Abstract
The title complex, [RuCl2(C25H26N2)]·CH2Cl2, is best thought of as containing an octahedrally coordinated Ru center with the arene occupying three sites. Two Ru—Cl bonds and one Ru–carbene bond complete the distorted octahedron. The carbene portion of the ligand is a benzimidazole ring. This ring is connected to the C6H2(CH3)3 arene group by a CH2 bridge. This leads to a system with very little apparent strain. A dichloromethane solvent molecule completes the crystal structure. Further stabilization is accomplished via C—H⋯N and C—H⋯Cl interactions.
Related literature
For synthesis, see: Yaşar et al. (2008 ▶); Çetinkaya et al. (2001 ▶, 2003 ▶); Özdemir et al. (2001 ▶, 2004 ▶). For general background, see: Herrmann (2002 ▶); Herrmann et al. (1995 ▶); Navarro et al. (2006 ▶); Arduengo & Krafczyc (1998 ▶). For related compounds, see: Begley et al. (1991 ▶); Steedman & Burrell (1997 ▶); Arslan et al. (2004 ▶, 2005 ▶, 2007 ▶).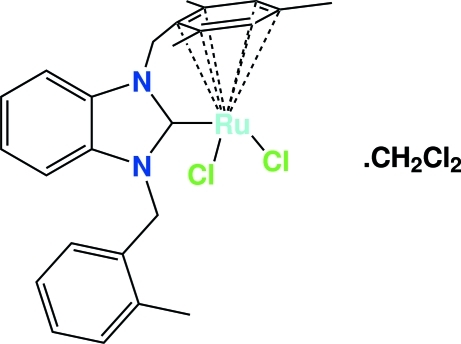
Experimental
Crystal data
[RuCl2(C25H26N2)]·CH2Cl2
M r = 611.37
Monoclinic,

a = 31.362 (6) Å
b = 8.1014 (16) Å
c = 20.484 (4) Å
β = 100.11 (3)°
V = 5123.8 (18) Å3
Z = 8
Mo Kα radiation
μ = 1.05 mm−1
T = 153 (2) K
0.46 × 0.14 × 0.06 mm
Data collection
Rigaku Mercury CCD diffractometer
Absorption correction: multi-scan (REQAB; Jacobson, 1998 ▶) T min = 0.644, T max = 0.940
15807 measured reflections
4501 independent reflections
3825 reflections with I > 2σ(I)
R int = 0.054
Refinement
R[F 2 > 2σ(F 2)] = 0.049
wR(F 2) = 0.115
S = 1.08
4501 reflections
298 parameters
H-atom parameters constrained
Δρmax = 0.92 e Å−3
Δρmin = −0.73 e Å−3
Data collection: CrystalClear (Rigaku/MSC, 2006 ▶); cell refinement: CrystalClear; data reduction: CrystalClear; program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: SHELXTL; software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S160053680900350X/at2716sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053680900350X/at2716Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C15—H15C⋯N2 | 0.98 | 2.60 | 3.244 (7) | 123 |
| C18—H18A⋯Cl2 | 0.99 | 2.67 | 3.468 (5) | 138 |
| C23—H23A⋯Cl1i | 0.95 | 2.78 | 3.730 (5) | 175 |
| C26—H26A⋯Cl2ii | 0.99 | 2.46 | 3.431 (6) | 168 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Acknowledgments
We thank the Technological and Scientific Research Council of Turkey TÜBİTAK-CNRS [grant No. TBAG-U/181(106 T716)] and İnönü University Research Fund (BAP 2007/39) for financial support.
supplementary crystallographic information
Comment
The ruthenium complexes of N-Heterocyclic carbenes have proved to be excellent catalysts for the Suzuki-Miyura, Sonogashira, Stille and Heck reactions (Herrmann et al., 1995; Herrmann, 2002; Navarro et al., 2006; Arduengo & Krafczyc, 1998). Expressive examples are found in various catalytic reactions with ruthenium catalysts for alken metathesis, cycloisomerization, and cyclopropanation reactions (Özdemir et al., 2004).
Previous work from our research groups in this area has focused on the elaboration of olefins as electron-rich heterocyclic carbene precursors which allow the formation of chelating carbenes, on the rapidly developing chemistry of η6-arene ruthenium(II) complexes containing substituted imidazolidin-2-ylidenes (Özdemir et al., 2001; Çetinkaya et al., 2001, 2003), and on the synthesis, characterization and uses of palladium, platinum and ruthenium N-heterocyclic carbene complexes as catalysts (Yaşar et al., 2008; Arslan et al., 2004, 2005, 2007, and references therein).
In the present study, we have synthesized and characterized a new ruthenium complex, (1-(2-methylbenzyl)-3-(2,4,6-trimethylbenzyl)-1H-benzo[d]imidazol-2(3H)-ylidene)ruthenium(II) dichloride. dichloromethane solvate, (I). The crystal structure of the title compound, (I), is depicted in Fig. 1.
The benzimidazol ring which has a carbene portion is connected to the C6H2(CH3)3 arene by a CH2 bridge. This leads to a system with very little apparent strain. The ruthenium atom in the title compound is best described as having an octahedral coordination environment, with the arene occupying three coordination sites. Two coordination sites are occupied by Cl ligands, while the sixth site is occupied by the carbene carbon of the benzimidazol ring.
The ruthenium atom is situated 1.6766 (19) Å from the ring centroid of the arene. While there are substantial differences in the C—C and Ru—C distances [Ru—C 92.099 (5), –C10 2.161 (4), –C11 2.246 (4), –C12 2.282 (5), –C13 2.203 (5), –C14 2.198 (5) Å] for the arene ring, there is no evidence of the alternating C—C bonds observed in some ruthenium-arene complexes (Begley et al., 1991).
The arene, the 2-methylbenzyl, imidazol and benzimidazol rings are almost planar with a maximum deviation of 0.038 (5) Å for atom C14, 0.015 (5) Å for atom C21, 0.004 (4) Å for atom N2, and 0.023 (5) Å for atom C5. The five-membered imidazole ring forms dihedral angles of 87.30 (4) ° and 78.53 (4) ° with the 2-methylbenzyl and 2,4,6-trimethylbenzyl rings, respectively.
