Abstract
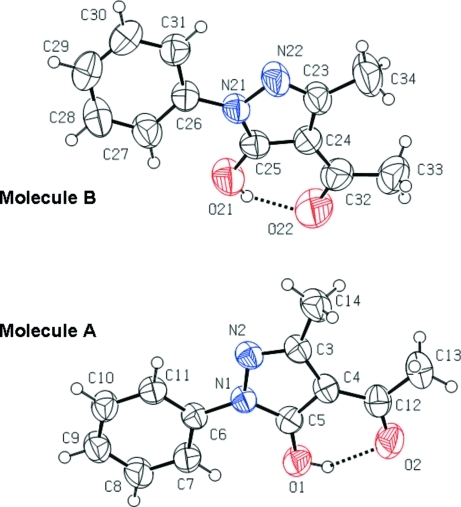
The title compound, C12H12N2O2, crystallized in the monolinic space group P21/n, with two independent molecules (A and B) in the asymmetric unit. This is in contrast to the first monoclinic polymorph reported [Cingolani et al. (2002 ▶). Inorg. Chem. 41, 1151–116], which crystallized in the space group C2/c with one independent molecule per asymmetric unit. The dihedral angles between the two rings differ slightly; in molecule A it is 4.90 (11)° and in molecule B it is 16.05 (13)°. In both molecules, there is an intramolecular O—H⋯O hydrogen bond involving the hydroxyl substituent and the carbonyl O atom of the adjacent acetyl group. In the crystal structure, molecules A and B are linked via a C—H⋯N interaction. There are also some weak C—H⋯π interactions involving the phenyl ring of molecule A and H atoms of the acetyl groups of both molecules.
Related literature
For early literature on pyrazoles, see: Knorr (1883 ▶). For information on the pharamceutical properties of pyrazoles, see: Grimmett (1970 ▶). For the monoclinic C2/c polymorph of the title compound, see: Cingolani et al. (2002 ▶).
Experimental
Crystal data
C12H12N2O2
M r = 216.24
Monoclinic,

a = 13.8735 (7) Å
b = 9.2037 (4) Å
c = 18.3702 (8) Å
β = 110.100 (2)°
V = 2202.78 (18) Å3
Z = 8
Mo Kα radiation
μ = 0.09 mm−1
T = 296 (2) K
0.34 × 0.22 × 0.16 mm
Data collection
Bruker Kappa APEXII CCD diffractometer
Absorption correction: none
24519 measured reflections
5500 independent reflections
2656 reflections with I > 2σ(I)
R int = 0.047
Refinement
R[F 2 > 2σ(F 2)] = 0.051
wR(F 2) = 0.149
S = 0.97
5500 reflections
292 parameters
H-atom parameters constrained
Δρmax = 0.18 e Å−3
Δρmin = −0.14 e Å−3
Data collection: APEX2 (Bruker, 2002 ▶); cell refinement: SAINT (Bruker, 2002 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: PLATON (Spek, 2003 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809001470/wn2305sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809001470/wn2305Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1O⋯O2 | 0.82 | 1.85 | 2.546 (2) | 142 |
| O21—H21O⋯O22 | 0.82 | 1.83 | 2.531 (3) | 142 |
| C8—H8⋯N22i | 0.93 | 2.56 | 3.489 (3) | 177 |
| C13—H13C⋯Cg2ii | 0.96 | 2.66 | 3.533 (3) | 150 |
| C33—H33B⋯Cg2ii | 0.96 | 2.98 | 3.774 (3) | 141 |
Symmetry codes: (i)  ; (ii)
; (ii)  . Cg2 is the centroid of the C6–C11 ring.
. Cg2 is the centroid of the C6–C11 ring.
