Abstract
The title compound (systematic name: 4-{[(2E)-5-hydroxy-3,7-dimethylocta-2,6-dien-1-yl]oxy}-7H-furo[3,2-g][1]benzopyran-7-one), C21H22O5, is a known furanocoumarin, which was isolated from the Chinese herbal product Radix seu Rhizoma Notopterygii. The crystal structure shows a weak O—H⋯O hydrogen bond.
Related literature
For the isolation, see: Yang et al. (1994 ▶); Xiao et al. (1994 ▶). For NMR shifts and coupling constants of related compounds, see: Hasegawa et al. (1999 ▶); Chemical Abstract Service (2009 ▶). 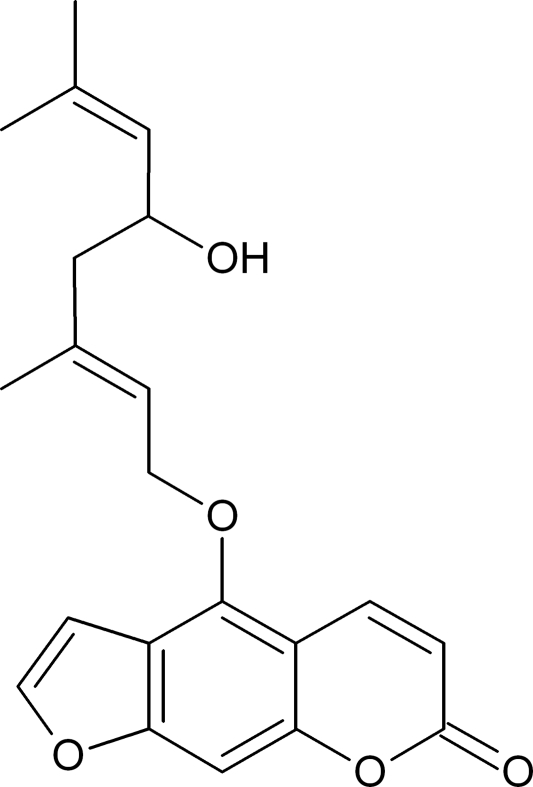
Experimental
Crystal data
C21H22O5
M r = 354.39
Triclinic,
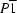
a = 6.4317 (10) Å
b = 8.0912 (16) Å
c = 17.206 (3) Å
α = 91.802 (15)°
β = 94.240 (13)°
γ = 97.473 (15)°
V = 884.6 (3) Å3
Z = 2
Mo Kα radiation
μ = 0.09 mm−1
T = 95 K
0.48 × 0.44 × 0.32 mm
Data collection
Stoe four-circle diffractometer
Absorption correction: none
4234 measured reflections
3462 independent reflections
2941 reflections with I > 2σ(I)
R int = 0.023
3 standard reflections every 100 reflections intensity decay: 0.4%
Refinement
R[F 2 > 2σ(F 2)] = 0.043
wR(F 2) = 0.113
S = 1.04
3462 reflections
249 parameters
Only H-atom displacement parameters refined
Δρmax = 0.38 e Å−3
Δρmin = −0.21 e Å−3
Data collection: local software; cell refinement: local software; data reduction: local software; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: modified ORTEP (Johnson, 1965 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809005030/bt2871sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809005030/bt2871Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O5—H51⋯O8i | 0.84 | 2.57 | 3.2193 (16) | 135 |
Symmetry code: (i) 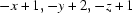 .
.
Acknowledgments
Funding provided by the Austrian Science Fund (FWF) within project S107 (Drugs from Nature Targeting Inflammation) is gratefully acknowledged.
supplementary crystallographic information
Comment
Rhizoma seu Radix Notopterygii is a herbal preparation commonly used in traditional Chinese medicine. It may either contain roots or rhizomes of N. incisum or N. forbesii (Apiaceae) or a mixture of both plants. The isolation of E-Notopterol (Fig.1) has been previously reported for both species (Yang et al., 1994; Xiao et al. 1994), which naturally occur in the region of South Central Asia, but the crystal structure has not been reported until now. In the process of structure elucidation, the title compound was primarily identified by 1H and 13C NMR experiments. Observed chemical shifts and coupling constants were in compliance with previously published literature (Hasegawa et al., 1999; Chemical Abstract Service, 2009). Further support was provided by the ESI-MS analysis, indicating the pseudo molecular ion at 377.29 m/z [M—Na+].
