Abstract
We report here the development of a regulated gene expression system for Dictyostelium discoideum based on the DNA-damage inducibility of the rnrB gene. rnrB, which codes for the small subunit of the enzyme ribonucleotide reductase, responds to DNA-damaging agents at all stages of the D.discoideum life cycle. Doses that have little effect on development have previously been shown to increase the level of the rnrB transcript by up to 15-fold. Here we show that all elements necessary for DNA-damage induction are contained in a 450 bp promoter fragment. We used a fusion of the rnrB promoter with the gene encoding GFP to demonstrate an up to 10-fold induction at the RNA level, which appears in all aspects similar to induction of the endogenous rnrB transcript. Using a fusion with the lacZ gene we observed an up to 7-fold induction at the protein level. These results indicate that the rnrB promoter can be used to regulate the expression of specific genes in D.discoideum. This controllable gene expression system provides the following new characteristics: the induction is rapid, taking place in the order of minutes, and the promoter is responsive at all stages of the D.discoideum life cycle.
INTRODUCTION
Inducible promoters are widely used for the analysis of gene function. Ideally, an inducible promoter vector should have the following characteristics: (i) low basal expression level; (ii) high expression following induction; (iii) rapid inducibility; (iv) inducibility under all physiological conditions. The cellular slime mold Dictyostelium discoideum is extensively exploited as a model organism for the analysis of developmental processes. However, very few regulated gene expression vectors are available. Presently, the most widely used regulated gene expression system in D.discoideum is based on repression of the discoidin Iγ promoter by the presence of folate (1). There are several limitations to the use of this system. The response to changes in folate levels is slow, with a full response observable only after ∼12 h (2; B.Wetterauer, personal communication). The discoidin Iγ promoter can be modulated by folate only during the growth phase (1). Moreover, since the discoidin Iγ promoter is insensitive to folate at high cell concentrations, cells must be held continuously at a low density (2).
Another conditional gene expression system has been adapted for D.discoideum that uses an inducible suppressor tRNA gene. However, this system also responds fairly slowly; furthermore, the coding sequence of the gene of interest must be mutated to generate the required amber codon (3).
Recently, the widely used tetracycline-regulated expression system from mammalian cells has been adapted for use in D.discoideum (4). Expression of the gene of interest is low in the presence of the antibiotic. Removal of tetracycline from the medium allows expression of the gene of interest to high levels. Similar to the discoidin system, however, the response is relatively slow and can only be triggered in vegetative cells.
The gene rnrB encodes the catalytic subunit of ribonucleotide reductase in D.discoideum (5). This enzyme reduces ribonucleotides to deoxyribonucleotides, the first reaction for de novo synthesis of deoxyribonucleotides. The genes encoding ribonucleotide reductase are up-regulated in the presence of DNA-damaging agents in all organisms studied up to now (6), including D.discoideum (7). We have identified promoter elements involved in the DNA-damage response of this gene (7). Several characteristics of the rnrB promoter suggest that it would be a good candidate to be used to regulate the expression of exogenous genes. The up-regulation of transcriptional activity is rapid; an increase in transcript level can be detected as early as 10 min after the addition of DNA-damaging agents. Cells at all stages of development are responsive in induction of the rnrB gene following exposure to DNA-damaging agents, a feature that presently none of the other systems allow. Also, the response can be modulated by adding different doses of drugs, which cause different levels of response (7). We report here the development of a regulated gene expression system for D.discoideum based on the DNA-damage inducibility of the rnrB gene.