Acknowledgments
TUS acknowledges the Government College of Education, Afzalpur, Azad Jammu & Kashmir, for granting permission for further studies and providing laboratory facilities.
supplementary crystallographic information
Comment
The history of pyrazoles began already in the late nineteenth century (Knorr, 1883). Pyrazole is isomeric with the biologically important imidazole ring system but, unlike imidazole, has fewer natural derivatives. The ring system is very stable and inert, and interest in such compounds stemmed from their applications as drugs, dyes and as anesthetics. They are also used as antioxidants in fuels but their major applications have been in the pharmaceutical (Grimmett, 1970) and agricultural industries. In view of the importance of pyrazole derivatives we have planned a systematic study of such compounds, and describe here the crystal structure of a new polymorph of the title compound.
It crystallized in the monoclinic space group P21/n, with two independent molecules (A and B) per asymmetric unit (Fig. 1). This is in contrast to an earlier reported monoclinic polymorph, (Cingolani et al., 2002), which crystallized in the space group C2/c with one independent molecule per asymmetric unit. The bond distances and angles in both polymorphs are very similar. The dihedral angles between the two rings differ slightly; in molecule A it is 4.90 (11)° and in molecule B it is 16.05 (13)°. The corresponding value in the other polymorph is 5.33 (10)°.
In both molecules (A and B), there is an intramolecular O—H···O hydrogen bond involving the hydroxyl substituent and the carbonyl O atom of the adjacent acetyl group (Table 1); this feature is also present in the C2/c polymorph. In the crystal structure, molecules A and B are linked via a C—H···N interaction (Fig. 2 and Table 1). There are also some weak C—H···π interactions involving the phenyl ring (centroid Cg2) of molecule A and some H atoms of the acetyl groups of both molecules (Table 1).
Experimental
1-Phenyl-3-methyl-5-pyrazolone (7.5 g) was dissolved by heating in tetrahydrofuran (80 ml). Calcium hydroxide (12 g) was added and acetyl chloride (4 ml) was then added dropwise over a period of 1 min. The temperature increased during the first few minutes and the reaction mixture became a thick paste. This mixture was then refluxed for 30 min. The calcium complex of the title compound that had formed in the flask was decomposed by pouring the mixture into a dilute solution of HCl (100 ml). A dark brownish-red organic layer was obtained which was extracted using dichloromethane. The solvent was then removed by vacuum distillation and the solid obtained was washed with a little water and THF. Crystals of the title compound, suitable for X-ray analysis, were obtained by recrystallization from methanol/water (1:1, v:v).
Refinement
The H atoms were included in calculated positions and treated as riding atoms: O—H = 0.83 Å, C—H = 0.93 - 0.96 Å with Uiso(H) = kUeq(parent atom), where k = 1.2 for aromatic H and 1.5 for all other H atoms. Methyl group C34 undergoes considerable thermal motion but splitting the atom did not improve the situation.
Figures
Fig. 1.
A view of the molecular structure of the two independent molecules (A and B), showing the displacement ellipsoids drawn at the 50% probability level and the intramolecular O—H···O hydrogen bonds as dashed lines. Hydrogen atoms are represented as spheres of arbitrary radius.
Fig. 2.
A view along the b axis of the crystal packing, showing the intramolecular O—H···H hydrogen bonds and the intermolecular C—H···N interactions as dashed lines. H atoms not involved in hydrogen bonding have been omitted for clarity.
