Abstract
In the crystal structure of the title compound, C18H18N2O4S, the thiazine ring adopts a distorted half-chair conformation. 1,2-Benzothiazines of this kind have a wide range of biological activities and are mainly used as medicines in the treatment of inflammation and rheumatoid arthritis. The enolic H atom is involved in an intramolecular O—H⋯O hydrogen bond, forming a six-membered ring. The molecules arrange themselves into centrosymmetric dimers by means of intermolecular N—H⋯O hydrogen bonds. A weak intermolcular C—H⋯O interaction is also present.
Related literature
For the synthesis of related molecules, see: Siddiqui, Ahmad, Khan et al. (2007 ▶); Zia-ur-Rehman et al. (2006 ▶); For the biological activity of 1,2-benzothiazine-1,1-dioxides, see: Zia-ur-Rehman et al. (2009 ▶). For related structures, see: Siddiqui et al. (2008 ▶); Siddiqui, Ahmad, Siddiqui et al. (2007 ▶). For the pharmacological background to 1,2-benzothiazine-3-carboxamide 1,1-dioxide derivatives, see Gennari et al. (1994 ▶); Lombardino & Wiseman (1972 ▶).
Experimental
Crystal data
C18H18N2O4S
M r = 358.40
Monoclinic,

a = 10.2461 (3) Å
b = 8.5421 (2) Å
c = 19.8944 (5) Å
β = 104.832 (1)°
V = 1683.20 (8) Å3
Z = 4
Mo Kα radiation
μ = 0.22 mm−1
T = 120 K
0.30 × 0.10 × 0.10 mm
Data collection
Bruker–Nonius CCD camera on κ-goniostat diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 2007 ▶) T min = 0.855, T max = 0.978
18371 measured reflections
3828 independent reflections
3000 reflections with I > 2σ(I)
R int = 0.055
Refinement
R[F 2 > 2σ(F 2)] = 0.045
wR(F 2) = 0.114
S = 1.06
3828 reflections
234 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.27 e Å−3
Δρmin = −0.57 e Å−3
Data collection: COLLECT (Hooft, 1998 ▶); cell refinement: DENZO (Otwinowski & Minor, 1997 ▶) and COLLECT; data reduction: DENZO and COLLECT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL and local programs.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809006837/bt2887sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809006837/bt2887Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1O⋯O2 | 0.84 | 1.79 | 2.5320 (18) | 147 |
| N2—H1N⋯O3i | 0.85 (2) | 2.26 (2) | 2.972 (2) | 141 (2) |
| C3—H3⋯O4ii | 0.95 | 2.49 | 3.352 (3) | 150 |
Symmetry codes: (i)  ; (ii)
; (ii) 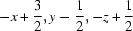 .
.
Acknowledgments
The authors are grateful to the Higher Education Commission of Pakistan for a grant.
supplementary crystallographic information
Comment
1,2-Benzothiazine-3-carboxamide 1,1-dioxide derivatives belonging to oxicams, a class of non-steroidal anti-inflammatory drugs (NSAIDs), are well known as analgesic and anti-inflammatory agents since the introduction of Piroxicam (Lombardino & Wiseman, 1972) in the United States in 1982 where it gained immediate acceptance and remained among the top fifty prescription drugs for several years. Besides having anti-inflammatory activity, these have also been found to be used for the treatment of rheumatoid arthritis, ankylosing spondylitis, osteoarthrosis and other inflammatory rheumatic and non- rheumatic processes, including onsets and traumatologic lesions (Gennari et al., 1994).
In continuation of our work on the synthesis (Siddiqui, Ahmad, Khan et al., 2007, Zia-ur-Rehman et al., 2006, biological activity (Zia-ur-Rehman et al, 2009) and crystal structures (Siddiqui, Ahmad, Siddiqui et al., 2007, Siddiqui et al., 2008) of various 1,2-benzothiazine-1,1-dioxides, we herein report the synthesis and crystal structure of the title compound (I) (Scheme and figure 1). The thiazine ring, involving two double bonds, exhibits a distorted half-chair conformation. The enolic hydrogen on O1 is involved in intramolecular hydrogen bonding giving rise to a six-membered hydrogen bond ring (Table 1). The molecules form centrosymmetric dimers through intermolecular N—H···O hydrogen bonds. In addition, the crystal packing is stabilized by weak C—H···O contacts.