MATERIALS AND METHODS
Plasmid construction and transformation of D.discoideum
Construction of the rnrB-ubi-S65TGFP and rnrB-ile-gal reporters has been described elsewhere (H.MacWilliams et al., submitted for publication). To make the rnrB-ubi-S65TGFP reporter construct the rnrB promoter was amplified from plasmid rnrB/lacZ (5) using the primers 5′-GATTTATTAATATCACCAAC-3′ (which binds upstream of the XbaI site of the rnrB promoter) and 5′-AGATCTCATTTTTATTTTTTATTTTTAAT-3′ (which overlaps the ATG codon of rnrB and adds a downstream BglII site). The 453 bp XbaI–BglII fragment was used to replace the PsA promoter in the PsA::ubi-S65TGFP plasmid (8), resulting in rnrB-ubi-S65TGFP.
The rnrB::ile-gal construct was made using the same PCR product to replace the PsA promoter in PsA::ile-gal (9). To construct rnrB::ile-αpgal the gene encoding ile-gal was amplified by PCR using PsA::ile-gal as template. The upstream primer (5′-AGATCTAAAATGCAGATTTTC-3′) starts at the BglII site and extends into the ubiquitin gene. The downstream primer (5′-AGATCTCTGCATTAATGAATCGGCC-3′) is complementary to the codons for ADSLMQ and adds a BglII site. This product was cloned into the BglII site of PsA::gal (9) to make PsA-ile-αpgal. The downstream BglII site was then eliminated with no change in coding by reamplification of the ubiquitin and destabilizing N-terminal sequences. This was carried out using as template a vector in which the PsA promoter had been replaced with an ecmA promoter; the 5′ primer (5′-AAAAAGAAATTAATTATACATACC-3′) recognized a sequence within this promoter, while the downstream primer (5′-AAGCTTGGTACCGCGATCTCTGCATTAATG-3′) mutated the downstream BglII site (at the underlined C) and extended to a HindIII site which, in the template, is immediately downstream of the destabilizing N-terminus. This second PCR product was cloned in pCR-script and opened at HindIII, and lacZ and Actin8 terminator sequences were added as a HindIII–HindIII fragment from PsA::gal. Finally, the ubi-ile-αpgal reporter gene was excised as a BglII–XhoI fragment, which was used to replace the ubi-S65TGFP fragment in rnrB-ubi-S65GFP to make rnrB-ile-αpgal.
To construct the RNRP vector, two oligonucleotides were synthesized that anneal in such a way that the duplex contains protruding sequences that anneal to a BglII restriction site at the 5′-end and a XhoI site at the 3′-end. The sequences of the oligonucleotides were as follows: sense strand, 5′-GATCTTAAGAATTCCTTCC-3′; antisense strand, 5′-TCGAGGAAGGAATTCTTAA-3′. The annealed oligonucleotides were used to replace the BglII–XhoI fragment containing the GFP gene in the rnrB-ubi-S65GFP construct.
To construct the pRNR-P vector, two oligonucleotides were synthesized that anneal in such a way that the duplex contains protruding sequences that potentially regenerate a BglII restriction site at the 5′-end and a XhoI site at the 3′-end. The sequences of the oligonucleotides were as follows: sense strand, 5′-GATCTTAAGAATTCCTTCC-3′; antisense strand, 5′-TCGAGGAAGGAATTCTTAA-3′. The annealed oligonucleotides were used to replace the BglII–XhoI fragment containing the GFP gene in the rnrB-ubi-S65TGFP construct.
Dictyostelium discoideum cells of the AX2 strain were transformed and selected on bacterial lawns according to Wetterauer et al. (10) using the ‘agar method’ as modified by Deichsel et al. (8).
Growth, development and drug treatment
Growth, development and drug treatment of D.discoideum cells were performed as described previously (7). Briefly, cells were grown axenically in HL-5 medium to mid log phase, washed in KKP buffer (20 mM KH2PO4/K2HPO4, pH 6.2) and developed at 107 cells/ml in KKP buffer or on polycarbonate filters at 2 × 106 cells/cm2. Methylmethane sulfonic acid (MMS) and 4-nitroquinoline 1-oxide (4NQO) were obtained from Sigma. For cell suspensions the drugs were added directly to the medium. For cells developed on filters the supporting pads were saturated with the drug at the appropriate concentration. UV treatments were performed with a UV cross-linker (Stratalinker 1800; Stratagene). Calibration of the UV lamp was verified using uridylic acid as described previously (7).