Crystal data
| C12H12N2O2 | F(000) = 912 |
| Mr = 216.24 | Dx = 1.304 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2yn | Cell parameters from 4106 reflections |
| a = 13.8735 (7) Å | θ = 2.3–22.4° |
| b = 9.2037 (4) Å | µ = 0.09 mm−1 |
| c = 18.3702 (8) Å | T = 296 K |
| β = 110.100 (2)° | Block, colorless |
| V = 2202.78 (18) Å3 | 0.34 × 0.22 × 0.16 mm |
| Z = 8 |
Data collection
| Bruker Kappa APEXII CCD diffractometer | 2656 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.047 |
| graphite | θmax = 28.6°, θmin = 2.4° |
| φ and ω scans | h = −18→18 |
| 24519 measured reflections | k = −12→12 |
| 5500 independent reflections | l = −24→24 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.051 | H-atom parameters constrained |
| wR(F2) = 0.149 | w = 1/[σ2(Fo2) + (0.0571P)2 + 0.4951P] where P = (Fo2 + 2Fc2)/3 |
| S = 0.97 | (Δ/σ)max = 0.001 |
| 5500 reflections | Δρmax = 0.18 e Å−3 |
| 292 parameters | Δρmin = −0.14 e Å−3 |
| 0 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.0059 (9) |
Special details
| Geometry. Bond distances, angles etc. have been calculated using the rounded fractional coordinates. All su's are estimated from the variances of the (full) variance-covariance matrix. The cell e.s.d.'s are taken into account in the estimation of distances, angles and torsion angles |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.66613 (12) | 0.62373 (18) | −0.03281 (8) | 0.0712 (6) | |
| O2 | 0.53649 (13) | 0.8021 (2) | −0.11913 (9) | 0.0837 (7) | |
| N1 | 0.62925 (12) | 0.57325 (17) | 0.08026 (8) | 0.0485 (5) | |
| N2 | 0.55538 (12) | 0.61513 (18) | 0.11211 (9) | 0.0540 (6) | |
| C3 | 0.49372 (15) | 0.7059 (2) | 0.06310 (11) | 0.0526 (6) | |
| C4 | 0.52380 (15) | 0.7271 (2) | −0.00295 (10) | 0.0512 (7) | |
| C5 | 0.61166 (15) | 0.6395 (2) | 0.01114 (10) | 0.0514 (7) | |
| C6 | 0.70789 (14) | 0.4754 (2) | 0.12184 (10) | 0.0480 (6) | |
| C7 | 0.77805 (16) | 0.4233 (3) | 0.09027 (12) | 0.0651 (8) | |
| C8 | 0.85362 (18) | 0.3279 (3) | 0.13225 (13) | 0.0744 (9) | |
| C9 | 0.85957 (17) | 0.2840 (3) | 0.20516 (12) | 0.0668 (8) | |
| C10 | 0.78986 (16) | 0.3365 (2) | 0.23610 (11) | 0.0617 (7) | |
| C11 | 0.71414 (15) | 0.4321 (2) | 0.19540 (10) | 0.0547 (7) | |
| C12 | 0.48768 (17) | 0.8096 (2) | −0.07168 (12) | 0.0612 (7) | |
| C13 | 0.39531 (18) | 0.9041 (3) | −0.09255 (13) | 0.0753 (9) | |