Experimental
A mixture of methyl 4-hydroxy-2-methyl-2H-1,2-benzothiazine-3-carboxylate-1,1-dioxide (2.693 g; 10.0 mmoles), 2,3-dimethyl aniline (1.818 g; 15.0 mmoles) and xylene (25.0 ml) was refluxed under nitrogen atmosphere in a Soxhlet apparatus having Linde type 4Å molecular sieves. Three fourth of the xylene was then distilled off and the remaining contents were allowed to stand overnight at room temperature. Settled solids were filtered off, washed with diethyl ether and crystallized from ethanol. Yield: 78%.
Refinement
All hydrogen atoms were identified in the difference map. Those bonded to O and C were fixed in ideal positions and treated as riding on their parent atoms. In the case of the methyl and hydroxyl H atoms the torsion angles were refined. The following distances were used: Methyl C—H 0.98 Å. ° Aromatic C—H 0.95 Å. ° Hydroxyl O—H 0.84 Å. U(H) was set to 1.2Ueq of the parent atoms or 1.5Ueq for methyl groups. The H atom bonded to N was freely refined.
Figures
Fig. 1.
The molecular structure of the title compound with displacement ellipsoids at the 50% probability level.
Fig. 2.
Perspective view of the three-dimensional crystal packing showing hydrogen-bonded interactions (dashed lines). H atoms not involved in hydrogen bonding have been omitted for clarity.
Crystal data
| C18H18N2O4S | F(000) = 752 |
| Mr = 358.40 | Dx = 1.414 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2yn | Cell parameters from 10340 reflections |
| a = 10.2461 (3) Å | θ = 2.9–27.5° |
| b = 8.5421 (2) Å | µ = 0.22 mm−1 |
| c = 19.8944 (5) Å | T = 120 K |
| β = 104.832 (1)° | Block, colourless |
| V = 1683.20 (8) Å3 | 0.30 × 0.10 × 0.10 mm |
| Z = 4 |
Data collection
| Bruker–Nonius CCD camera on κ-goniostat diffractometer | 3828 independent reflections |
| Radiation source: Bruker Nonius FR591 Rotating Anode | 3000 reflections with I > 2σ(I) |
| graphite | Rint = 0.055 |
| Detector resolution: 9.091 pixels mm-1 | θmax = 27.5°, θmin = 3.2° |
| φ and ω scans to fill the asymmetric unit | h = −13→11 |
| Absorption correction: multi-scan (SADABS; Sheldrick, 2007) | k = −11→9 |
| Tmin = 0.855, Tmax = 0.978 | l = −25→24 |
| 18371 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.045 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.114 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.06 | w = 1/[σ2(Fo2) + (0.0495P)2 + 0.8901P] where P = (Fo2 + 2Fc2)/3 |
| 3828 reflections | (Δ/σ)max = 0.001 |
| 234 parameters | Δρmax = 0.27 e Å−3 |
| 0 restraints | Δρmin = −0.57 e Å−3 |
Special details
| Experimental. SADABS was used to perform the Absorption correction Parameter refinement on 11612 reflections reduced R(int) from 0.0681 to 0.0328 Ratio of minimum to maximum apparent transmission: 0.867291 The given Tmin and Tmax were generated using the SHELX SIZE command |
