O···O4i | 0.84 | 1.85 | 2.659 (3) | 161 |
| O4—H4O···O3i | 0.84 | 1.83 | 2.659 (3) | 171 |
Symmetry codes: (i) −x+2, −y+1, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HG2473).
References
- Ahmad, H. B., Rama, N. H., Hussain, M., Hussain, M. T., Qasim, M. M., Hameed, S., Malana, M. A. & Malik, A. (2003). Indian J. Chem. Sect. B, 42, 611–615.
- Altomare, A., Burla, M. C., Camalli, M., Cascarano, G. L., Giacovazzo, C., Guagliardi, A., Moliterni, A. G. G., Polidori, G. & Spagna, R. (1999). J. Appl. Cryst.32, 115–119.
- Aranzazu, M.-G., Ernesto, P. & Antonio, B. (2006). Polymer, 47, 2080–2090.
- Gabert, A. J., Verploegen, E., Hammond, P. T. & Schrock, R. R. (2006). Macromolecules, 39, 3993–4000.
- Hameed, S. & Rama, N. H. (2004). J. Chem. Soc. Pak.26, 157–162.
- Hartung, H., Hoffmann, F. & Weissflog, W. (1997). J. Mol. Struct.415, 205–214.
- Hussain, M. T., Rama, N. H., Hameed, S., Malik, A. & Khan, K. M. (2005). Nat. Prod. Res.19, 41–51. [DOI] [PubMed]
- Johnson, C. K. (1976). ORTEPII Report ORNL-5138. Oak Ridge National Laboratory, Tennessee, USA.
- Molecular Structure Corporation & Rigaku (2001). CrystalClear MSC, The Woodlands, Texas, USA, and Rigaku Corporation, Tokyo, Japan.
- Molecular Structure Corporation & Rigaku (2004). TEXSAN MSC, The Woodlands, Texas, USA, and Rigaku Corporation, Tokyo, Japan.
- Muhammad, K., Khawar Rauf, M., Ebihara, M. & Hameed, S. (2008). Acta Cryst. E64, o1251. [DOI] [PMC free article] [PubMed]
- Naoum, M. M., Fahmi, A. A. & Alaasar, M. A. (2008). Mol. Cryst. Liq. Cryst.487, 74–91.
- Nazir, S., Khawar Rauf, M., Ebihara, M. & Hameed, S. (2008a). Acta Cryst. E64, o423. [DOI] [PMC free article] [PubMed]
- Nazir, S., Muhammad, K., Khawar Rauf, M., Ebihara, M. & Hameed, S. (2008b). Acta Cryst. E64, o1013. [DOI] [PMC free article] [PubMed]
- Ribeiro, G., Benadiba, M., Colquhoun, A. & Silva, D. D. (2008). Polyhedron, 27, 1131–1137.
- Shafiq, Z., Arfan, M., Rama, N. H., Hameed, S., Abbas, G. & Hussain, M. T. (2005). Turk. J. Chem.29, 321–325.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809003298/hg2473sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809003298/hg2473Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report