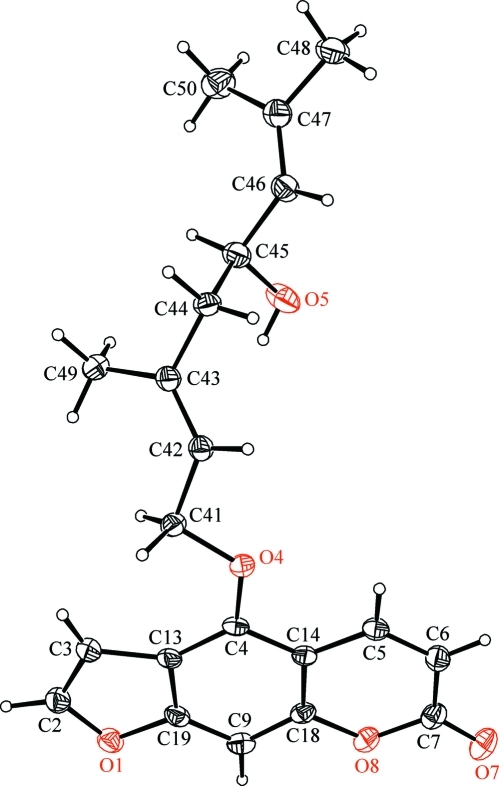
The crystal structure analysis confirmed the proposed structure as 4-{[(2E)-5-hydroxy-3,7-dimethyl- octa-2,6-dien-1-yl]oxy}-7H-furo[3,2-g][1]benzopyran-7-one. All the atoms are lying on general positions. In Fig. 1 the S-enantiomer of the racemate is shown. The least-squares plane through the non-H atoms around the double bond C42=C43 enclose an angle of 12.36 (9)° with the plane through the atoms around C46=C47, and further an angle of 41.35 (6)° with the plane through the tricycle. The packing is dominated by an antiparallel stacking of the tricycles (s. Fig. 2) preventing the formation of a strong hydrogen bond of the OH group [O5—H51···.O8i 134.5°, O5···.O8i 3.2193 (16) Å; symmetry transformation used to generate the equivalent atom: (i) 1 - x, 2 - y, 1 - z].
Experimental
Plant Material: Rhizoma seu Radix Notopterygii was purchased from Plantasia (Oberndorf, Austria).
Isolation and purification: Plant material (2000 g) was grinded and extracted with 11 L dichloromethane at room temperature, yielding 381.87 g of dried extract after vacuum evaporation. Next 100 g dried extract were fractionated on a Silicagel 60 column (230–400 mesh, Merck, Darmstadt, Germany; size: 8.5 x 20 cm). In the process a stepwise gradient elution starting from 100% hexane (Hex) and going to 100% ethylacetate (EtOAc) was performed. For each step (2400 ml) the content of Hex in the eluent was reduced by 16.7 vol.%, and 6–8 subfractions were collected. Subfraction D7 collected between 2030 ml and 2400 ml from eluent D (Hex:EtOAc: 50:50, v:v) yielded in the isolation of 86.49 mg crystallized 1. Finally the colourless block crystals were washed with eluent C (Hex:EtOAc: 67:33, v:v) before being analyzed.
NMR analysis: 1H and 13C NMR spectra were recorded at 293 K on a Varian UnityInova 400 NMR spectrometer with a 5 mm broadband probe. E-Notpterol was dissolved in chloroform-d (Aldrich, USA) and TMS was used as internal standard.