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.65769 (5) | 0.38059 (5) | 0.08727 (2) | 0.01787 (14) | |
| O2 | 0.85778 (13) | 0.51574 (15) | −0.10128 (6) | 0.0190 (3) | |
| O1 | 0.93907 (13) | 0.27205 (16) | −0.03046 (7) | 0.0208 (3) | |
| H1O | 0.9310 | 0.3360 | −0.0633 | 0.031* | |
| N2 | 0.70234 (16) | 0.67387 (18) | −0.06965 (8) | 0.0162 (3) | |
| N1 | 0.73387 (15) | 0.51959 (17) | 0.05471 (8) | 0.0155 (3) | |
| O3 | 0.55860 (13) | 0.31248 (15) | 0.03050 (7) | 0.0207 (3) | |
| O4 | 0.61656 (14) | 0.44317 (17) | 0.14534 (7) | 0.0255 (3) | |
| C6 | 0.88260 (18) | 0.2273 (2) | 0.07631 (9) | 0.0173 (4) | |
| C9 | 0.78894 (18) | 0.5530 (2) | −0.05975 (9) | 0.0154 (4) | |
| C10 | 0.67298 (17) | 0.7818 (2) | −0.12596 (9) | 0.0166 (4) | |
| C7 | 0.87323 (18) | 0.3265 (2) | 0.01508 (9) | 0.0162 (4) | |
| C1 | 0.78842 (19) | 0.2430 (2) | 0.11619 (9) | 0.0187 (4) | |
| C18 | 0.8113 (2) | 0.6356 (2) | 0.10495 (10) | 0.0226 (4) | |
| H18A | 0.8911 | 0.5851 | 0.1348 | 0.034* | |
| H18B | 0.7542 | 0.6764 | 0.1336 | 0.034* | |
| H18C | 0.8397 | 0.7220 | 0.0795 | 0.034* | |
| C5 | 0.98373 (19) | 0.1138 (2) | 0.09600 (10) | 0.0216 (4) | |
| H5 | 1.0480 | 0.1000 | 0.0695 | 0.026* | |
| C8 | 0.80079 (18) | 0.4615 (2) | 0.00417 (9) | 0.0151 (4) | |
| C14 | 0.61045 (19) | 1.0447 (2) | −0.16562 (11) | 0.0226 (4) | |
| C16 | 0.6568 (2) | 0.9831 (2) | −0.03662 (10) | 0.0233 (4) | |
| H16A | 0.5896 | 0.9260 | −0.0190 | 0.035* | |
| H16B | 0.6410 | 1.0959 | −0.0344 | 0.035* | |
| H16C | 0.7475 | 0.9578 | −0.0082 | 0.035* | |
| C4 | 0.9901 (2) | 0.0215 (2) | 0.15404 (11) | 0.0266 (5) | |
| H4 | 1.0594 | −0.0549 | 0.1672 | 0.032* | |
| C15 | 0.64525 (18) | 0.9363 (2) | −0.11082 (10) | 0.0181 (4) | |
| C12 | 0.6398 (2) | 0.8437 (2) | −0.24637 (10) | 0.0246 (4) | |
| H12 | 0.6402 | 0.8140 | −0.2923 | 0.030* | |
| C11 | 0.67017 (19) | 0.7340 (2) | −0.19320 (9) | 0.0203 (4) | |
| H11 | 0.6887 | 0.6283 | −0.2025 | 0.024* | |
| C3 | 0.8966 (2) | 0.0393 (2) | 0.19319 (11) | 0.0294 (5) | |
| H3 | 0.9024 | −0.0246 | 0.2329 | 0.035* | |
| C13 | 0.6091 (2) | 0.9960 (2) | −0.23262 (10) | 0.0248 (4) | |
| H13 | 0.5865 | 1.0692 | −0.2697 | 0.030* | |
| C2 | 0.7944 (2) | 0.1507 (2) | 0.17439 (10) | 0.0258 (4) | |
| H2 | 0.7299 | 0.1633 | 0.2009 | 0.031* | |
| C17 | 0.5769 (2) | 1.2125 (3) | −0.15299 (12) | 0.0350 (5) | |
| H17A | 0.5529 | 1.2691 | −0.1973 | 0.052* | |
| H17B | 0.6555 | 1.2624 | −0.1217 | 0.052* | |
| H17C | 0.5006 | 1.2151 | −0.1317 | 0.052* | |
| H1N | 0.657 (2) | 0.683 (3) | −0.0397 (12) | 0.026 (6)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0187 (2) | 0.0193 (3) | 0.0181 (2) | −0.00080 (18) | 0.00918 (18) | −0.00048 (18) |