The small steric demand of the benzimidazole ligand is reflected in the Cl—Ru—C1 angles, which are 87.51 (12) ° and 97.42 (12) °. These are significantly larger than the angles in the pyridine substituted complexes [RuCl2(py)(η6-arene)] (Steedman & Burrell, 1997), and agree with Arslan results, (Arslan et al., 2004, 2005, 2007, and references therein). On the other side, the Ru—Cl distances in the coordination sphere are equal within experimental error [Ru—Cl1 = 2.4167 (12) Å and Ru—Cl2 = 2.4175 (13) Å]. The Cl—Ru—Cl angle is 88.52 (5) °.
The components of the title compound are assembled by two intermolecular C—H···Cl hydrogen bonds, to form a three-dimensional framework (Fig. 2 and Table 1). The intramolecular contacts, C—H···N and C—H···Cl, are also listed in Table 1.
Experimental
A suspension of 1-(2-methylbenzyl)-3-(2,4,6-trimethylbenzyl)benzimidazolium chloride (1.00 g, 2.56 mmol), Cs2CO3 (0.84 g, 2.56 mmol), [RuCl2(p-cymene)]2 (0.78 g, 1.28 mmol) and molecular sieves was heated under reflux in degassed dry toluene (20 ml) for 12 h. The reaction mixture was then filtered while hot, and the volume was reduced to about 10 ml before addition of n-hexane (10 ml). The precipitate formed was crystallized from CH2Cl2:hexane (5:10 ml) to give crystal product (Figure 3). Yield 0.58 g (86%), M.p.: 549–550 K. FT—IR (KBr pellet, cm-1): νCN 1424 cm-1. 1H NMR (δ, 399.9 MHz, CDCl3): 2.18 and 2.34 [s, 9H, CH2C6H2(CH3)3-2,4,6]; 2.39 [s, 3H, CH2C6H4(CH3)-2]; 5.08 [s, 2H, CH2C6H4(CH3)-2]; 5.59 [s, 2H, CH2C6H2(CH3)3-2,4,6]; 5.76 [s, 2H, CH2C6H2(CH3)3-2,4,6]; 6.80–7.50 [m, 8H, NC6H4N and CH2C6H4(CH3)-2]. 13C{H} NMR (δ, 100.5 MHz, CDCl3): 17.0 and 17.4 [CH2C6H2(CH3)3-2,4,6]; 19.5 [CH2C6H4(CH3)-2]; 49.7 [CH2C6H2(CH3)3-2,4,6]; 53.5 [CH2C6H4(CH3)-2]; 90.0, 92.8, 98.6, 101.6, 110.0, 112.5, 123.4, 123.8, 126.0, 127.0, 127.1, 130.2, 133.2, 134.8, 135.0 and 135.4 [CH2C6H2(CH3)3-2,4,6; NC6H4N and CH2C6H4(CH3)-2]; 185.9 [Ccarbene].
Refinement
H atoms were geometrically fixed and allowed to ride on the parent atom with C—H = 0.95 - 0.99 Å, and Uiso(H) = 1.5Ueq(C) for methyl H atoms and 1.2Ueq(C) for other H atoms.
Figures
Fig. 1.
The molecular structure of the title compound, showing the atom-numbering scheme and displacement ellipsoids drawn at the 50% probability level.
Fig. 2.
A packing diagram for (I). [Symmetry codes: A = 1 - x, y, 0.5 - z; B = -1/2 + x, 1/2 + y, z; C = 1.5 - x, 1/2 + y, 0.5 - z; D = 1 - x, 1 - y, 1 - z; E = x, 1 - y, 1/2 + z; F = 1.5 - x, 0.5 - y, 1 - z; G = -1/2 + x, 0.5 - y, 1/2 + z; H = x, -1 + y, z ].
Fig. 3.
Synthesis of Ru(NHC) complex.
Crystal data
| [RuCl2(C25H26N2)]·CH2Cl2 | F(000) = 2480 |
| Mr = 611.37 | Dx = 1.585 Mg m−3 |
| Monoclinic, C2/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -C 2yc | Cell parameters from 5683 reflections |
| a = 31.362 (6) Å | θ = 2.8–26.0° |
| b = 8.1014 (16) Å | µ = 1.05 mm−1 |
| c = 20.484 (4) Å | T = 153 K |
| β = 100.11 (3)° | Rod, orange |
| V = 5123.8 (18) Å3 | 0.46 × 0.14 × 0.06 mm |
| Z = 8 |
Data collection
| Rigaku Mercury CCD (2x2 bin mode) diffractometer | 4501 independent reflections |
| Radiation source: Sealed Tube | 3825 reflections with I > 2σ(I) |
| Graphite Monochromator | Rint = 0.054 |
| Detector resolution: 14.6306 pixels mm-1 | θmax = 25.2°, θmin = 2.8° |
| ω scans | h = −28→37 |
| Absorption correction: multi-scan (REQAB; Jacobson, 1998) | k = −9→9 |
| Tmin = 0.644, Tmax = 0.940 | l = −23→24 |