NMR results: 1H NMR (CDCl3, 400 MHz) δ 1.70 (3H, d, J = 1.2, H-50), 1.72 (3H, d, J = 1.2, H-48), 1.77 (3H, s, H-49), 2.22 (1H, dd, J = 13.7, 5.0, H-44b), 2.31 (1H, dd, J = 13.7, 8.2, H-44a), 4.52 (1H, m, H-45), 4.98 (2H, d, J = 6.9, H-41), 5.18 (1H, pseudo td, J = 8.4, 1.3, H-46), 5.65 (1H, pseudo dt, J = 6.7, 1.0, H-42), 6.28 (1H, d, J = 9.8, H-6), 6.98 (1H, dd, J = 2.4, 0.8, H-3), 7.16 (1H, s, H-9), 7.60 (1H, d, J = 2.4, H-2), 8.16 (1H, d, J = 9.8, H-5); 13C NMR (CDCl3, 400 MHz) δ 17.0 (CH3, C-49), 18.2 (CH3, C-50), 25.7 (CH3, C-48), 47.7 (CH2, C-44), 66.4 (CH, C-45), 69.4 (CH2, C-41), 94.3 (CH, C-9), 105.0 (CH, C-3), 107.4 (C, C-14), 112.6 (CH, C-6), 114.0 (C, C-13), 122.4 (CH, C-42), 127.3 (CH, C-46), 135.6 (C, C-47), 139.5 (C, C-43), 139.5 (CH, C-5), 145.0 (CH, C-2), 148.7 (C, C-4), 152.6 (C, C-18), 158.1 (C, C-19), 161.3 (C, C-7).
LC—MS-analysis: Experiments were performed on a Thermo Finnigan Surveyor liquid chromatograph interfaced with a LCQTM Deca XPPLUS mass detector. E-Notopterol was dissolved in methanol (HPLC grade, Merck, Darmstadt, Germany) and analyzed in the ESI+ mode under following conditions: Sheath gas flow: 70 units, ionization voltage: 5,50 kV, capillary voltage 15 V, tube lens offset: 50 V, capillary temperature: 623 K.
Refinement
H atoms were located in a difference map, but geometrically positioned with Caromatic-H = 0.95Å, Cmethyl-H = 0.98Å, Cmethylene-H = 0.99Å and O-H = 0.84Å. The torsion angle about the C-O bond was also refined. The U values of the H atoms were refined using the same U values for similar H atoms.
Figures
Fig. 1.
ORTEP plot (Johnson, 1965) showing the atomic numbering scheme. The probability ellipsoids are drawn at the 50% probability level.
Fig. 2.
Stereoscopic ORTEP plot (Johnson, 1965) of the packing. The atoms are drawn with arbitrary radii.
Crystal data
| C21H22O5 | Z = 2 |
| Mr = 354.39 | F(000) = 376 |
| Triclinic, P1 | Dx = 1.330 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71069 Å |
| a = 6.4317 (10) Å | Cell parameters from 60 reflections |
| b = 8.0912 (16) Å | θ = 16.0–19.8° |
| c = 17.206 (3) Å | µ = 0.09 mm−1 |
| α = 91.802 (15)° | T = 95 K |
| β = 94.240 (13)° | Block, colourless |
| γ = 97.473 (15)° | 0.48 × 0.44 × 0.32 mm |
| V = 884.6 (3) Å3 |
Data collection
| Stoe four-circle diffractometer | Rint = 0.023 |
| Radiation source: fine-focus sealed tube | θmax = 26.0°, θmin = 2.5° |
| graphite | h = −7→7 |
| ω–2θ scans | k = −9→1 |
| 4234 measured reflections | l = −21→21 |
| 3462 independent reflections | 3 standard reflections every 100 reflections |