| O2 | 0.0210 (7) | 0.0221 (7) | 0.0160 (6) | 0.0015 (5) | 0.0085 (5) | 0.0000 (5) |
| O1 | 0.0218 (7) | 0.0229 (7) | 0.0208 (7) | 0.0045 (6) | 0.0112 (6) | 0.0014 (5) |
| N2 | 0.0159 (8) | 0.0182 (8) | 0.0155 (8) | 0.0018 (6) | 0.0062 (6) | 0.0012 (6) |
| N1 | 0.0162 (8) | 0.0170 (8) | 0.0151 (7) | −0.0005 (6) | 0.0073 (6) | −0.0023 (6) |
| O3 | 0.0156 (7) | 0.0230 (7) | 0.0245 (7) | −0.0022 (5) | 0.0072 (5) | −0.0032 (6) |
| O4 | 0.0325 (8) | 0.0269 (8) | 0.0232 (7) | 0.0000 (6) | 0.0183 (6) | −0.0027 (6) |
| C6 | 0.0168 (9) | 0.0173 (9) | 0.0163 (9) | −0.0034 (7) | 0.0018 (7) | −0.0016 (7) |
| C9 | 0.0157 (9) | 0.0163 (9) | 0.0136 (8) | −0.0038 (7) | 0.0028 (7) | −0.0017 (7) |
| C10 | 0.0121 (8) | 0.0192 (9) | 0.0170 (9) | −0.0021 (7) | 0.0013 (7) | 0.0026 (7) |
| C7 | 0.0130 (8) | 0.0187 (9) | 0.0171 (9) | −0.0023 (7) | 0.0041 (7) | −0.0018 (7) |
| C1 | 0.0223 (9) | 0.0172 (9) | 0.0161 (9) | −0.0019 (7) | 0.0037 (7) | −0.0020 (7) |
| C18 | 0.0263 (10) | 0.0227 (10) | 0.0197 (10) | −0.0050 (8) | 0.0075 (8) | −0.0057 (8) |
| C5 | 0.0192 (10) | 0.0194 (10) | 0.0243 (10) | 0.0010 (7) | 0.0020 (8) | −0.0008 (8) |
| C8 | 0.0147 (8) | 0.0179 (9) | 0.0134 (9) | −0.0015 (7) | 0.0049 (7) | −0.0016 (7) |
| C14 | 0.0180 (9) | 0.0190 (10) | 0.0273 (10) | −0.0014 (8) | −0.0003 (8) | 0.0024 (8) |
| C16 | 0.0267 (11) | 0.0192 (10) | 0.0250 (10) | 0.0004 (8) | 0.0083 (9) | −0.0035 (8) |
| C4 | 0.0292 (11) | 0.0210 (10) | 0.0246 (10) | 0.0028 (8) | −0.0023 (9) | 0.0033 (8) |
| C15 | 0.0140 (9) | 0.0193 (9) | 0.0199 (9) | −0.0028 (7) | 0.0025 (7) | −0.0007 (7) |
| C12 | 0.0275 (11) | 0.0277 (11) | 0.0163 (9) | −0.0036 (8) | 0.0015 (8) | 0.0010 (8) |
| C11 | 0.0235 (10) | 0.0196 (10) | 0.0168 (9) | −0.0011 (8) | 0.0034 (8) | −0.0005 (7) |
| C3 | 0.0429 (13) | 0.0229 (11) | 0.0187 (10) | −0.0008 (9) | 0.0013 (9) | 0.0054 (8) |
| C13 | 0.0250 (11) | 0.0249 (11) | 0.0211 (10) | −0.0024 (8) | −0.0005 (8) | 0.0073 (8) |
| C2 | 0.0353 (12) | 0.0248 (10) | 0.0190 (10) | −0.0030 (9) | 0.0101 (9) | 0.0017 (8) |
| C17 | 0.0428 (14) | 0.0220 (11) | 0.0361 (13) | 0.0039 (9) | 0.0029 (11) | 0.0019 (9) |
Geometric parameters (Å, °)
| S1—O4 | 1.4308 (13) | C5—C4 | 1.386 (3) |
| S1—O3 | 1.4345 (14) | C5—H5 | 0.9500 |
| S1—N1 | 1.6424 (15) | C14—C13 | 1.393 (3) |
| S1—C1 | 1.7639 (19) | C14—C15 | 1.405 (3) |
| O2—C9 | 1.257 (2) | C14—C17 | 1.510 (3) |
| O1—C7 | 1.344 (2) | C16—C15 | 1.504 (3) |
| O1—H1O | 0.8400 | C16—H16A | 0.9800 |
| N2—C9 | 1.343 (2) | C16—H16B | 0.9800 |
| N2—C10 | 1.422 (2) | C16—H16C | 0.9800 |
| N2—H1N | 0.85 (2) | C4—C3 | 1.389 (3) |
| N1—C8 | 1.442 (2) | C4—H4 | 0.9500 |
| N1—C18 | 1.485 (2) | C12—C13 | 1.383 (3) |
| C6—C5 | 1.400 (3) | C12—C11 | 1.388 (3) |
| C6—C1 | 1.404 (3) | C12—H12 | 0.9500 |
| C6—C7 | 1.467 (3) | C11—H11 | 0.9500 |