RNA analysis
Total RNA was extracted from 2 × 107 cells in microfuge tubes (11). The RNA was quantified spectrophotometrically and 10 µg RNA were resolved on formaldehyde gels as described (12). Nucleic acids were transferred onto Nytran membranes (Schleicher & Shuell) in 10× SSC by capillarity and cross-linked using a UV Stratalinker 1800 (Stratagene). Probes were made by random priming following the manufacturer’s instructions (Pharmacia) with [α-32P]dCTP (Amersham). Hybridizations were done in Denhardt’s hybridization solution with 50% formamide at 40°C (13). Stringency washes were done at 65°C in 1× SSC, 0.1% SDS. Blots were exposed to Kodak X-Omat films with intensifying screens. The blots were quantified directly using a PhosphorImager GS-363 and Molecular Analyst® software (Bio-Rad) or X-ray films were scanned with the GeneGenius BioImaging system and quantified with SynGene Tools v.3.00.13 (SynGene Laboratories).
Assay for β-galactosidase activity
β-Galactosidase activity was assayed using the substrate Galacton-Plus™ (Tropix) following the manufacturer’s protocol using 60 µl of substrate and 10 µl of sample, containing 500 µg/ml protein. After a reaction time of 30 min the chemiluminescent product was detected with a Berthold Lumat LB9501 luminometer.
RESULTS AND DISCUSSION
Construction of a vector for regulated gene expression
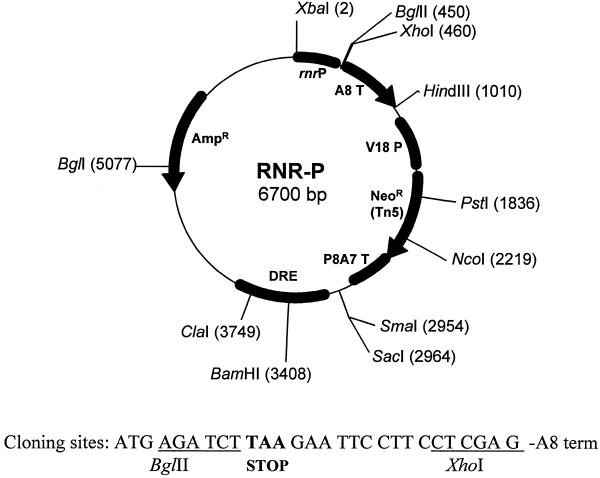
We have constructed a plasmid in which any gene can be cloned under control of the rnrB promoter, called the pRNR-P vector (Fig. 1). This vector contains a 450 bp rnrB promoter fragment that provides DNA-damage controlled gene expression. The vector contains a BglII site immediately downstream from the ATG codon for cloning. Other features of the plasmid include the neomycin phosphotransferase gene under control of the V18 promoter, which allows transformation of non-axenic strains, as well as a fragment of the retrotransposable element DRE that may promote integration in transcriptionally active regions of the genome (14).
Figure 1.
Map of the pRNR-P vector. Unique restriction endonuclease sites are indicated. Other features of the vector are described in the text.