| 2941 reflections with I > 2σ(I) | intensity decay: 0.4% |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.043 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.113 | Only H-atom displacement parameters refined |
| S = 1.04 | w = 1/[σ2(Fo2) + (0.0482P)2 + 0.2588P] where P = (Fo2 + 2Fc2)/3 |
| 3462 reflections | (Δ/σ)max < 0.001 |
| 249 parameters | Δρmax = 0.38 e Å−3 |
| 0 restraints | Δρmin = −0.21 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger.The non-hydrogen atoms were refined with anisotropic displacement parameters without any constraints. The H atoms of the 7H-Furo[3,2-g][1]benzopyran-7-one ring system were put at the external bisector of the C—C—C angle at a C—H distance of 0.95 Å and a common isotropic displacement parameter was refined (AFIX 43 of SHELXL97). The H atoms of the CH2 groups were refined with common isotropic displacement parameters for the H atoms of the same group and idealized geometry with approximately tetrahedral angles and C—H distances of 0.99 Å (AFIX 23 of SHELXL97). The H atoms bonded to a C atom of a C=C double bond were put at the external bisector of the C—C—C angle at a C—H distance of 0.95 Å but the individual isotropic displacement parameters are free to refine (AFIX 43 of SHELXL97). The H atom of the tertiary C—H group was refined with an individual isotropic displacement parameter and all X—C—H angles equal at a C—H distance of 1.00 Å (AFIX 13 of SHELXL97). The H atoms of the methyl groups were refined with common isotropic displacement parameters for the H atoms of the same group and idealized geometry with tetrahedral angles, enabling rotation around the X—C bond, and C—H distances of 0.98 Å (AFIX 137 of SHELXL97). The H atom of the OH group was refined with a tetrahedral C—O—H angle, enabling rotation around the C—O bond, O—H distance of 0.84 Å, and with an individual isotropic displacement parameter (AFIX 147 of SHELXL97). |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.08010 (16) | 1.43591 (14) | 0.41654 (6) | 0.0244 (3) | |
| C2 | 0.0927 (2) | 1.43677 (19) | 0.33658 (9) | 0.0246 (3) | |
| H2 | −0.0037 | 1.4827 | 0.3016 | 0.028 (2)* | |
| C3 | 0.2568 (2) | 1.36524 (19) | 0.31406 (9) | 0.0226 (3) | |
| H3 | 0.2957 | 1.3524 | 0.2622 | 0.028 (2)* | |
| C4 | 0.5403 (2) | 1.23232 (17) | 0.40535 (8) | 0.0177 (3) | |
| C5 | 0.7767 (2) | 1.13743 (18) | 0.51262 (9) | 0.0202 (3) | |
| H5 | 0.8670 | 1.1018 | 0.4760 | 0.028 (2)* | |
| C6 | 0.8224 (2) | 1.11882 (19) | 0.58937 (9) | 0.0225 (3) | |
| H6 | 0.9445 | 1.0707 | 0.6059 | 0.028 (2)* | |