| C9—C8 | 1.471 (2) | C3—C2 | 1.394 (3) |
| C10—C11 | 1.392 (3) | C3—H3 | 0.9500 |
| C10—C15 | 1.399 (3) | C13—H13 | 0.9500 |
| C7—C8 | 1.359 (3) | C2—H2 | 0.9500 |
| C1—C2 | 1.389 (3) | C17—H17A | 0.9800 |
| C18—H18A | 0.9800 | C17—H17B | 0.9800 |
| C18—H18B | 0.9800 | C17—H17C | 0.9800 |
| C18—H18C | 0.9800 | ||
| O4—S1—O3 | 119.46 (8) | C7—C8—C9 | 120.66 (16) |
| O4—S1—N1 | 108.44 (8) | N1—C8—C9 | 118.12 (15) |
| O3—S1—N1 | 107.14 (8) | C13—C14—C15 | 118.96 (18) |
| O4—S1—C1 | 109.98 (9) | C13—C14—C17 | 119.71 (18) |
| O3—S1—C1 | 108.17 (8) | C15—C14—C17 | 121.33 (18) |
| N1—S1—C1 | 102.26 (8) | C15—C16—H16A | 109.5 |
| C7—O1—H1O | 109.5 | C15—C16—H16B | 109.5 |
| C9—N2—C10 | 127.78 (16) | H16A—C16—H16B | 109.5 |
| C9—N2—H1N | 115.6 (16) | C15—C16—H16C | 109.5 |
| C10—N2—H1N | 116.6 (16) | H16A—C16—H16C | 109.5 |
| C8—N1—C18 | 115.56 (14) | H16B—C16—H16C | 109.5 |
| C8—N1—S1 | 112.65 (12) | C5—C4—C3 | 120.83 (19) |
| C18—N1—S1 | 116.30 (12) | C5—C4—H4 | 119.6 |
| C5—C6—C1 | 118.36 (17) | C3—C4—H4 | 119.6 |
| C5—C6—C7 | 121.28 (17) | C10—C15—C14 | 118.67 (17) |
| C1—C6—C7 | 120.36 (17) | C10—C15—C16 | 119.51 (17) |
| O2—C9—N2 | 123.99 (16) | C14—C15—C16 | 121.80 (17) |
| O2—C9—C8 | 119.87 (16) | C13—C12—C11 | 120.17 (18) |
| N2—C9—C8 | 116.14 (15) | C13—C12—H12 | 119.9 |
| C11—C10—C15 | 121.91 (17) | C11—C12—H12 | 119.9 |
| C11—C10—N2 | 120.98 (17) | C12—C11—C10 | 118.68 (18) |
| C15—C10—N2 | 117.10 (16) | C12—C11—H11 | 120.7 |
| O1—C7—C8 | 122.29 (16) | C10—C11—H11 | 120.7 |
| O1—C7—C6 | 114.99 (16) | C4—C3—C2 | 120.22 (19) |
| C8—C7—C6 | 122.71 (16) | C4—C3—H3 | 119.9 |
| C2—C1—C6 | 121.79 (18) | C2—C3—H3 | 119.9 |
| C2—C1—S1 | 121.16 (15) | C12—C13—C14 | 121.55 (18) |
| C6—C1—S1 | 117.01 (14) | C12—C13—H13 | 119.2 |
| N1—C18—H18A | 109.5 | C14—C13—H13 | 119.2 |
| N1—C18—H18B | 109.5 | C1—C2—C3 | 118.73 (19) |
| H18A—C18—H18B | 109.5 | C1—C2—H2 | 120.6 |
| N1—C18—H18C | 109.5 | C3—C2—H2 | 120.6 |
| H18A—C18—H18C | 109.5 | C14—C17—H17A | 109.5 |
| H18B—C18—H18C | 109.5 | C14—C17—H17B | 109.5 |
| C4—C5—C6 | 120.06 (18) | H17A—C17—H17B | 109.5 |
| C4—C5—H5 | 120.0 | C14—C17—H17C | 109.5 |
| C6—C5—H5 | 120.0 | H17A—C17—H17C | 109.5 |
| C7—C8—N1 | 121.22 (16) | H17B—C17—H17C | 109.5 |
| O4—S1—N1—C8 | 169.35 (12) | C6—C7—C8—C9 | −176.92 (16) |
| O3—S1—N1—C8 | −60.45 (14) | C18—N1—C8—C7 | 94.4 (2) |
| C1—S1—N1—C8 | 53.19 (14) | S1—N1—C8—C7 | −42.6 (2) |
| O4—S1—N1—C18 | 32.66 (15) | C18—N1—C8—C9 | −85.4 (2) |
| O3—S1—N1—C18 | 162.85 (13) | S1—N1—C8—C9 | 137.60 (14) |
| C1—S1—N1—C18 | −83.51 (14) | O2—C9—C8—C7 | −7.7 (3) |
| C10—N2—C9—O2 | −1.0 (3) | N2—C9—C8—C7 | 172.23 (16) |
| C10—N2—C9—C8 | 179.07 (16) | O2—C9—C8—N1 | 172.09 (16) |
| C9—N2—C10—C11 | 35.7 (3) | N2—C9—C8—N1 | −8.0 (2) |