| C14 | 0.40749 (17) | 0.7731 (3) | 0.08202 (13) | 0.0771 (9) | |
| O21 | 0.23044 (14) | 0.0334 (2) | 0.01074 (9) | 0.0987 (8) | |
| O22 | 0.18315 (17) | 0.1293 (3) | −0.12625 (11) | 0.1230 (10) | |
| N21 | 0.11930 (13) | 0.1209 (2) | 0.07053 (9) | 0.0632 (7) | |
| N22 | 0.03141 (16) | 0.2078 (3) | 0.04976 (11) | 0.0871 (9) | |
| C23 | 0.01369 (18) | 0.2479 (3) | −0.02204 (13) | 0.0760 (9) | |
| C24 | 0.08731 (16) | 0.1905 (2) | −0.05120 (11) | 0.0617 (8) | |
| C25 | 0.15261 (17) | 0.1086 (2) | 0.01064 (12) | 0.0622 (8) | |
| C26 | 0.15670 (17) | 0.0590 (2) | 0.14600 (11) | 0.0591 (7) | |
| C27 | 0.2554 (2) | 0.0047 (3) | 0.17540 (14) | 0.0830 (10) | |
| C28 | 0.2890 (2) | −0.0560 (3) | 0.24905 (15) | 0.0940 (11) | |
| C29 | 0.2273 (2) | −0.0600 (3) | 0.29305 (14) | 0.0882 (10) | |
| C30 | 0.1312 (2) | −0.0014 (3) | 0.26406 (14) | 0.0818 (10) | |
| C31 | 0.09504 (19) | 0.0573 (3) | 0.19051 (12) | 0.0706 (8) | |
| C32 | 0.1070 (2) | 0.2010 (3) | −0.12067 (13) | 0.0786 (10) | |
| C33 | 0.0467 (2) | 0.2899 (3) | −0.18792 (13) | 0.0988 (11) | |
| C34 | −0.0776 (2) | 0.3424 (4) | −0.06259 (17) | 0.1267 (14) | |
| H1O | 0.64280 | 0.67500 | −0.07140 | 0.1070* | |
| H7 | 0.77450 | 0.45230 | 0.04090 | 0.0780* | |
| H8 | 0.90090 | 0.29290 | 0.11090 | 0.0890* | |
| H9 | 0.91040 | 0.21950 | 0.23310 | 0.0800* | |
| H10 | 0.79360 | 0.30710 | 0.28550 | 0.0740* | |
| H11 | 0.66750 | 0.46730 | 0.21730 | 0.0660* | |
| H13A | 0.38720 | 0.95360 | −0.14030 | 0.1130* | |
| H13B | 0.40330 | 0.97420 | −0.05220 | 0.1130* | |
| H13C | 0.33570 | 0.84560 | −0.09860 | 0.1130* | |
| H14A | 0.34390 | 0.75280 | 0.04120 | 0.1160* | |
| H14B | 0.41750 | 0.87630 | 0.08720 | 0.1160* | |
| H14C | 0.40560 | 0.73360 | 0.12990 | 0.1160* | |
| H21O | 0.23880 | 0.04250 | −0.03110 | 0.1480* | |
| H27 | 0.29860 | 0.00880 | 0.14640 | 0.1000* | |
| H28 | 0.35480 | −0.09470 | 0.26880 | 0.1130* | |
| H29 | 0.25040 | −0.10200 | 0.34210 | 0.1060* | |
| H30 | 0.08950 | −0.00120 | 0.29430 | 0.0980* | |
| H31 | 0.02910 | 0.09560 | 0.17120 | 0.0850* | |
| H33A | 0.06630 | 0.26630 | −0.23180 | 0.1480* | |
| H33B | 0.05960 | 0.39110 | −0.17560 | 0.1480* | |
| H33C | −0.02510 | 0.27020 | −0.20000 | 0.1480* | |
| H34A | −0.11410 | 0.36330 | −0.02790 | 0.1910* | |
| H34B | −0.12220 | 0.29250 | −0.10740 | 0.1910* | |