Induction of reporter genes under control of the rnrB promoter by DNA-damaging agents
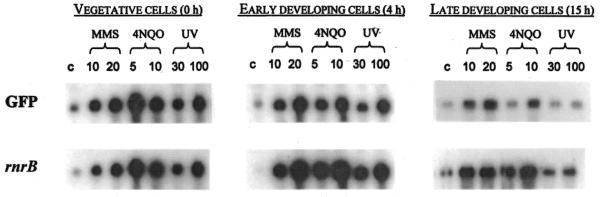
We have used two reporter genes, encoding GFP and an unstable version of β-galactosidase, respectively, to examine the DNA-damage response of the pRNR-P vector. Figure 2 shows the levels of the rnrB and GFP transcripts expressed by transformed cells bearing the rnrB-ubi-S65TGFP fusion construct following treatment with 10 or 20 mM MMS, 5 or 10 µg/ml 4NQO or 30 or 100 J/m2 UV irradiation. The level of GFP transcript increased in the presence of DNA-damaging agents. The response of the rnrB-ubi-S65TGFP fusion gene appears similar to that of the endogenous rnrB gene. We observed induction at all stages of the life cycle tested, vegetatively growing cells as well as cells developed for 4 or 15 h. Moreover, the response was dose dependent up to 100 J/m2 UV, 15 mM MMS and 10 µg/ml 4NQO (7). Irradiation with UV produced a much weaker response in late developing cells, presumably due to the failure of UV light to penetrate through the slime sheath.
Figure 2.
Effect of DNA-damaging agents on accumulation of the rnrB and GFP transcripts during growth and development. AX2 cells transformed with the RnrB-ubi-S65TGFP construct were treated during vegetative growth (0 h) or developed for 4 or 15 h before treatment. Cells were exposed for 1 h to 10 or 20 mM MMS or 5 or 10 µg/ml 4NQO. For UV irradiation cells were treated with 30 or 100 J/m2 and incubated for 1 h before a sample was collected. Shown here are autoradiographs of blots probed for GFP and rnrB transcripts. c, untreated control.
The highest inductions were observed for cells treated at 4 h; these cells also showed the lowest basal expression levels. Quantification of the blots revealed that induction of the GFP transcript ranged from 2- to 10-fold, while that of rnrB was between 4- and 89-fold. The induction factor depends strongly on the expression level in untreated cells. The induction factor for the rnrB transcript is very high because the level of rnrB message is barely above background in untreated cells.
rnrB-driven gene induction is rapid
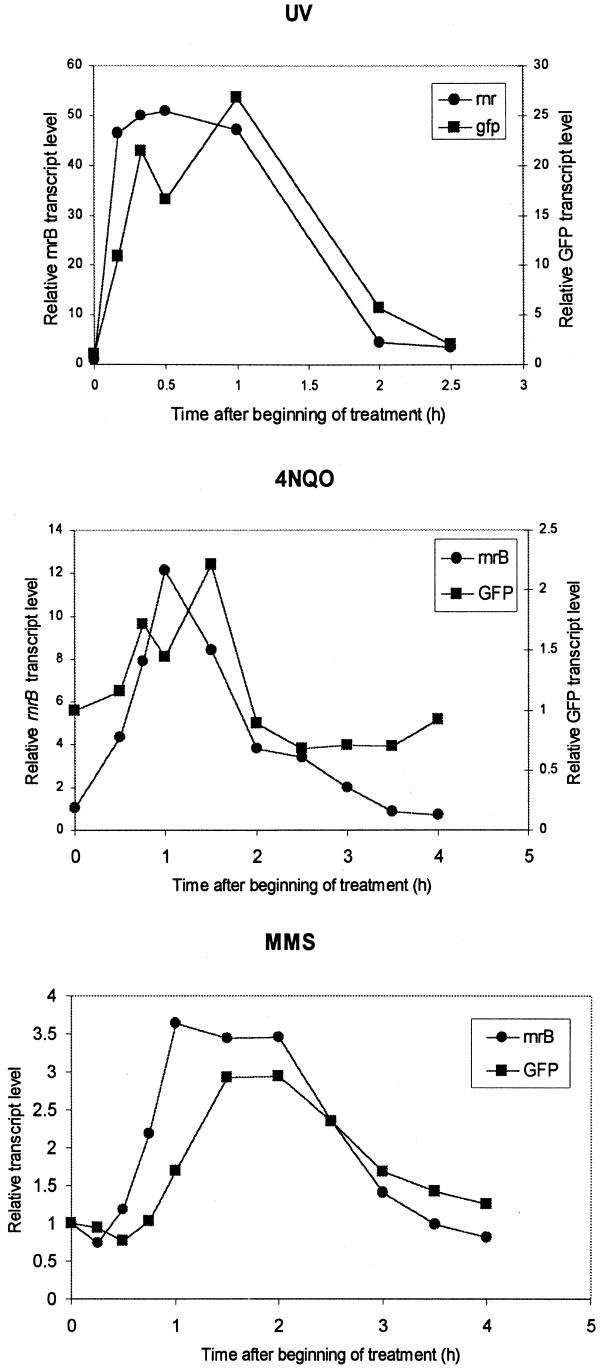
We have examined the kinetics of induction of the reporter gene using low doses of DNA-damaging agents. All agents elicited a rapid and transient response. Ten minutes after treatment with 30 J/m2 UV the levels of GFP and rnrB transcripts increased by 10- and 45-fold, respectively. The increased levels persisted for at least 1.5 h. By 2.5 h the levels of rnrB and GFP transcripts were ∼2- to 3-fold higher than in untreated cells.