| C9—N2—C10—C15 | −145.22 (18) | C6—C5—C4—C3 | 0.3 (3) |
| C5—C6—C7—O1 | 18.7 (2) | C11—C10—C15—C14 | 1.6 (3) |
| C1—C6—C7—O1 | −160.50 (16) | N2—C10—C15—C14 | −177.46 (16) |
| C5—C6—C7—C8 | −162.85 (18) | C11—C10—C15—C16 | −176.85 (17) |
| C1—C6—C7—C8 | 18.0 (3) | N2—C10—C15—C16 | 4.1 (2) |
| C5—C6—C1—C2 | 0.7 (3) | C13—C14—C15—C10 | −2.1 (3) |
| C7—C6—C1—C2 | 179.90 (17) | C17—C14—C15—C10 | 178.91 (18) |
| C5—C6—C1—S1 | −176.97 (14) | C13—C14—C15—C16 | 176.29 (18) |
| C7—C6—C1—S1 | 2.2 (2) | C17—C14—C15—C16 | −2.7 (3) |
| O4—S1—C1—C2 | 32.47 (18) | C13—C12—C11—C10 | −2.0 (3) |
| O3—S1—C1—C2 | −99.61 (17) | C15—C10—C11—C12 | 0.5 (3) |
| N1—S1—C1—C2 | 147.51 (16) | N2—C10—C11—C12 | 179.45 (17) |
| O4—S1—C1—C6 | −149.86 (14) | C5—C4—C3—C2 | 0.1 (3) |
| O3—S1—C1—C6 | 78.06 (16) | C11—C12—C13—C14 | 1.4 (3) |
| N1—S1—C1—C6 | −34.82 (16) | C15—C14—C13—C12 | 0.6 (3) |
| C1—C6—C5—C4 | −0.7 (3) | C17—C14—C13—C12 | 179.7 (2) |
| C7—C6—C5—C4 | −179.94 (17) | C6—C1—C2—C3 | −0.2 (3) |
| O1—C7—C8—N1 | −178.38 (15) | S1—C1—C2—C3 | 177.33 (16) |
| C6—C7—C8—N1 | 3.3 (3) | C4—C3—C2—C1 | −0.2 (3) |
| O1—C7—C8—C9 | 1.4 (3) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1O···O2 | 0.84 | 1.79 | 2.5320 (18) | 147 |
| N2—H1N···O3i | 0.85 (2) | 2.26 (2) | 2.972 (2) | 141 (2) |
| C3—H3···O4ii | 0.95 | 2.49 | 3.352 (3) | 150 |
Symmetry codes: (i) −x+1, −y+1, −z; (ii) −x+3/2, y−1/2, −z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: BT2887).
References
- Gennari, C., Salom, B., Potenza, D. & Williams, A. (1994). Angew. Chem. Int. Ed. Engl 33, 2067–2069.
- Hooft, R. W. W. (1998). COLLECT Nonius BV, Delft, The Netherlands.
- Lombardino, J. G. & Wiseman, E. H. (1972). J. Med. Chem.15, 848–849. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods in Enzymology, Vol. 276, Macromolecular Crystallography, Part A, edited by C. W. Carter Jr & R. M. Sweet, pp. 307–326. New York: Academic Press.
- Sheldrick, G. M. (2007). SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Siddiqui, W. A., Ahmad, S., Khan, I. U., Siddiqui, H. L. & Weaver, G. W. (2007). Synth. Commun 37, 767–773.
- Siddiqui, W. A., Ahmad, S., Siddiqui, H. L., Tariq, M. I. & Parvez, M. (2007). Acta Cryst. E63, o4585.
- Siddiqui, W. A., Ahmad, S., Tariq, M. I., Siddiqui, H. L. & Parvez, M. (2008). Acta Cryst. C64, o4-o6. [DOI] [PubMed]
- Zia-ur-Rehman, M., Anwar, J., Ahmad, S. & Siddiqui, H. L. (2006). Chem. Pharm. Bull 54, 1175–1178. [DOI] [PubMed]
- Zia-ur-Rehman, M., Choudary, J. A., Elsegood, M. R. J., Siddiqui, H. L. & Khan, K. M. (2009). Eur. J. Med. Chem 44, 1311–1316. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809006837/bt2887sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809006837/bt2887Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