| H34C | −0.05460 | 0.43150 | −0.07830 | 0.1910* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0762 (10) | 0.0911 (12) | 0.0547 (8) | 0.0196 (9) | 0.0331 (8) | 0.0127 (8) |
| O2 | 0.0920 (12) | 0.1028 (14) | 0.0569 (9) | 0.0153 (10) | 0.0262 (9) | 0.0176 (8) |
| N1 | 0.0489 (9) | 0.0560 (10) | 0.0417 (8) | 0.0082 (8) | 0.0170 (7) | −0.0040 (7) |
| N2 | 0.0520 (10) | 0.0624 (11) | 0.0501 (9) | 0.0100 (9) | 0.0207 (8) | −0.0060 (8) |
| C3 | 0.0503 (11) | 0.0557 (12) | 0.0498 (10) | 0.0059 (10) | 0.0145 (9) | −0.0072 (10) |
| C4 | 0.0511 (12) | 0.0513 (12) | 0.0461 (10) | 0.0036 (10) | 0.0101 (9) | −0.0066 (9) |
| C5 | 0.0543 (12) | 0.0567 (12) | 0.0429 (10) | −0.0004 (10) | 0.0165 (9) | −0.0060 (9) |
| C6 | 0.0478 (11) | 0.0500 (11) | 0.0460 (10) | 0.0046 (9) | 0.0157 (9) | −0.0041 (9) |
| C7 | 0.0678 (14) | 0.0804 (16) | 0.0544 (11) | 0.0209 (13) | 0.0305 (11) | 0.0094 (11) |
| C8 | 0.0685 (15) | 0.0919 (18) | 0.0725 (14) | 0.0327 (14) | 0.0366 (12) | 0.0133 (13) |
| C9 | 0.0623 (14) | 0.0737 (15) | 0.0630 (13) | 0.0189 (12) | 0.0199 (11) | 0.0076 (11) |
| C10 | 0.0686 (14) | 0.0690 (14) | 0.0478 (10) | 0.0116 (12) | 0.0203 (10) | 0.0043 (10) |
| C11 | 0.0574 (12) | 0.0613 (13) | 0.0498 (10) | 0.0076 (11) | 0.0239 (9) | −0.0034 (9) |
| C12 | 0.0645 (14) | 0.0599 (13) | 0.0499 (11) | −0.0007 (11) | 0.0078 (11) | −0.0041 (10) |
| C13 | 0.0745 (16) | 0.0677 (15) | 0.0679 (14) | 0.0120 (13) | 0.0041 (12) | 0.0042 (11) |
| C14 | 0.0697 (15) | 0.0915 (18) | 0.0737 (14) | 0.0268 (14) | 0.0293 (12) | −0.0001 (13) |
| O21 | 0.0956 (13) | 0.1380 (17) | 0.0771 (11) | 0.0562 (12) | 0.0485 (10) | 0.0161 (11) |
| O22 | 0.1323 (17) | 0.178 (2) | 0.0862 (12) | 0.0505 (16) | 0.0727 (13) | 0.0229 (13) |
| N21 | 0.0616 (11) | 0.0788 (13) | 0.0554 (10) | 0.0240 (10) | 0.0282 (9) | 0.0052 (9) |
| N22 | 0.0824 (14) | 0.1190 (18) | 0.0700 (12) | 0.0519 (13) | 0.0393 (11) | 0.0221 (12) |
| C23 | 0.0722 (15) | 0.0956 (18) | 0.0634 (13) | 0.0270 (14) | 0.0275 (12) | 0.0125 (13) |
| C24 | 0.0601 (13) | 0.0746 (15) | 0.0528 (11) | 0.0049 (12) | 0.0226 (10) | 0.0014 (10) |
| C25 | 0.0618 (13) | 0.0703 (15) | 0.0607 (12) | 0.0113 (12) | 0.0292 (11) | −0.0019 (11) |
| C26 | 0.0674 (14) | 0.0600 (13) | 0.0524 (11) | 0.0121 (11) | 0.0239 (11) | −0.0007 (10) |
| C27 | 0.0782 (17) | 0.102 (2) | 0.0700 (15) | 0.0283 (15) | 0.0272 (13) | 0.0093 (14) |
| C28 | 0.0888 (19) | 0.103 (2) | 0.0754 (17) | 0.0238 (17) | 0.0092 (16) | 0.0120 (15) |