| 15807 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.049 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.115 | H-atom parameters constrained |
| S = 1.08 | w = 1/[σ2(Fo2) + (0.0431P)2 + 37.6344P] where P = (Fo2 + 2Fc2)/3 |
| 4501 reflections | (Δ/σ)max = 0.001 |
| 298 parameters | Δρmax = 0.92 e Å−3 |
| 0 restraints | Δρmin = −0.73 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Ru1 | 0.859197 (11) | 0.28327 (4) | 0.574586 (17) | 0.01982 (13) | |
| Cl1 | 0.91585 (4) | 0.08062 (14) | 0.60413 (6) | 0.0281 (3) | |
| Cl2 | 0.82324 (4) | 0.09300 (16) | 0.49193 (6) | 0.0355 (3) | |
| N1 | 0.91793 (11) | 0.3570 (4) | 0.46538 (17) | 0.0191 (7) | |
| N2 | 0.91101 (12) | 0.5520 (4) | 0.53600 (17) | 0.0214 (8) | |
| C1 | 0.89796 (14) | 0.3939 (5) | 0.5173 (2) | 0.0201 (9) | |
| C2 | 0.94310 (13) | 0.4900 (5) | 0.4509 (2) | 0.0201 (9) | |
| C3 | 0.96901 (14) | 0.5119 (6) | 0.4034 (2) | 0.0241 (10) | |
| H3A | 0.9716 | 0.4287 | 0.3716 | 0.029* | |
| C4 | 0.99086 (14) | 0.6595 (6) | 0.4043 (2) | 0.0262 (10) | |
| H4A | 1.0093 | 0.6778 | 0.3728 | 0.031* | |
| C5 | 0.98659 (14) | 0.7840 (6) | 0.4505 (2) | 0.0252 (10) | |
| H5A | 1.0026 | 0.8834 | 0.4500 | 0.030* | |
| C6 | 0.95983 (14) | 0.7658 (6) | 0.4967 (2) | 0.0238 (10) | |
| H6A | 0.9562 | 0.8509 | 0.5271 | 0.029* | |
| C7 | 0.93834 (13) | 0.6142 (5) | 0.4960 (2) | 0.0205 (9) | |
| C8 | 0.89772 (16) | 0.6340 (6) | 0.5930 (2) | 0.0276 (10) | |
| H8A | 0.8851 | 0.7437 | 0.5802 | 0.033* | |
| H8B | 0.9228 | 0.6485 | 0.6293 | 0.033* | |
| C9 | 0.86420 (15) | 0.5220 (6) | 0.6150 (2) | 0.0242 (10) | |
| C10 | 0.87528 (15) | 0.4087 (6) | 0.6688 (2) | 0.0249 (10) | |
| C11 | 0.84433 (15) | 0.2836 (6) | 0.6779 (2) | 0.0265 (10) | |
| H11A | 0.8549 | 0.1881 | 0.7073 | 0.032* | |
| C12 | 0.80490 (15) | 0.2661 (6) | 0.6353 (2) | 0.0274 (10) | |
| C13 | 0.79482 (15) | 0.3822 (6) | 0.5822 (2) | 0.0292 (11) | |
| H13A | 0.7701 | 0.3561 | 0.5457 | 0.035* | |
| C14 | 0.82217 (15) | 0.5141 (6) | 0.5727 (2) | 0.0277 (10) | |
| C15 | 0.80849 (18) | 0.6362 (7) | 0.5183 (3) | 0.0381 (12) | |
| H15A | 0.7922 | 0.7258 | 0.5346 | 0.057* | |
| H15B | 0.7901 | 0.5812 | 0.4810 | 0.057* | |
| H15C | 0.8342 | 0.6816 | 0.5036 | 0.057* | |
| C16 | 0.91763 (16) | 0.4145 (6) | 0.7169 (2) | 0.0330 (11) | |
| H16A | 0.9142 | 0.4825 | 0.7553 | 0.049* | |
| H16B | 0.9401 | 0.4624 | 0.6950 | 0.049* | |
| H16C | 0.9261 | 0.3024 | 0.7318 | 0.049* | |
| C17 | 0.77421 (17) | 0.1265 (7) | 0.6407 (3) | 0.0396 (13) | |
| H17A | 0.7546 | 0.1577 | 0.6710 | 0.059* | |
| H17B | 0.7907 | 0.0282 | 0.6578 | 0.059* | |
| H17C | 0.7573 | 0.1024 | 0.5968 | 0.059* | |
| C18 | 0.91506 (14) | 0.2017 (5) | 0.4291 (2) | 0.0203 (9) | |
| H18A | 0.8997 | 0.1196 | 0.4523 | 0.024* | |
| H18B | 0.9446 | 0.1596 | 0.4287 | 0.024* | |
| C19 | 0.89144 (14) | 0.2196 (5) | 0.3583 (2) | 0.0212 (9) | |
| C20 | 0.85252 (15) | 0.3085 (6) | 0.3461 (2) | 0.0264 (10) | |
| H20A | 0.8427 | 0.3619 | 0.3820 | 0.032* | |
| C21 | 0.82819 (16) | 0.3203 (6) | 0.2835 (2) | 0.0320 (11) | |
| H21A | 0.8023 | 0.3838 | 0.2762 | 0.038* | |
| C22 | 0.84158 (18) | 0.2394 (6) | 0.2313 (2) | 0.0339 (12) | |
| H22A | 0.8243 | 0.2426 | 0.1884 | 0.041* | |
| C23 | 0.88025 (16) | 0.1539 (6) | 0.2418 (2) | 0.0296 (11) | |
| H23A | 0.8896 | 0.1011 | 0.2054 | 0.036* | |
| C24 | 0.90593 (16) | 0.1429 (6) | 0.3047 (2) | 0.0262 (10) | |
| C25 | 0.94799 (17) | 0.0499 (7) | 0.3128 (3) | 0.0372 (12) | |
| H25A | 0.9520 | 0.0060 | 0.2697 | 0.056* | |
| H25B | 0.9474 | −0.0415 | 0.3439 | 0.056* | |