| C7 | 0.6889 (2) | 1.17088 (19) | 0.64707 (9) | 0.0228 (3) | |
| O7 | 0.71375 (18) | 1.15883 (16) | 0.71723 (6) | 0.0305 (3) | |
| O8 | 0.51411 (17) | 1.24050 (14) | 0.62003 (6) | 0.0231 (3) | |
| C9 | 0.2897 (2) | 1.33880 (19) | 0.52396 (9) | 0.0216 (3) | |
| H9 | 0.2045 | 1.3732 | 0.5626 | 0.028 (2)* | |
| C13 | 0.3631 (2) | 1.31129 (18) | 0.38445 (8) | 0.0185 (3) | |
| C14 | 0.5942 (2) | 1.21012 (17) | 0.48527 (8) | 0.0179 (3) | |
| C18 | 0.4657 (2) | 1.26325 (18) | 0.54174 (8) | 0.0189 (3) | |
| C19 | 0.2464 (2) | 1.36067 (18) | 0.44558 (9) | 0.0200 (3) | |
| O4 | 0.66912 (16) | 1.17002 (14) | 0.35580 (6) | 0.0223 (3) | |
| C41 | 0.6201 (2) | 1.17328 (19) | 0.27226 (8) | 0.0206 (3) | |
| H411 | 0.6516 | 1.2880 | 0.2540 | 0.022 (3)* | |
| H412 | 0.4695 | 1.1328 | 0.2584 | 0.022 (3)* | |
| C42 | 0.7567 (2) | 1.05985 (18) | 0.23652 (8) | 0.0197 (3) | |
| H42 | 0.8956 | 1.0633 | 0.2601 | 0.030 (5)* | |
| C43 | 0.7031 (2) | 0.95436 (18) | 0.17484 (8) | 0.0198 (3) | |
| C44 | 0.8569 (2) | 0.84090 (19) | 0.14954 (9) | 0.0215 (3) | |
| H441 | 0.9936 | 0.8725 | 0.1801 | 0.031 (3)* | |
| H442 | 0.8797 | 0.8576 | 0.0939 | 0.031 (3)* | |
| C45 | 0.7817 (3) | 0.6553 (2) | 0.16021 (9) | 0.0247 (4) | |
| H45 | 0.6569 | 0.6179 | 0.1228 | 0.026 (4)* | |
| O5 | 0.7265 (2) | 0.62481 (16) | 0.23818 (7) | 0.0342 (3) | |
| H51 | 0.6537 | 0.6973 | 0.2528 | 0.068 (8)* | |
| C46 | 0.9538 (3) | 0.5518 (2) | 0.14538 (9) | 0.0246 (3) | |
| H46 | 1.0863 | 0.5869 | 0.1731 | 0.022 (4)* | |
| C47 | 0.9430 (3) | 0.4165 (2) | 0.09793 (9) | 0.0251 (3) | |
| C48 | 1.1328 (3) | 0.3278 (2) | 0.08976 (10) | 0.0322 (4) | |
| H481 | 1.2543 | 0.3891 | 0.1207 | 0.042 (3)* | |
| H482 | 1.1628 | 0.3221 | 0.0348 | 0.042 (3)* | |
| H483 | 1.1048 | 0.2146 | 0.1085 | 0.042 (3)* | |
| C49 | 0.4927 (3) | 0.9353 (2) | 0.12808 (9) | 0.0248 (3) | |
| H491 | 0.4120 | 1.0230 | 0.1448 | 0.043 (3)* | |
| H492 | 0.4150 | 0.8258 | 0.1365 | 0.043 (3)* | |
| H493 | 0.5146 | 0.9448 | 0.0725 | 0.043 (3)* | |
| C50 | 0.7499 (3) | 0.3374 (2) | 0.04907 (10) | 0.0335 (4) | |
| H501 | 0.6298 | 0.3945 | 0.0608 | 0.058 (4)* | |
| H502 | 0.7204 | 0.2194 | 0.0609 | 0.058 (4)* | |
| H503 | 0.7737 | 0.3470 | −0.0063 | 0.058 (4)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0215 (5) | 0.0223 (6) | 0.0295 (6) | 0.0061 (4) | −0.0006 (4) | −0.0023 (5) |
| C2 | 0.0264 (8) | 0.0193 (8) | 0.0272 (8) | 0.0028 (6) | −0.0037 (6) | 0.0002 (6) |
| C3 | 0.0268 (8) | 0.0172 (7) | 0.0236 (8) | 0.0043 (6) | −0.0013 (6) | −0.0002 (6) |