When cells were treated with 5 µg/ml 4NQO we detected a 4.5-fold induction 30 min after the beginning of treatment for the endogenous rnrB transcript, while a 2-fold induction was detected 45 min after the beginning of treatment for the GFP transcript (Fig. 3). The maximal response was observed at 1 h for the endogenous rnrB transcript (12-fold) and at 1.5 h for the GFP transcript (2-fold). After 3 h both the rnrB and GFP transcript levels returned to basal levels.
Figure 3.
Kinetics of activation of gene expression controlled by the rnrB promoter. Cells were developed for 4 h before addition of the drugs. Cells were treated with 10 mM MMS, 5 µg/ml 4NQO or 30 J/m2 UV. Samples were collected at the indicated times after treatment and processed for RNA analysis. The 0 time point corresponds to drug addition or irradiation. To correct for loading, blots were probed for expression of the capA gene, whose expression is not affected by the presence of mutagens (7). RNA levels were normalized by dividing the GFP or rnrB transcript levels by that of the capA transcript.
The increase in GFP transcript level was slower upon treatment with 10 mM MMS. A 3-fold increase in transcript level was observed 1.5 h after the beginning of treatment. The response was sustained for at least 2 h and the decrease in transcript level was more gradual as compared to treatment with 4NQO or UV. The endogenous rnrB transcript level following treatment with MMS reached a plateau in 1 h and remained relatively constant for 2 h.
The level of induction of the reporter gene was lower than that of the endogenous rnrB gene. This could in part be due to the relatively high basal level of the GFP transcript. However, it is important to point out that for the three DNA-damaging agents tested the profiles of induction of the reporter gene closely resembled that of the endogenous rnrB gene.
rnrB-driven gene induction results in up-regulation at the protein level
To be useful, inducible gene expression systems should result in the production of functional proteins. We therefore investigated whether the increased GFP transcript level resulted in an increased level of GFP protein. Despite the striking induction at the transcript level, no large changes in GFP fluorescence were seen. We attribute this to the fact that GFP is a relatively stable protein. In particular, ubi-S65TGFP has been reported to have a half-life of 7 h in early developing cells (8). GFP can thus accumulate over time, so a significant level of fluorescence will be present in uninduced cells as a result of basal promoter activity. The amount produced during the short induction period may not result in a large relative increase. To address this problem, we used an unstable β-galactosidase reporter with a half-life of ∼30 min (H.MacWilliams et al., submitted for publication).