| H25C | 0.9720 | 0.1244 | 0.3299 | 0.056* | |
| C26 | 0.81998 (16) | 0.8842 (7) | 0.3454 (3) | 0.0353 (12) | |
| H26A | 0.8182 | 0.9577 | 0.3835 | 0.042* | |
| H26B | 0.8224 | 0.9543 | 0.3066 | 0.042* | |
| Cl3 | 0.77287 (6) | 0.7648 (3) | 0.32788 (10) | 0.0729 (6) | |
| Cl4 | 0.86627 (5) | 0.7586 (2) | 0.36431 (8) | 0.0496 (4) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Ru1 | 0.0249 (2) | 0.0163 (2) | 0.0197 (2) | −0.00305 (13) | 0.00788 (14) | −0.00191 (13) |
| Cl1 | 0.0343 (6) | 0.0203 (6) | 0.0332 (6) | 0.0028 (4) | 0.0156 (5) | 0.0037 (5) |
| Cl2 | 0.0372 (7) | 0.0366 (7) | 0.0346 (7) | −0.0150 (5) | 0.0119 (5) | −0.0159 (5) |
| N1 | 0.0201 (18) | 0.0165 (18) | 0.0207 (18) | −0.0012 (14) | 0.0035 (14) | −0.0015 (15) |
| N2 | 0.031 (2) | 0.0133 (18) | 0.0213 (19) | −0.0040 (15) | 0.0094 (15) | −0.0024 (14) |
| C1 | 0.026 (2) | 0.017 (2) | 0.017 (2) | −0.0001 (17) | 0.0010 (17) | 0.0010 (17) |
| C2 | 0.021 (2) | 0.017 (2) | 0.022 (2) | −0.0026 (17) | 0.0024 (17) | 0.0032 (17) |
| C3 | 0.027 (2) | 0.023 (2) | 0.023 (2) | 0.0029 (18) | 0.0048 (18) | 0.0029 (18) |
| C4 | 0.023 (2) | 0.028 (3) | 0.030 (2) | −0.0005 (19) | 0.0099 (19) | 0.010 (2) |
| C5 | 0.022 (2) | 0.020 (2) | 0.033 (3) | −0.0061 (18) | 0.0033 (19) | 0.0048 (19) |
| C6 | 0.023 (2) | 0.021 (2) | 0.026 (2) | −0.0019 (18) | 0.0015 (18) | 0.0011 (19) |
| C7 | 0.021 (2) | 0.019 (2) | 0.021 (2) | −0.0009 (17) | 0.0037 (17) | 0.0026 (17) |
| C8 | 0.039 (3) | 0.020 (2) | 0.026 (2) | −0.006 (2) | 0.013 (2) | −0.0064 (19) |
| C9 | 0.031 (2) | 0.020 (2) | 0.023 (2) | 0.0011 (19) | 0.0112 (19) | −0.0055 (18) |
| C10 | 0.032 (2) | 0.022 (2) | 0.023 (2) | 0.0023 (19) | 0.0114 (19) | −0.0065 (18) |
| C11 | 0.033 (3) | 0.021 (2) | 0.027 (2) | 0.0008 (19) | 0.012 (2) | −0.0024 (19) |
| C12 | 0.029 (2) | 0.029 (3) | 0.029 (3) | −0.002 (2) | 0.017 (2) | −0.004 (2) |
| C13 | 0.029 (2) | 0.031 (3) | 0.028 (2) | 0.006 (2) | 0.008 (2) | −0.005 (2) |
| C14 | 0.030 (2) | 0.024 (2) | 0.031 (3) | 0.0044 (19) | 0.012 (2) | −0.003 (2) |
| C15 | 0.042 (3) | 0.031 (3) | 0.039 (3) | 0.005 (2) | 0.000 (2) | 0.005 (2) |
| C16 | 0.038 (3) | 0.031 (3) | 0.030 (3) | −0.002 (2) | 0.007 (2) | −0.003 (2) |
| C17 | 0.038 (3) | 0.035 (3) | 0.049 (3) | −0.010 (2) | 0.017 (2) | −0.001 (2) |
| C18 | 0.024 (2) | 0.016 (2) | 0.022 (2) | −0.0007 (17) | 0.0056 (17) | −0.0036 (17) |
| C19 | 0.029 (2) | 0.015 (2) | 0.020 (2) | −0.0061 (17) | 0.0062 (18) | −0.0014 (17) |
| C20 | 0.031 (2) | 0.024 (2) | 0.024 (2) | −0.0016 (19) | 0.0067 (19) | −0.0007 (19) |
| C21 | 0.030 (3) | 0.034 (3) | 0.031 (3) | −0.003 (2) | 0.003 (2) | 0.005 (2) |
| C22 | 0.044 (3) | 0.034 (3) | 0.022 (3) | −0.012 (2) | 0.002 (2) | 0.005 (2) |
| C23 | 0.047 (3) | 0.025 (3) | 0.019 (2) | −0.007 (2) | 0.012 (2) | −0.0028 (19) |
| C24 | 0.039 (3) | 0.018 (2) | 0.023 (2) | −0.004 (2) | 0.010 (2) | 0.0002 (18) |
| C25 | 0.049 (3) | 0.032 (3) | 0.033 (3) | 0.005 (2) | 0.013 (2) | −0.004 (2) |
| C26 | 0.041 (3) | 0.034 (3) | 0.030 (3) | −0.001 (2) | 0.003 (2) | −0.003 (2) |
| Cl3 | 0.0424 (9) | 0.0820 (13) | 0.0869 (13) | −0.0229 (9) | −0.0094 (9) | 0.0202 (10) |
| Cl4 | 0.0440 (8) | 0.0547 (9) | 0.0488 (8) | 0.0042 (7) | 0.0051 (6) | 0.0140 (7) |
Geometric parameters (Å, °)
| Ru1—C1 | 2.039 (4) | C12—C17 | 1.502 (7) |
| Ru1—C9 | 2.099 (4) | C13—C14 | 1.405 (7) |
| Ru1—C10 | 2.162 (4) | C13—H13A | 1.0000 |
| Ru1—C14 | 2.198 (5) | C14—C15 | 1.496 (7) |
| Ru1—C13 | 2.203 (5) | C15—H15A | 0.9800 |
| Ru1—C11 | 2.246 (5) | C15—H15B | 0.9800 |