| C29 | 0.112 (2) | 0.0838 (19) | 0.0616 (14) | −0.0010 (17) | 0.0206 (16) | 0.0095 (13) |
| C30 | 0.107 (2) | 0.0783 (17) | 0.0679 (15) | −0.0045 (16) | 0.0400 (15) | 0.0041 (13) |
| C31 | 0.0774 (16) | 0.0741 (15) | 0.0651 (13) | 0.0103 (13) | 0.0306 (12) | 0.0031 (12) |
| C32 | 0.0825 (17) | 0.0944 (19) | 0.0616 (14) | −0.0036 (15) | 0.0283 (13) | −0.0008 (13) |
| C33 | 0.111 (2) | 0.118 (2) | 0.0601 (14) | −0.0131 (19) | 0.0201 (14) | 0.0143 (15) |
| C34 | 0.114 (2) | 0.177 (3) | 0.093 (2) | 0.084 (2) | 0.0405 (18) | 0.042 (2) |
Geometric parameters (Å, °)
| O1—C5 | 1.290 (3) | C11—H11 | 0.9300 |
| O2—C12 | 1.276 (3) | C13—H13B | 0.9600 |
| O1—H1O | 0.8200 | C13—H13A | 0.9600 |
| O21—C25 | 1.282 (3) | C13—H13C | 0.9600 |
| O22—C32 | 1.279 (4) | C14—H14C | 0.9600 |
| O21—H21O | 0.8200 | C14—H14B | 0.9600 |
| N1—C5 | 1.353 (2) | C14—H14A | 0.9600 |
| N1—C6 | 1.419 (2) | C23—C24 | 1.409 (3) |
| N1—N2 | 1.398 (2) | C23—C34 | 1.506 (4) |
| N2—C3 | 1.308 (3) | C24—C25 | 1.404 (3) |
| N21—C25 | 1.337 (3) | C24—C32 | 1.397 (3) |
| N21—C26 | 1.422 (2) | C26—C31 | 1.372 (3) |
| N21—N22 | 1.397 (3) | C26—C27 | 1.381 (4) |
| N22—C23 | 1.309 (3) | C27—C28 | 1.388 (4) |
| C3—C14 | 1.490 (3) | C28—C29 | 1.365 (4) |
| C3—C4 | 1.426 (3) | C29—C30 | 1.365 (4) |
| C4—C12 | 1.410 (3) | C30—C31 | 1.380 (3) |
| C4—C5 | 1.409 (3) | C32—C33 | 1.478 (3) |
| C6—C11 | 1.383 (2) | C27—H27 | 0.9300 |
| C6—C7 | 1.379 (3) | C28—H28 | 0.9300 |
| C7—C8 | 1.382 (4) | C29—H29 | 0.9300 |
| C8—C9 | 1.374 (3) | C30—H30 | 0.9300 |
| C9—C10 | 1.367 (3) | C31—H31 | 0.9300 |
| C10—C11 | 1.377 (3) | C33—H33A | 0.9600 |
| C12—C13 | 1.486 (3) | C33—H33B | 0.9600 |
| C7—H7 | 0.9300 | C33—H33C | 0.9600 |
| C8—H8 | 0.9300 | C34—H34A | 0.9600 |
| C9—H9 | 0.9300 | C34—H34B | 0.9600 |
| C10—H10 | 0.9300 | C34—H34C | 0.9600 |
| C5—O1—H1O | 110.00 | H14A—C14—H14B | 109.00 |
| C25—O21—H21O | 110.00 | H14A—C14—H14C | 110.00 |
| N2—N1—C5 | 110.30 (16) | C3—C14—H14B | 109.00 |
| C5—N1—C6 | 130.45 (17) | C3—C14—H14C | 109.00 |
| N2—N1—C6 | 119.25 (14) | C3—C14—H14A | 109.00 |
| N1—N2—C3 | 106.75 (15) | N22—C23—C24 | 111.6 (2) |
| C25—N21—C26 | 130.94 (19) | N22—C23—C34 | 119.8 (2) |
| N22—N21—C26 | 119.17 (17) | C24—C23—C34 | 128.7 (2) |
| N22—N21—C25 | 109.87 (17) | C23—C24—C25 | 104.13 (19) |
| N21—N22—C23 | 106.3 (2) | C23—C24—C32 | 135.7 (2) |
| C4—C3—C14 | 129.42 (18) | C25—C24—C32 | 120.1 (2) |