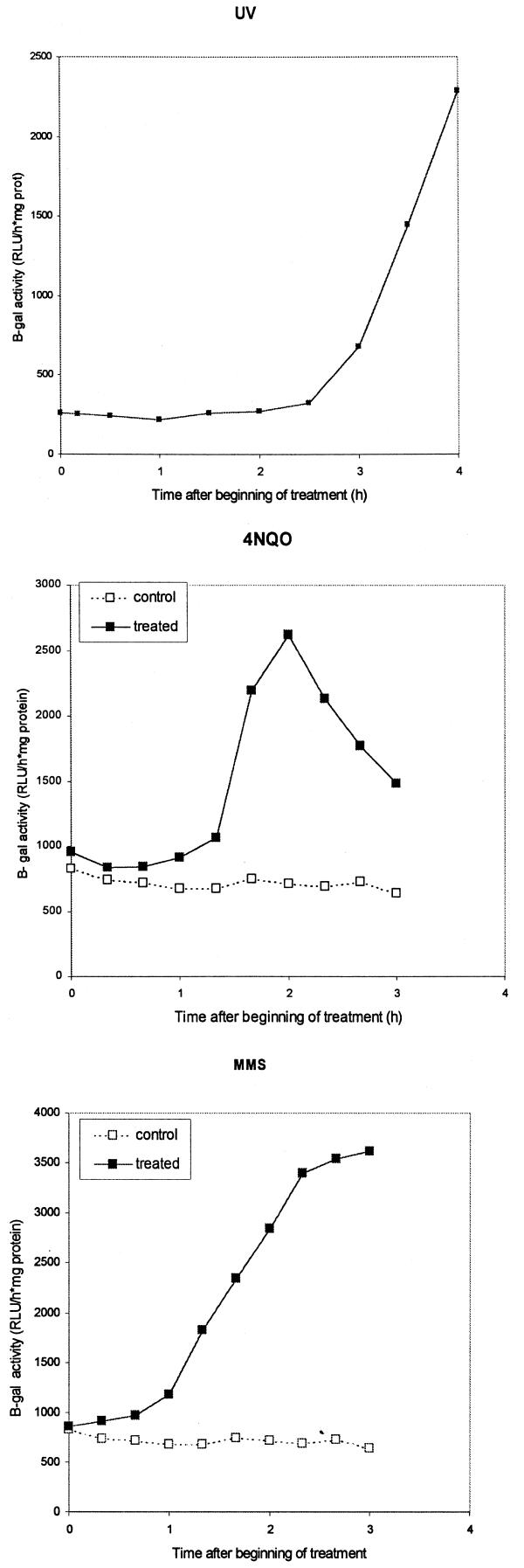
Using this RnrB-ile-αpgal fusion construct we have been able to detect increases in β-galactosidase activity of up to 8.7-fold over untreated cells for 30 J/m2 UV, 5.6-fold for 10 mM MMS and 3.6-fold for 5 µg/ml 4NQO (Fig. 4). For 4NQO the maximum induction was from 1.5 to 2.5 h after the beginning of treatment, lagging the increase in transcript level by ∼1 h. When cells were treated with MMS the increase in β-galactosidase activity was also observed ∼1 h after the first increase in transcript level. However, the response appeared to be more persistent, with high β-galactosidase activity observable 3 h after the beginning of treatment.
Figure 4.
β-Galactosidase activity of AX2 cells transformed with a rnrB-ile-αpgal fusion construct upon treatment with 10 mM MMS, 5 µg/ml 4NQO or 30 J/m2 UV. Cells were treated 4 h after initiation of development. Protein samples were taken periodically and assayed for β-galactosidase activity.
In the case of UV irradiation the induction of β-galactosidase activity was lower than expected based on induction of the endogenous rnrB message. Also, induction of β-galactosidase activity was slower than induction of rnrB transcript, being first detectable only 2 h after irradiation, while a significant increase in transcript level can be seen only 10 min after irradiation (Fig. 3). This might be due to partial inhibition of translation after UV exposure (15,16). Consistent with this idea, both MMS and 4NQO caused a faster response than UV in induction of β-galactosidase activity (Fig. 4). These results show that lacZ induction by DNA-damaging agents results in the production of a functional enzyme. Moreover, the pRNR-P vector has been used successfully to overexpress rasG protein and activate a potential rasG downstream signaling pathway (D.M.Secko et al., submitted for publication).