| Ru1—C12 | 2.282 (5) | C15—H15C | 0.9800 |
| Ru1—Cl1 | 2.4167 (12) | C16—H16A | 0.9800 |
| Ru1—Cl2 | 2.4175 (13) | C16—H16B | 0.9800 |
| N1—C1 | 1.359 (6) | C16—H16C | 0.9800 |
| N1—C2 | 1.398 (5) | C17—H17A | 0.9800 |
| N1—C18 | 1.456 (5) | C17—H17B | 0.9800 |
| N2—C1 | 1.378 (5) | C17—H17C | 0.9800 |
| N2—C7 | 1.381 (6) | C18—C19 | 1.515 (6) |
| N2—C8 | 1.466 (6) | C18—H18A | 0.9900 |
| C2—C3 | 1.384 (6) | C18—H18B | 0.9900 |
| C2—C7 | 1.392 (6) | C19—C20 | 1.401 (6) |
| C3—C4 | 1.377 (6) | C19—C24 | 1.405 (6) |
| C3—H3A | 0.9500 | C20—C21 | 1.377 (6) |
| C4—C5 | 1.407 (7) | C20—H20A | 0.9500 |
| C4—H4A | 0.9500 | C21—C22 | 1.380 (8) |
| C5—C6 | 1.378 (7) | C21—H21A | 0.9500 |
| C5—H5A | 0.9500 | C22—C23 | 1.380 (7) |
| C6—C7 | 1.399 (6) | C22—H22A | 0.9500 |
| C6—H6A | 0.9500 | C23—C24 | 1.397 (7) |
| C8—C9 | 1.515 (6) | C23—H23A | 0.9500 |
| C8—H8A | 0.9900 | C24—C25 | 1.503 (7) |
| C8—H8B | 0.9900 | C25—H25A | 0.9800 |
| C9—C10 | 1.430 (6) | C25—H25B | 0.9800 |
| C9—C14 | 1.446 (6) | C25—H25C | 0.9800 |
| C10—C11 | 1.438 (6) | C26—Cl3 | 1.750 (5) |
| C10—C16 | 1.509 (7) | C26—Cl4 | 1.760 (5) |
| C11—C12 | 1.390 (7) | C26—H26A | 0.9900 |
| C11—H11A | 1.0000 | C26—H26B | 0.9900 |
| C12—C13 | 1.430 (7) | ||
| C1—Ru1—C9 | 79.10 (17) | C16—C10—Ru1 | 129.7 (3) |
| C1—Ru1—C10 | 103.77 (17) | C12—C11—C10 | 122.4 (4) |
| C9—Ru1—C10 | 39.19 (17) | C12—C11—Ru1 | 73.5 (3) |
| C1—Ru1—C14 | 89.00 (17) | C10—C11—Ru1 | 67.8 (2) |
| C9—Ru1—C14 | 39.24 (17) | C12—C11—H11A | 117.6 |
| C10—Ru1—C14 | 69.83 (17) | C10—C11—H11A | 117.6 |
| C1—Ru1—C13 | 121.82 (18) | Ru1—C11—H11A | 117.6 |
| C9—Ru1—C13 | 69.08 (18) | C11—C12—C13 | 117.7 (4) |
| C10—Ru1—C13 | 80.71 (18) | C11—C12—C17 | 122.8 (5) |
| C14—Ru1—C13 | 37.24 (18) | C13—C12—C17 | 119.5 (4) |
| C1—Ru1—C11 | 141.73 (17) | C11—C12—Ru1 | 70.7 (3) |
| C9—Ru1—C11 | 68.95 (17) | C13—C12—Ru1 | 68.4 (3) |
| C10—Ru1—C11 | 38.02 (17) | C17—C12—Ru1 | 129.4 (3) |
| C14—Ru1—C11 | 79.57 (17) | C14—C13—C12 | 123.1 (4) |
| C13—Ru1—C11 | 65.68 (18) | C14—C13—Ru1 | 71.2 (3) |
| C1—Ru1—C12 | 156.54 (17) | C12—C13—Ru1 | 74.4 (3) |
| C9—Ru1—C12 | 81.52 (17) | C14—C13—H13A | 117.9 |
| C10—Ru1—C12 | 67.76 (17) | C12—C13—H13A | 117.9 |
| C14—Ru1—C12 | 67.59 (17) | Ru1—C13—H13A | 117.9 |
| C13—Ru1—C12 | 37.12 (18) | C13—C14—C9 | 117.7 (4) |
| C11—Ru1—C12 | 35.75 (17) | C13—C14—C15 | 120.3 (4) |
| C1—Ru1—Cl1 | 87.51 (12) | C9—C14—C15 | 121.9 (4) |
| C9—Ru1—Cl1 | 121.77 (13) | C13—C14—Ru1 | 71.6 (3) |
| C10—Ru1—Cl1 | 92.85 (13) | C9—C14—Ru1 | 66.7 (2) |
| C14—Ru1—Cl1 | 160.95 (13) | C15—C14—Ru1 | 131.1 (3) |
| C13—Ru1—Cl1 | 150.67 (13) | C14—C15—H15A | 109.5 |
| C11—Ru1—Cl1 | 91.71 (13) | C14—C15—H15B | 109.5 |
| C12—Ru1—Cl1 | 114.09 (13) | H15A—C15—H15B | 109.5 |
| C1—Ru1—Cl2 | 97.42 (12) | C14—C15—H15C | 109.5 |
| C9—Ru1—Cl2 | 149.00 (13) | H15A—C15—H15C | 109.5 |
| C10—Ru1—Cl2 | 158.81 (13) | H15B—C15—H15C | 109.5 |
| C14—Ru1—Cl2 | 110.51 (13) | C10—C16—H16A | 109.5 |
| C13—Ru1—Cl2 | 87.77 (13) | C10—C16—H16B | 109.5 |
| C11—Ru1—Cl2 | 120.82 (12) | H16A—C16—H16B | 109.5 |
| C12—Ru1—Cl2 | 92.40 (12) | C10—C16—H16C | 109.5 |
| Cl1—Ru1—Cl2 | 88.52 (5) | H16A—C16—H16C | 109.5 |
| C1—N1—C2 | 110.6 (3) | H16B—C16—H16C | 109.5 |
| C1—N1—C18 | 126.4 (4) | C12—C17—H17A | 109.5 |
| C2—N1—C18 | 123.0 (3) | C12—C17—H17B | 109.5 |
| C1—N2—C7 | 111.1 (4) | H17A—C17—H17B | 109.5 |
| C1—N2—C8 | 122.0 (4) | C12—C17—H17C | 109.5 |
| C7—N2—C8 | 126.9 (4) | H17A—C17—H17C | 109.5 |
| N1—C1—N2 | 105.5 (4) | H17B—C17—H17C | 109.5 |