| N2—C3—C14 | 119.48 (18) | O21—C25—C24 | 126.9 (2) |
| N2—C3—C4 | 111.08 (18) | N21—C25—C24 | 108.1 (2) |
| C3—C4—C5 | 104.66 (16) | O21—C25—N21 | 125.02 (19) |
| C3—C4—C12 | 135.9 (2) | N21—C26—C27 | 120.5 (2) |
| C5—C4—C12 | 119.44 (19) | C27—C26—C31 | 120.0 (2) |
| N1—C5—C4 | 107.21 (17) | N21—C26—C31 | 119.5 (2) |
| O1—C5—C4 | 127.25 (17) | C26—C27—C28 | 118.9 (2) |
| O1—C5—N1 | 125.54 (18) | C27—C28—C29 | 121.3 (3) |
| N1—C6—C11 | 119.18 (17) | C28—C29—C30 | 119.0 (2) |
| C7—C6—C11 | 119.64 (19) | C29—C30—C31 | 121.0 (3) |
| N1—C6—C7 | 121.18 (17) | C26—C31—C30 | 119.8 (2) |
| C6—C7—C8 | 119.7 (2) | O22—C32—C33 | 117.7 (2) |
| C7—C8—C9 | 120.7 (2) | C24—C32—C33 | 124.7 (2) |
| C8—C9—C10 | 119.2 (2) | O22—C32—C24 | 117.6 (2) |
| C9—C10—C11 | 121.10 (19) | C26—C27—H27 | 121.00 |
| C6—C11—C10 | 119.65 (19) | C28—C27—H27 | 121.00 |
| O2—C12—C13 | 117.93 (19) | C27—C28—H28 | 119.00 |
| O2—C12—C4 | 118.3 (2) | C29—C28—H28 | 119.00 |
| C4—C12—C13 | 123.8 (2) | C28—C29—H29 | 120.00 |
| C8—C7—H7 | 120.00 | C30—C29—H29 | 120.00 |
| C6—C7—H7 | 120.00 | C29—C30—H30 | 120.00 |
| C9—C8—H8 | 120.00 | C31—C30—H30 | 120.00 |
| C7—C8—H8 | 120.00 | C26—C31—H31 | 120.00 |
| C10—C9—H9 | 120.00 | C30—C31—H31 | 120.00 |
| C8—C9—H9 | 120.00 | C32—C33—H33A | 110.00 |
| C9—C10—H10 | 119.00 | C32—C33—H33B | 109.00 |
| C11—C10—H10 | 119.00 | C32—C33—H33C | 109.00 |
| C10—C11—H11 | 120.00 | H33A—C33—H33B | 109.00 |
| C6—C11—H11 | 120.00 | H33A—C33—H33C | 110.00 |
| C12—C13—H13B | 109.00 | H33B—C33—H33C | 109.00 |
| C12—C13—H13C | 109.00 | C23—C34—H34A | 109.00 |
| H13A—C13—H13C | 110.00 | C23—C34—H34B | 109.00 |
| C12—C13—H13A | 109.00 | C23—C34—H34C | 109.00 |
| H13A—C13—H13B | 109.00 | H34A—C34—H34B | 109.00 |
| H13B—C13—H13C | 109.00 | H34A—C34—H34C | 110.00 |
| H14B—C14—H14C | 109.00 | H34B—C34—H34C | 110.00 |
| C5—N1—N2—C3 | −0.1 (2) | C3—C4—C5—N1 | 0.3 (2) |
| C6—N1—N2—C3 | 179.20 (16) | C12—C4—C5—O1 | −0.2 (3) |
| N2—N1—C5—O1 | 179.96 (19) | C12—C4—C5—N1 | 179.92 (16) |
| N2—N1—C5—C4 | −0.1 (2) | C3—C4—C12—O2 | 179.4 (2) |
| C6—N1—C5—O1 | 0.8 (3) | N1—C6—C7—C8 | −179.9 (2) |
| C6—N1—C5—C4 | −179.31 (18) | C11—C6—C7—C8 | 0.3 (3) |
| N2—N1—C6—C7 | 175.42 (19) | N1—C6—C11—C10 | 179.64 (18) |
| N2—N1—C6—C11 | −4.7 (3) | C7—C6—C11—C10 | −0.5 (3) |
| C5—N1—C6—C7 | −5.4 (3) | C6—C7—C8—C9 | 0.1 (4) |