| C4 | 0.0202 (7) | 0.0113 (7) | 0.0212 (7) | 0.0004 (6) | 0.0036 (6) | −0.0027 (6) |
| C5 | 0.0224 (8) | 0.0142 (7) | 0.0234 (8) | 0.0002 (6) | 0.0025 (6) | −0.0003 (6) |
| C6 | 0.0241 (8) | 0.0178 (7) | 0.0247 (8) | 0.0007 (6) | −0.0012 (6) | 0.0025 (6) |
| C7 | 0.0239 (8) | 0.0191 (8) | 0.0227 (8) | −0.0053 (6) | −0.0015 (6) | 0.0015 (6) |
| O7 | 0.0328 (6) | 0.0363 (7) | 0.0199 (6) | −0.0044 (5) | −0.0006 (5) | 0.0032 (5) |
| O8 | 0.0251 (6) | 0.0251 (6) | 0.0183 (5) | 0.0006 (5) | 0.0026 (4) | −0.0005 (4) |
| C9 | 0.0199 (7) | 0.0190 (8) | 0.0254 (8) | −0.0001 (6) | 0.0053 (6) | −0.0041 (6) |
| C13 | 0.0212 (7) | 0.0118 (7) | 0.0217 (7) | 0.0011 (6) | 0.0005 (6) | −0.0018 (6) |
| C14 | 0.0201 (7) | 0.0120 (7) | 0.0208 (7) | −0.0001 (6) | 0.0014 (6) | −0.0016 (6) |
| C18 | 0.0224 (7) | 0.0144 (7) | 0.0185 (7) | −0.0026 (6) | 0.0014 (6) | −0.0013 (6) |
| C19 | 0.0180 (7) | 0.0131 (7) | 0.0283 (8) | 0.0016 (6) | 0.0001 (6) | −0.0014 (6) |
| O4 | 0.0263 (6) | 0.0255 (6) | 0.0168 (5) | 0.0100 (5) | 0.0020 (4) | −0.0023 (4) |
| C41 | 0.0266 (8) | 0.0184 (7) | 0.0167 (7) | 0.0033 (6) | 0.0014 (6) | 0.0002 (6) |
| C42 | 0.0217 (7) | 0.0186 (7) | 0.0192 (7) | 0.0032 (6) | 0.0022 (6) | 0.0020 (6) |
| C43 | 0.0255 (8) | 0.0163 (7) | 0.0181 (7) | 0.0032 (6) | 0.0037 (6) | 0.0038 (6) |
| C44 | 0.0277 (8) | 0.0195 (8) | 0.0180 (7) | 0.0043 (6) | 0.0043 (6) | −0.0008 (6) |
| C45 | 0.0281 (8) | 0.0212 (8) | 0.0256 (8) | 0.0058 (7) | 0.0039 (6) | 0.0000 (6) |
| O5 | 0.0427 (7) | 0.0292 (7) | 0.0366 (7) | 0.0163 (6) | 0.0186 (6) | 0.0099 (5) |
| C46 | 0.0267 (8) | 0.0224 (8) | 0.0253 (8) | 0.0058 (7) | 0.0028 (6) | 0.0017 (6) |
| C47 | 0.0329 (9) | 0.0202 (8) | 0.0233 (8) | 0.0049 (7) | 0.0062 (6) | 0.0053 (6) |
| C48 | 0.0444 (10) | 0.0266 (9) | 0.0290 (9) | 0.0141 (8) | 0.0090 (7) | 0.0005 (7) |
| C49 | 0.0306 (9) | 0.0237 (8) | 0.0200 (7) | 0.0047 (7) | −0.0004 (6) | −0.0013 (6) |
| C50 | 0.0424 (10) | 0.0243 (9) | 0.0322 (9) | −0.0008 (8) | 0.0030 (8) | −0.0013 (7) |
Geometric parameters (Å, °)
| O1—C19 | 1.3688 (18) | C41—H412 | 0.99 |
| O1—C2 | 1.3843 (19) | C42—C43 | 1.336 (2) |
| C2—C3 | 1.343 (2) | C42—H42 | 0.95 |
| C2—H2 | 0.95 | C43—C49 | 1.509 (2) |
| C3—C13 | 1.454 (2) | C43—C44 | 1.512 (2) |
| C3—H3 | 0.95 | C44—C45 | 1.537 (2) |
| C4—O4 | 1.3613 (18) | C44—H441 | 0.99 |
| C4—C13 | 1.407 (2) | C44—H442 | 0.99 |
| C4—C14 | 1.416 (2) | C45—O5 | 1.4336 (19) |
| C5—C6 | 1.349 (2) | C45—C46 | 1.504 (2) |
| C5—C14 | 1.436 (2) | C45—H45 | 1.00 |
| C5—H5 | 0.95 | O5—H51 | 0.84 |
| C6—C7 | 1.447 (2) | C46—C47 | 1.337 (2) |