Both the basal level and the level of induction of enzyme activity varied in different clones (data not shown). This was also observed for another construct made using this system (D.M.Secko and G.Weeks, personal communication). The basis of these differences is not understood, although it is perhaps relevant that a clone exhibiting low damage-inducibility also had a high rate of spontaneous vector loss while another clone that was better at responding to the drugs did not. This suggests differences in integration locus, which could modulate the accessibility of DNA to various transcription factors. Similar differences in induction levels for different clones have been reported for the tetracycline-regulated system (4). Therefore, clones should be selected that express low basal levels and high induced levels to ensure the best results.
The fact that the drugs used for the control of gene expression with the pRNR-P system are DNA-damaging agents raises concern about the possible induction of mutations. However, the number of mutations caused is relatively low. Over 95% of AX2 cells survive exposure to 30 J/m2 UV light (7), which has been reported to result in a <2-fold increase in the number of mutations compared to the spontaneous mutation level (17). For a 1 h treatment with 10 mM MMS the survival rate was ∼70% (7), which corresponds to an ∼3.5-fold higher mutation frequency than the spontaneous mutation level (17).
We have investigated the effects of DNA-damaging agents on development and found that the doses required for induction caused little or no detectable effects on the progress of development and on the number and size of fruiting bodies. No adverse effects were observed when cells were treated with 30 J/m2 UV. A <1 h delay in development was observed when cells were treated with 100 J/m2. At 10 µg/ml or lower concentrations 4NQO did not alter the profile of development, even when the treatment was sustained throughout the 24 h development cycle. Treatment with 10 mM MMS during the first 2 h of development caused a significant delay in development. However, no defects were observed when the cells were treated after this critical period for a duration of <2 h.
The main limitation with the pRNR-P inducible expression system is that the rnrB promoter fragment used here confers basal expression in vegetative cells and is also expressed in prespore cells from 12 to 20 h of development (5,18; unpublished data). Expression from the promoter is minimal, however, in the first few hours of development, when substantial changes in signal transduction occur, and it appears ideal for examining the consequences of perturbed gene expression during this period. Moreover, the rapid and significant response to UV irradiation may make this type of treatment particularly useful for antisense RNA mutagenesis.
The pRNR-P vector is an important new tool for the study of D.discoideum development, because it provides solutions to several limitations encountered in other controllable gene expression systems: it is inducible at all stages of the D.discoideum life cycle; induction is rapid; the level of induction can be modulated by using different doses of drugs.
Our results with reporters with different half-lives highlight the importance of protein half-life in experiments in which high induction factors are desired. When inducible systems such as this one are used to study physiological processes it may be helpful to attach destabilizing signals to the proteins under investigation.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Muriel Herrington for helpful advice and Dr Gerald Weeks for comments on the manuscript. This work was supported by NSERC of Canada and the Deutsche Forschungsgemeinschaft. P.G. is a recipient of a FCAR post-graduate scholarship.
References
- 1.Blusch J., Morandini,P. and Nellen,W. (1992) Transcriptional regulation by folate: inducible gene expression in Dictyostelium transformants during growth and early development. Nucleic Acids Res., 20, 6235–6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wetterauer B.W., Salger,K., Carballo-Metzner,C. and MacWilliams,H.K. (1995) Cell-density-dependent repression of discoidin in Dictyostelium discoideum. Differentiation, 59, 289–297. [DOI] [PubMed] [Google Scholar]
- 3.Dingermann T., Werner,H., Schutz,A., Zundorf,I., Nerke,K., Knecht,D. and Marschalek,R. (1992) Establishment of a system for conditional gene expression using an inducible tRNA suppressor gene. Mol. Cell. Biol., 12, 4038–4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blaauw M., Linskens,M.H.K. and van Haastert,P.J.M. (2000) Efficient control of gene expression by a tetracycline-dependent transactivator in single Dictyostelium discoideum cells. Gene, 252, 71–82. [DOI] [PubMed] [Google Scholar]
- 5.Tsang A., Bonfils,C., Czaika,G., Shtevi,A. and Grant,C. (1996) A prespore-specific gene of Dictyostelium discoideum encodes the small subunit of ribonucleotide reductase. Biochim. Biophys. Acta, 1309, 100–108. [DOI] [PubMed] [Google Scholar]
- 6.Friedberg E.C, Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC.