| C5—N1—C6—C11 | 174.41 (19) | C7—C8—C9—C10 | −0.3 (4) |
| N1—N2—C3—C4 | 0.3 (2) | C8—C9—C10—C11 | 0.1 (4) |
| N1—N2—C3—C14 | −178.50 (18) | C9—C10—C11—C6 | 0.4 (3) |
| N22—N21—C25—O21 | −178.4 (2) | N22—C23—C24—C25 | 0.5 (3) |
| N22—N21—C25—C24 | 1.0 (2) | N22—C23—C24—C32 | −177.7 (3) |
| C26—N21—C25—O21 | −0.3 (4) | C34—C23—C24—C25 | −178.6 (3) |
| C26—N21—C25—C24 | 179.2 (2) | C34—C23—C24—C32 | 3.1 (5) |
| N22—N21—C26—C27 | −163.5 (2) | C23—C24—C25—O21 | 178.5 (2) |
| C25—N21—N22—C23 | −0.7 (3) | C23—C24—C25—N21 | −1.0 (2) |
| C26—N21—N22—C23 | −179.1 (2) | C32—C24—C25—O21 | −2.9 (3) |
| C25—N21—C26—C31 | −163.4 (2) | C32—C24—C25—N21 | 177.6 (2) |
| N22—N21—C26—C31 | 14.6 (3) | C23—C24—C32—O22 | −179.3 (3) |
| C25—N21—C26—C27 | 18.5 (3) | C23—C24—C32—C33 | 1.3 (5) |
| N21—N22—C23—C24 | 0.1 (3) | C25—C24—C32—O22 | 2.7 (4) |
| N21—N22—C23—C34 | 179.3 (2) | C25—C24—C32—C33 | −176.8 (2) |
| C14—C3—C4—C5 | 178.3 (2) | N21—C26—C27—C28 | −179.5 (2) |
| C14—C3—C4—C12 | −1.3 (4) | C31—C26—C27—C28 | 2.4 (4) |
| N2—C3—C4—C12 | −179.9 (2) | N21—C26—C31—C30 | −179.4 (2) |
| N2—C3—C4—C5 | −0.3 (2) | C27—C26—C31—C30 | −1.3 (4) |
| C3—C4—C5—O1 | −179.81 (19) | C26—C27—C28—C29 | −1.4 (4) |
| C3—C4—C12—C13 | −1.2 (4) | C27—C28—C29—C30 | −0.8 (4) |
| C5—C4—C12—O2 | −0.1 (3) | C28—C29—C30—C31 | 2.0 (4) |
| C5—C4—C12—C13 | 179.3 (2) | C29—C30—C31—C26 | −0.9 (4) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1O···O2 | 0.82 | 1.85 | 2.546 (2) | 142 |
| O21—H21O···O22 | 0.82 | 1.83 | 2.531 (3) | 142 |
| C8—H8···N22i | 0.93 | 2.56 | 3.489 (3) | 177 |
| C13—H13C···Cg2ii | 0.96 | 2.66 | 3.533 (3) | 150 |
| C33—H33B···Cg2ii | 0.96 | 2.98 | 3.774 (3) | 141 |
Symmetry codes: (i) x+1, y, z; (ii) −x+1, −y+1, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: WN2305).
References
- Bruker (2002). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Cingolani, A., Effendy, Marchetti, F., Pettinari, C., Skelton, B. W. & White, A. H. (2002). Inorg. Chem.41, 1151–1161. [DOI] [PubMed]
- Grimmett, M. R. (1970). Adv. Heterocycl. Chem.12, 103–183.
- Knorr, L. (1883). Ber. Dtsch. Chem. Ges.16, 2593–2596.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2003). J. Appl. Cryst.36, 7–13.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809001470/wn2305sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809001470/wn2305Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report