| C6—H6 | 0.95 | C46—H46 | 0.95 |
| C7—O7 | 1.2145 (19) | C47—C50 | 1.504 (2) |
| C7—O8 | 1.3788 (19) | C47—C48 | 1.507 (2) |
| O8—C18 | 1.3834 (18) | C48—H481 | 0.98 |
| C9—C18 | 1.375 (2) | C48—H482 | 0.98 |
| C9—C19 | 1.378 (2) | C48—H483 | 0.98 |
| C9—H9 | 0.95 | C49—H491 | 0.98 |
| C13—C19 | 1.413 (2) | C49—H492 | 0.98 |
| C14—C18 | 1.413 (2) | C49—H493 | 0.98 |
| O4—C41 | 1.4504 (17) | C50—H501 | 0.98 |
| C41—C42 | 1.499 (2) | C50—H502 | 0.98 |
| C41—H411 | 0.99 | C50—H503 | 0.98 |
| C19—O1—C2 | 105.94 (12) | C43—C42—H42 | 116.7 |
| C3—C2—O1 | 112.26 (14) | C41—C42—H42 | 116.7 |
| C3—C2—H2 | 123.9 | C42—C43—C49 | 124.47 (14) |
| O1—C2—H2 | 123.9 | C42—C43—C44 | 119.56 (14) |
| C2—C3—C13 | 106.62 (14) | C49—C43—C44 | 115.95 (13) |
| C2—C3—H3 | 126.7 | C43—C44—C45 | 113.08 (13) |
| C13—C3—H3 | 126.7 | C43—C44—H441 | 109.0 |
| O4—C4—C13 | 126.59 (13) | C45—C44—H441 | 109.0 |
| O4—C4—C14 | 114.47 (13) | C43—C44—H442 | 109.0 |
| C13—C4—C14 | 118.94 (13) | C45—C44—H442 | 109.0 |
| C6—C5—C14 | 121.15 (14) | H441—C44—H442 | 107.8 |
| C6—C5—H5 | 119.4 | O5—C45—C46 | 106.58 (13) |
| C14—C5—H5 | 119.4 | O5—C45—C44 | 111.88 (13) |
| C5—C6—C7 | 121.23 (15) | C46—C45—C44 | 110.45 (13) |
| C5—C6—H6 | 119.4 | O5—C45—H45 | 109.3 |
| C7—C6—H6 | 119.4 | C46—C45—H45 | 109.3 |
| O7—C7—O8 | 116.20 (14) | C44—C45—H45 | 109.3 |
| O7—C7—C6 | 126.77 (15) | C45—O5—H51 | 109.5 |
| O8—C7—C6 | 117.03 (13) | C47—C46—C45 | 127.87 (15) |
| C7—O8—C18 | 122.82 (12) | C47—C46—H46 | 116.1 |
| C18—C9—C19 | 114.72 (14) | C45—C46—H46 | 116.1 |
| C18—C9—H9 | 122.6 | C46—C47—C50 | 125.44 (16) |
| C19—C9—H9 | 122.6 | C46—C47—C48 | 120.82 (15) |
| C4—C13—C19 | 117.12 (13) | C50—C47—C48 | 113.73 (15) |
| C4—C13—C3 | 138.18 (14) | C47—C48—H481 | 109.5 |
| C19—C13—C3 | 104.68 (13) | C47—C48—H482 | 109.5 |
| C18—C14—C4 | 119.34 (13) | H481—C48—H482 | 109.5 |
| C18—C14—C5 | 117.56 (13) | C47—C48—H483 | 109.5 |
| C4—C14—C5 | 123.09 (13) | H481—C48—H483 | 109.5 |
| C9—C18—O8 | 116.11 (13) | H482—C48—H483 | 109.5 |
| C9—C18—C14 | 123.71 (14) | C43—C49—H491 | 109.5 |
| O8—C18—C14 | 120.18 (13) | C43—C49—H492 | 109.5 |
| O1—C19—C9 | 123.36 (14) | H491—C49—H492 | 109.5 |
| O1—C19—C13 | 110.49 (13) | C43—C49—H493 | 109.5 |
| C9—C19—C13 | 126.15 (14) | H491—C49—H493 | 109.5 |
| C4—O4—C41 | 119.53 (11) | H492—C49—H493 | 109.5 |
| O4—C41—C42 | 105.47 (12) | C47—C50—H501 | 109.5 |
| O4—C41—H411 | 110.6 | C47—C50—H502 | 109.5 |
| C42—C41—H411 | 110.6 | H501—C50—H502 | 109.5 |
| O4—C41—H412 | 110.6 | C47—C50—H503 | 109.5 |
| C42—C41—H412 | 110.6 | H501—C50—H503 | 109.5 |