- 7.Gaudet P. and Tsang,A. (1999) Regulation of the ribonucleotide reductase small subunit gene by DNA-damaging agents in Dictyostelium discoideum. Nucleic Acids Res., 27, 3042–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deichsel H., Friedel,S., Detterbeck,A., Coyne,C., Hamker,U. and MacWilliams,H.K. (1999) Green fluorescent proteins with short half-lives as reporters in Dictyostelium discoideum. Dev. Genes Evol., 209, 63–68. [DOI] [PubMed] [Google Scholar]
- 9.Detterbeck S, Morandini,P., Wetterauer,B., Bachmair,A., Fischer,K. and MacWilliams,H.K. (1994) The ‘prespore-like cells’ of Dictyostelium have ceased to express a prespore gene: analysis using short-lived β-galactosidases as reporters. Development, 120, 2847–2855. [DOI] [PubMed] [Google Scholar]
- 10.Wetterauer B., Morandini,P., Hribar,I., Murgia-Morandini,I., Hamker,U., Singleton,C. and MacWilliams,H.K. (1996) Wild-type strains of Dictyostelium discoideum can be transformed using a novel selection cassette driven by the promoter of the ribosomal V18 gene. Plasmid, 36, 169–181. [DOI] [PubMed] [Google Scholar]
- 11.Franke J., Podgorski,G.J. and Kessin,R.H. (1987) The expression of two transcripts of the phosphodiesterase gene during the development of Dictyostelium discoideum. Dev. Biol., 124, 504–511. [DOI] [PubMed] [Google Scholar]
- 12.Fourney R.M., Miyakoshi,J., Day,R.S. and Paterson,M.S. (1988) Northern blotting: efficient RNA staining and transfer. Focus, 10, 5–6. [Google Scholar]
- 13.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 14.Marschalek R., Hofmann,J., Schumann,G., Gösseringer,R. and Dingermann,T. (1992) Two distinct subforms of the retrotransposable DRE element in NC4 strains of Dictyostelium discoideum. Mol. Cell. Biol., 12, 229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumari P.A. and Nair,V.R. (1984) Mitotic delays and macromolecular synthesis in G2 phase-irradiated plasmodia of Physarum polycephalum. Exp. Cell Res., 151, 104–111. [DOI] [PubMed] [Google Scholar]
- 16.Hyodo M., Fujita,H., Sizuki,K., Yoshino,K., Matuso,I. and Ohkido,M. (1982) DNA replication and cell-cycle progression of cultured mouse FM3a cells after treatment with 8-methoxypsoralen plus near UV-radiation. Mutat. Res., 94, 199–211. [DOI] [PubMed] [Google Scholar]
- 17.Pogdorski G. and Deering,R.A. (1980) Quantitation of induced mutation in Dictyostelium discoideum: characterization and use of a methanol-resistance mutation assay. Mutat. Res., 74, 459–468. [DOI] [PubMed] [Google Scholar]
- 18.Bonfils C., Gaudet,P. and Tsang,A. (1999) Identification of cis-regulating elements and trans-acting factors regulating the expression of the gene encoding the small subunit of ribonucleotide reductase in Dictyostelium discoideum. J. Biol. Chem., 274, 20384–20390. [DOI] [PubMed] [Google Scholar]