| N1—C1—Ru1 | 139.2 (3) | N1—C18—C19 | 112.5 (3) |
| N2—C1—Ru1 | 115.2 (3) | N1—C18—H18A | 109.1 |
| C3—C2—C7 | 121.3 (4) | C19—C18—H18A | 109.1 |
| C3—C2—N1 | 132.1 (4) | N1—C18—H18B | 109.1 |
| C7—C2—N1 | 106.6 (4) | C19—C18—H18B | 109.1 |
| C4—C3—C2 | 117.0 (4) | H18A—C18—H18B | 107.8 |
| C4—C3—H3A | 121.5 | C20—C19—C24 | 118.6 (4) |
| C2—C3—H3A | 121.5 | C20—C19—C18 | 118.9 (4) |
| C3—C4—C5 | 121.7 (4) | C24—C19—C18 | 122.4 (4) |
| C3—C4—H4A | 119.1 | C21—C20—C19 | 121.7 (4) |
| C5—C4—H4A | 119.1 | C21—C20—H20A | 119.2 |
| C6—C5—C4 | 121.8 (4) | C19—C20—H20A | 119.2 |
| C6—C5—H5A | 119.1 | C20—C21—C22 | 119.6 (5) |
| C4—C5—H5A | 119.1 | C20—C21—H21A | 120.2 |
| C5—C6—C7 | 115.9 (4) | C22—C21—H21A | 120.2 |
| C5—C6—H6A | 122.0 | C21—C22—C23 | 119.7 (5) |
| C7—C6—H6A | 122.0 | C21—C22—H22A | 120.1 |
| N2—C7—C2 | 106.3 (4) | C23—C22—H22A | 120.1 |
| N2—C7—C6 | 131.5 (4) | C22—C23—C24 | 121.6 (4) |
| C2—C7—C6 | 122.2 (4) | C22—C23—H23A | 119.2 |
| N2—C8—C9 | 105.9 (4) | C24—C23—H23A | 119.2 |
| N2—C8—H8A | 110.6 | C23—C24—C19 | 118.7 (4) |
| C9—C8—H8A | 110.6 | C23—C24—C25 | 119.1 (4) |
| N2—C8—H8B | 110.6 | C19—C24—C25 | 122.3 (4) |
| C9—C8—H8B | 110.6 | C24—C25—H25A | 109.5 |
| H8A—C8—H8B | 108.7 | C24—C25—H25B | 109.5 |
| C10—C9—C14 | 120.4 (4) | H25A—C25—H25B | 109.5 |
| C10—C9—C8 | 121.7 (4) | C24—C25—H25C | 109.5 |
| C14—C9—C8 | 117.1 (4) | H25A—C25—H25C | 109.5 |
| C10—C9—Ru1 | 72.8 (3) | H25B—C25—H25C | 109.5 |
| C14—C9—Ru1 | 74.1 (3) | Cl3—C26—Cl4 | 111.1 (3) |
| C8—C9—Ru1 | 116.3 (3) | Cl3—C26—H26A | 109.4 |
| C9—C10—C11 | 118.3 (4) | Cl4—C26—H26A | 109.4 |
| C9—C10—C16 | 123.4 (4) | Cl3—C26—H26B | 109.4 |
| C11—C10—C16 | 118.3 (4) | Cl4—C26—H26B | 109.4 |
| C9—C10—Ru1 | 68.0 (2) | H26A—C26—H26B | 108.0 |
| C11—C10—Ru1 | 74.2 (3) | ||
| C2—N1—C1—N2 | −0.4 (4) | C14—Ru1—C11—C12 | 65.9 (3) |
| C18—N1—C1—N2 | 178.7 (4) | C13—Ru1—C11—C12 | 29.6 (3) |
| C2—N1—C1—Ru1 | −176.3 (4) | Cl1—Ru1—C11—C12 | −131.1 (3) |
| C18—N1—C1—Ru1 | 2.8 (7) | Cl2—Ru1—C11—C12 | −41.8 (3) |
| C7—N2—C1—N1 | 0.7 (5) | C1—Ru1—C11—C10 | 4.3 (4) |
| C8—N2—C1—N1 | −177.2 (4) | C9—Ru1—C11—C10 | −31.1 (3) |
| C7—N2—C1—Ru1 | 177.7 (3) | C14—Ru1—C11—C10 | −70.5 (3) |
| C8—N2—C1—Ru1 | −0.1 (5) | C13—Ru1—C11—C10 | −106.9 (3) |
| C9—Ru1—C1—N1 | −178.4 (5) | C12—Ru1—C11—C10 | −136.4 (4) |
| C10—Ru1—C1—N1 | 150.9 (5) | Cl1—Ru1—C11—C10 | 92.4 (3) |
| C14—Ru1—C1—N1 | −140.1 (5) | Cl2—Ru1—C11—C10 | −178.3 (2) |
| C13—Ru1—C1—N1 | −121.6 (5) | C10—C11—C12—C13 | −2.4 (7) |
| C11—Ru1—C1—N1 | 148.2 (4) | Ru1—C11—C12—C13 | −51.5 (4) |
| C12—Ru1—C1—N1 | −143.5 (5) | C10—C11—C12—C17 | 174.2 (4) |
| Cl1—Ru1—C1—N1 | 58.6 (5) | Ru1—C11—C12—C17 | 125.1 (5) |
| Cl2—Ru1—C1—N1 | −29.6 (5) | C10—C11—C12—Ru1 | 49.1 (4) |
| C9—Ru1—C1—N2 | 6.0 (3) | C1—Ru1—C12—C11 | −100.0 (5) |
| C10—Ru1—C1—N2 | −24.7 (3) | C9—Ru1—C12—C11 | −65.5 (3) |
| C14—Ru1—C1—N2 | 44.2 (3) | C10—Ru1—C12—C11 | −27.3 (3) |
| C13—Ru1—C1—N2 | 62.8 (4) | C14—Ru1—C12—C11 | −103.7 (3) |
| C11—Ru1—C1—N2 | −27.4 (5) | C13—Ru1—C12—C11 | −131.9 (4) |
| C12—Ru1—C1—N2 | 40.8 (6) | Cl1—Ru1—C12—C11 | 55.6 (3) |
| Cl1—Ru1—C1—N2 | −117.0 (3) | Cl2—Ru1—C12—C11 | 145.0 (3) |
| Cl2—Ru1—C1—N2 | 154.8 (3) | C1—Ru1—C12—C13 | 31.8 (6) |
| C1—N1—C2—C3 | 180.0 (4) | C9—Ru1—C12—C13 | 66.3 (3) |
| C18—N1—C2—C3 | 0.9 (7) | C10—Ru1—C12—C13 | 104.6 (3) |
| C1—N1—C2—C7 | 0.0 (5) | C14—Ru1—C12—C13 | 28.1 (3) |
| C18—N1—C2—C7 | −179.1 (4) | C11—Ru1—C12—C13 | 131.9 (4) |
| C7—C2—C3—C4 | 2.1 (6) | Cl1—Ru1—C12—C13 | −172.6 (2) |