| H411—C41—H412 | 108.8 | H502—C50—H503 | 109.5 |
| C43—C42—C41 | 126.62 (14) | ||
| C19—O1—C2—C3 | 0.41 (17) | C4—C14—C18—O8 | 179.09 (12) |
| O1—C2—C3—C13 | 0.19 (18) | C5—C14—C18—O8 | −1.7 (2) |
| C14—C5—C6—C7 | 0.2 (2) | C2—O1—C19—C9 | 178.89 (14) |
| C5—C6—C7—O7 | 179.33 (15) | C2—O1—C19—C13 | −0.87 (16) |
| C5—C6—C7—O8 | −0.2 (2) | C18—C9—C19—O1 | −178.88 (13) |
| O7—C7—O8—C18 | 179.60 (13) | C18—C9—C19—C13 | 0.9 (2) |
| C6—C7—O8—C18 | −0.8 (2) | C4—C13—C19—O1 | 179.76 (12) |
| O4—C4—C13—C19 | 177.50 (13) | C3—C13—C19—O1 | 0.98 (16) |
| C14—C4—C13—C19 | −1.4 (2) | C4—C13—C19—C9 | 0.0 (2) |
| O4—C4—C13—C3 | −4.3 (3) | C3—C13—C19—C9 | −178.78 (14) |
| C14—C4—C13—C3 | 176.88 (16) | C13—C4—O4—C41 | −3.8 (2) |
| C2—C3—C13—C4 | −179.08 (17) | C14—C4—O4—C41 | 175.09 (12) |
| C2—C3—C13—C19 | −0.70 (16) | C4—O4—C41—C42 | −166.21 (12) |
| O4—C4—C14—C18 | −177.18 (12) | O4—C41—C42—C43 | 141.31 (15) |
| C13—C4—C14—C18 | 1.8 (2) | C41—C42—C43—C49 | 2.1 (2) |
| O4—C4—C14—C5 | 3.7 (2) | C41—C42—C43—C44 | −176.19 (14) |
| C13—C4—C14—C5 | −177.31 (13) | C42—C43—C44—C45 | 113.54 (16) |
| C6—C5—C14—C18 | 0.8 (2) | C49—C43—C44—C45 | −64.89 (17) |
| C6—C5—C14—C4 | 179.90 (14) | C43—C44—C45—O5 | −53.12 (17) |
| C19—C9—C18—O8 | 179.60 (12) | C43—C44—C45—C46 | −171.68 (13) |
| C19—C9—C18—C14 | −0.4 (2) | O5—C45—C46—C47 | 109.83 (18) |
| C7—O8—C18—C9 | −178.14 (13) | C44—C45—C46—C47 | −128.43 (17) |
| C7—O8—C18—C14 | 1.8 (2) | C45—C46—C47—C50 | −0.7 (3) |
| C4—C14—C18—C9 | −0.9 (2) | C45—C46—C47—C48 | 179.69 (15) |
| C5—C14—C18—C9 | 178.22 (14) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O5—H51···O8i | 0.84 | 2.57 | 3.2193 (16) | 135 |
Symmetry codes: (i) −x+1, −y+2, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: BT2871).
References
- Chemical Abstract Service (2009). The SciFinder database. Chemical Abstracts Service, Columbus, Ohio, USA.
- Hasegawa, A., Nakamura, H., Watanabe, T., Okuyama, E., Ohmori, S., Ishikawa, T. & Kitada, M. (1999). Biol. Pharm. Bull 22, 725–726. [DOI] [PubMed]
- Johnson, C. K. (1965). ORTEP Report ORNL-3794. Oak Ridge National Laboratory, Tennessee, USA.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Xiao, Y., Sun, Y. & Liu, K. (1994). Zhongguo Zhongyao Zazhi, 19, 421–422. [PubMed]
- Yang, X., Yan, Z., Gu, Z., Zhou, G., Hattori, M. & Namba, T. (1994). Zhongguo Yaoxue Zazhi, 29, 141–143.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809005030/bt2871sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809005030/bt2871Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report