| N1—C2—C3—C4 | −177.9 (4) | Cl2—Ru1—C12—C13 | −83.1 (3) |
| C2—C3—C4—C5 | −1.0 (6) | C1—Ru1—C12—C17 | 142.9 (5) |
| C3—C4—C5—C6 | −1.0 (7) | C9—Ru1—C12—C17 | 177.4 (5) |
| C4—C5—C6—C7 | 2.0 (6) | C10—Ru1—C12—C17 | −144.3 (5) |
| C1—N2—C7—C2 | −0.7 (5) | C14—Ru1—C12—C17 | 139.2 (5) |
| C8—N2—C7—C2 | 177.1 (4) | C13—Ru1—C12—C17 | 111.1 (6) |
| C1—N2—C7—C6 | −179.0 (4) | C11—Ru1—C12—C17 | −117.0 (6) |
| C8—N2—C7—C6 | −1.2 (8) | Cl1—Ru1—C12—C17 | −61.5 (5) |
| C3—C2—C7—N2 | −179.6 (4) | Cl2—Ru1—C12—C17 | 28.0 (5) |
| N1—C2—C7—N2 | 0.4 (4) | C11—C12—C13—C14 | −1.9 (7) |
| C3—C2—C7—C6 | −1.1 (6) | C17—C12—C13—C14 | −178.5 (4) |
| N1—C2—C7—C6 | 178.9 (4) | Ru1—C12—C13—C14 | −54.4 (4) |
| C5—C6—C7—N2 | 177.1 (4) | C11—C12—C13—Ru1 | 52.5 (4) |
| C5—C6—C7—C2 | −0.9 (6) | C17—C12—C13—Ru1 | −124.1 (4) |
| C1—N2—C8—C9 | −8.3 (6) | C1—Ru1—C13—C14 | −31.7 (3) |
| C7—N2—C8—C9 | 174.2 (4) | C9—Ru1—C13—C14 | 29.9 (3) |
| N2—C8—C9—C10 | 98.1 (5) | C10—Ru1—C13—C14 | 68.8 (3) |
| N2—C8—C9—C14 | −71.9 (5) | C11—Ru1—C13—C14 | 105.4 (3) |
| N2—C8—C9—Ru1 | 12.9 (5) | C12—Ru1—C13—C14 | 134.0 (4) |
| C1—Ru1—C9—C10 | −128.3 (3) | Cl1—Ru1—C13—C14 | 147.9 (2) |
| C14—Ru1—C9—C10 | 129.7 (4) | Cl2—Ru1—C13—C14 | −129.1 (3) |
| C13—Ru1—C9—C10 | 101.3 (3) | C1—Ru1—C13—C12 | −165.7 (3) |
| C11—Ru1—C9—C10 | 30.3 (3) | C9—Ru1—C13—C12 | −104.1 (3) |
| C12—Ru1—C9—C10 | 65.0 (3) | C10—Ru1—C13—C12 | −65.2 (3) |
| Cl1—Ru1—C9—C10 | −48.2 (3) | C14—Ru1—C13—C12 | −134.0 (4) |
| Cl2—Ru1—C9—C10 | 145.4 (2) | C11—Ru1—C13—C12 | −28.5 (3) |
| C1—Ru1—C9—C14 | 102.0 (3) | Cl1—Ru1—C13—C12 | 13.9 (4) |
| C10—Ru1—C9—C14 | −129.7 (4) | Cl2—Ru1—C13—C12 | 96.9 (3) |
| C13—Ru1—C9—C14 | −28.4 (3) | C12—C13—C14—C9 | 6.2 (7) |
| C11—Ru1—C9—C14 | −99.5 (3) | Ru1—C13—C14—C9 | −49.7 (4) |
| C12—Ru1—C9—C14 | −64.7 (3) | C12—C13—C14—C15 | −176.7 (4) |
| Cl1—Ru1—C9—C14 | −177.9 (2) | Ru1—C13—C14—C15 | 127.4 (4) |
| Cl2—Ru1—C9—C14 | 15.7 (4) | C12—C13—C14—Ru1 | 55.9 (4) |
| C1—Ru1—C9—C8 | −10.9 (3) | C10—C9—C14—C13 | −6.4 (6) |
| C10—Ru1—C9—C8 | 117.4 (5) | C8—C9—C14—C13 | 163.8 (4) |
| C14—Ru1—C9—C8 | −112.8 (4) | Ru1—C9—C14—C13 | 52.0 (4) |
| C13—Ru1—C9—C8 | −141.3 (4) | C10—C9—C14—C15 | 176.6 (4) |
| C11—Ru1—C9—C8 | 147.7 (4) | C8—C9—C14—C15 | −13.3 (6) |
| C12—Ru1—C9—C8 | −177.6 (4) | Ru1—C9—C14—C15 | −125.0 (4) |
| Cl1—Ru1—C9—C8 | 69.3 (4) | C10—C9—C14—Ru1 | −58.4 (4) |
| Cl2—Ru1—C9—C8 | −97.1 (4) | C8—C9—C14—Ru1 | 111.8 (4) |
| C14—C9—C10—C11 | 2.5 (6) | C1—Ru1—C14—C13 | 153.4 (3) |
| C8—C9—C10—C11 | −167.2 (4) | C9—Ru1—C14—C13 | −132.7 (4) |
| Ru1—C9—C10—C11 | −56.5 (4) | C10—Ru1—C14—C13 | −101.5 (3) |
| C14—C9—C10—C16 | −177.0 (4) | C11—Ru1—C14—C13 | −63.3 (3) |
| C8—C9—C10—C16 | 13.3 (7) | C12—Ru1—C14—C13 | −28.0 (3) |
| Ru1—C9—C10—C16 | 124.0 (4) | Cl1—Ru1—C14—C13 | −127.2 (4) |
| C14—C9—C10—Ru1 | 59.0 (4) | Cl2—Ru1—C14—C13 | 55.9 (3) |
| C8—C9—C10—Ru1 | −110.7 (4) | C1—Ru1—C14—C9 | −73.9 (3) |
| C1—Ru1—C10—C9 | 52.5 (3) | C10—Ru1—C14—C9 | 31.2 (3) |
| C14—Ru1—C10—C9 | −31.2 (3) | C13—Ru1—C14—C9 | 132.7 (4) |
| C13—Ru1—C10—C9 | −68.2 (3) | C11—Ru1—C14—C9 | 69.4 (3) |
| C11—Ru1—C10—C9 | −130.2 (4) | C12—Ru1—C14—C9 | 104.6 (3) |
| C12—Ru1—C10—C9 | −104.4 (3) | Cl1—Ru1—C14—C9 | 5.5 (5) |
| Cl1—Ru1—C10—C9 | 140.6 (2) | Cl2—Ru1—C14—C9 | −171.4 (2) |
| Cl2—Ru1—C10—C9 | −126.1 (3) | C1—Ru1—C14—C15 | 39.0 (5) |
| C1—Ru1—C10—C11 | −177.3 (3) | C9—Ru1—C14—C15 | 112.8 (6) |
| C9—Ru1—C10—C11 | 130.2 (4) | C10—Ru1—C14—C15 | 144.0 (5) |
| C14—Ru1—C10—C11 | 99.0 (3) | C13—Ru1—C14—C15 | −114.5 (6) |
| C13—Ru1—C10—C11 | 62.1 (3) | C11—Ru1—C14—C15 | −177.8 (5) |
| C12—Ru1—C10—C11 | 25.8 (3) | C12—Ru1—C14—C15 | −142.5 (5) |
| Cl1—Ru1—C10—C11 | −89.1 (3) | Cl1—Ru1—C14—C15 | 118.4 (5) |
| Cl2—Ru1—C10—C11 | 4.1 (5) | Cl2—Ru1—C14—C15 | −58.6 (5) |
| C1—Ru1—C10—C16 | −63.4 (4) | C1—N1—C18—C19 | 112.6 (5) |
| C9—Ru1—C10—C16 | −115.9 (5) | C2—N1—C18—C19 | −68.4 (5) |
| C14—Ru1—C10—C16 | −147.1 (5) | N1—C18—C19—C20 | −45.8 (5) |
| C13—Ru1—C10—C16 | 175.9 (5) | N1—C18—C19—C24 | 137.6 (4) |
| C11—Ru1—C10—C16 | 113.9 (5) | C24—C19—C20—C21 | 0.8 (7) |
| C12—Ru1—C10—C16 | 139.7 (5) | C18—C19—C20—C21 | −175.9 (4) |
| Cl1—Ru1—C10—C16 | 24.7 (4) | C19—C20—C21—C22 | 1.7 (7) |
| Cl2—Ru1—C10—C16 | 118.0 (4) | C20—C21—C22—C23 | −3.0 (8) |
| C9—C10—C11—C12 | 2.0 (7) | C21—C22—C23—C24 | 1.7 (7) |
| C16—C10—C11—C12 | −178.5 (4) | C22—C23—C24—C19 | 0.9 (7) |
| Ru1—C10—C11—C12 | −51.5 (4) | C22—C23—C24—C25 | −179.2 (5) |
| C9—C10—C11—Ru1 | 53.5 (3) | C20—C19—C24—C23 | −2.1 (6) |
| C16—C10—C11—Ru1 | −127.0 (4) | C18—C19—C24—C23 | 174.5 (4) |
| C1—Ru1—C11—C12 | 140.7 (3) | C20—C19—C24—C25 | 177.9 (4) |
| C9—Ru1—C11—C12 | 105.3 (3) | C18—C19—C24—C25 | −5.5 (7) |
| C10—Ru1—C11—C12 | 136.4 (4) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C15—H15C···N2 | 0.98 | 2.60 | 3.244 (7) | 123 |
| C18—H18A···Cl2 | 0.99 | 2.67 | 3.468 (5) | 138 |
| C23—H23A···Cl1i | 0.95 | 2.78 | 3.730 (5) | 175 |
| C26—H26A···Cl2ii | 0.99 | 2.46 | 3.431 (6) | 168 |
Symmetry codes: (i) x, −y, z−1/2; (ii) x, y+1, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: AT2716).
References
- Arduengo, A. J. & Krafczyc, R. (1998). Chem. Ztg, 32, 6–14.
- Arslan, H., VanDerveer, D., Özdemir, İ., Çetinkaya, B. & Demir, S. (2004). Z. Kristallogr. New Cryst. Struct.219, 377–378.
- Arslan, H., VanDerveer, D., Özdemir, I., Yaşar, S. & Çetinkaya, B. (2005). Acta Cryst. E61, m1873–m1875.
- Arslan, H., VanDerveer, D., Yaşar, S., Özdemir, İ. & Çetinkaya, B. (2007). Acta Cryst. E63, m1001–m1003.
- Begley, M. J., Harrison, S. & Wright, A. H. (1991). Acta Cryst. C47, 318–320.
- Çetinkaya, B., Demir, S., Özdemir, İ., Toupet, L., Semeril, D., Bruneau, C. & Dixneuf, P. H. (2001). New J. Chem.25, 519–521. [DOI] [PubMed]
- Çetinkaya, B., Demir, S., Özdemir, İ., Toupet, L., Semeril, D., Bruneau, C. & Dixneuf, P. H. (2003). Chem. Eur. J.9, 2323–2330. [DOI] [PubMed]
- Herrmann, W. A. (2002). Angew. Chem. Int. Ed.41, 1290–1309.
- Herrmann, W. A., Elison, M., Fischer, J., Köcher, C. & Artus, G. R. J. (1995). Angew. Chem. Int. Ed. Engl.34, 2371–2374.
- Jacobson, R. (1998). REQAB. Molecular Structure Corporation, The Woodlands, Texas, USA.
- Navarro, O., Marion, N., Oonishi, Y., Kelly, R. A. & Nolan, S. P. (2006). J. Org. Chem.71, 685–692. [DOI] [PubMed]
- Özdemir, İ., Alıcı, B., Gürbüz, N., Çetinkaya, E. & Çetinkaya, B. (2004). J. Mol. Catal. A Chem.217, 37–40.
- Özdemir, İ., Yiğit, B., Çetinkaya, B., Ülkü, D., Tahir, M. N. & Arici, C. (2001). J. Organomet. Chem.633, 27–32.
- Rigaku/MSC (2006). CrystalClear Rigaku/MSC, The Woodlands, Texas, USA.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Steedman, A. J. & Burrell, A. K. (1997). Acta Cryst. C53, 864–866.
- Yaşar, S., Özdemir, İ., Çetinkaya, B., Renaud, J. L. & Bruneau, C. (2008). Eur. J. Org. Chem.12, 2142–2149.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S160053680900350X/at2716sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053680900350X/at2716Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report





