Abstract
Context
Pneumonia is the leading cause of childhood death in sub-Saharan Africa. Comparative estimates of the contribution of causative pathogens to the burden of disease are essential for targeted vaccine development.
Objective
To determine the viral etiology of severe pneumonia among infants and children at a rural Kenyan hospital using comprehensive and sensitive molecular diagnostic techniques.
Design, Setting & Participants
Prospective observational and case control study during 2007 in a rural Kenyan district hospital. We recruited children age 1 day to 12 years who were resident in a systematically enumerated catchment area: i) those admitted to Kilifi District Hospital meeting WHO clinical criteria for ‘severe pneumonia’ or ‘very severe pneumonia’; ii) those presenting with mild upper respiratory tract infection but not admitted and iii) well infants and children attending for immunization.
Main Outcome Measures
The presence of respiratory viruses and the odds ratio for admission with severe disease.
Results
759/922 (83%) eligible admissions were sampled (median age 9 months). One or more respiratory viruses were detected in 425/759 (56%, 95% CI 52 to 60%). Respiratory syncytial virus (RSV) was detected in 260 (34%, 95% CI 31 to 38%) and other respiratory viruses in 219 (29%, 95% CI 26 to 32%), the commonest being human coronavirus 229E (n=51, 6.7%, 95% CI 5.0 to 8.7%), influenza type A (n=44, 5.8%, 95% CI 4.2 to 7.7%), parainfluenza type 3 (n=29, 3.8%, 95% CI 2.6 to 5.4%), adenovirus (n=29, 3.8%, 95% CI 2.6 to 5.4%) and human metapneumovirus (n=23, 3.0%, 95% CI 1.9 to 4.5%). Compared to well controls, detection of RSV was associated with severe disease (4% in controls, adjusted odds Ratio 6.11 [95% CI 1.65 to 22.6]) whilst collectively, other respiratory viruses were not (23% in controls, adjusted odds Ratio 1.27 [95% CI 0.64 to 2.52]).
Conclusions
In a sample of Kenyan infants and children admitted with severe pneumonia to a rural hospital, RSV was the predominant viral pathogen.
Introduction
Pneumonia is the leading cause of childhood death in sub-Saharan Africa. The main direct means for controlling disease and death due to pneumonia are infant vaccination and case management. Thus, establishing the relative contribution to severe disease of individual pathogens in infancy and vaccine efficacy in this age group are essential to reducing the burden of disease.
Observational studies including microbial culture of blood or lung aspirates, and vaccine probe studies, have identified Streptococcus pneumoniae and Haemophilus influenzae as the commonest bacterial causes.1, 2 Both are preventable by currently available conjugate vaccines. Consequently, other bacteria; Mycobacterium tuberculosis; Pneumocystis jiroveci and respiratory viruses are becoming of increased importance.3 Vaccines currently exist, or are in development for several respiratory viruses including respiratory syncytial virus (RSV), influenza type A and parainfluenza type 3, including combinations.4-6 In previous studies in sub-Saharan Africa, viral etiology of pneumonia has been examined by antigen detection, serology and viral culture.7-11
The development of molecular methods has led to two advances in viral diagnosis: increased sensitivity and the discovery of new viruses of clinical importance. However, the high sensitivity of molecular diagnostics raises questions about specificity since viral nucleic acid sequences may be detected in healthy children and may persist after illness.12 The detection of multiple respiratory viruses by molecular methods in individual cases further underlines this etiological quandary.13 To date, no published studies have reported comprehensive viral etiology of pneumonia among children in sub-Saharan Africa using sensitive molecular diagnostic methods. We aimed to determine viral etiology, incidence and clinical features among infants and children meeting World Health Organisation (WHO) clinical criteria for ‘severe pneumonia’ or ‘very severe pneumonia’ admitted to a rural Kenyan district hospital.
Methods
Location
The study was conducted at Kilifi District Hospital, in a rural area on the Kenyan coast. The hospital serves a rural agrarian population, mainly comprising the Mjikenda tribe. Humidity is high throughout the year and there are two annual rainy seasons, April-July and November-December.14 The area is endemic for malaria, with declining transmission over the last 10 years. Haemophilus influenzae type b conjugate vaccine, given with Diptheria-Tetanus-Pertussis at 6, 10 and 14 weeks was introduced in 2001. Coverage of these vaccines (3rd dose) and measles vaccine at 9 months are >90%.15 Conjugate pneumococcal vaccine had not been introduced at the time of the study.
The Collaborative Research Programme between the Kenya Medical Research Institute (KEMRI) and the Wellcome Trust runs the hospital pediatric wards and a demographic surveillance system covering an area of ~900km2 within 50km north and south, and 30km west of the hospital including a population of ~240,000. The area was mapped in 2000 and every building registered by global positioning system (GPS). The population register is updated for births, deaths and migration events, which are monitored by household visits with three re-enumeration rounds each year.16 All field based data is checked, double entered and verified within 48 hours of acquisition. Since 2002, patients admitted to hospital are individually identified on the population register. In 2007, about 65% of paediatric admissions were confirmed residents of the census area.
Participants and Clinical Methods
We included all children age 1 day to 12 years, residing in the census area who were admitted to the pediatric wards between 1st January and 31st December 2007 and met WHO criteria for the clinical syndromes of ‘severe pneumonia’ (cough or difficult breathing plus lower chest wall indrawing and no signs of ‘very severe pneumonia’) or ‘very severe pneumonia’ (cough or difficult breathing plus at least one of: hypoxia, defined as an oxygen saturation <90% by fingertip pulse oximetry (Nellcor, USA), inability to drink or breast feed, inability to sit or impaired consciousness) at admission,17 including infants aged <2 months. Clinicians were trained in the recognition of the clinical signs used in the WHO syndromic criteria for pneumonia through teaching sessions that included videos and practical demonstrations, with additional bedside training on the wards. Clinical findings were individually recorded for each item of history or examination on a database during the admission assessment. Then, a standardized set of investigations were performed including: complete blood count (Beckman/Coulter, UK), blood film for malaria parasites, blood culture (Bactec PedsPlus, Becton Dickinson, New Jersey, USA) processed by standard methods,18 and a nasal wash sample.19 Nasal washes were conducted between 8am and 10pm daily. For safety, nasal wash was not performed on infants and children with severe respiratory or cardio-vascular compromise.
HIV testing was performed according to the Kenyan national policy for all pediatric hospital admissions using two rapid antibody tests: Determine (Inverness Medical, Florida, USA) and Unigold (Trinity Biotech, Bray, Ireland). Children, and their families, who tested positive were given further post-test counseling and referred to the hospital comprehensive HIV care clinic, which provided HIV care before and after discharge according to national recommendations.
For this analysis, a single sign, capillary refill time of 3 seconds or more was used as a marker of circulatory shock. Severe anemia was defined as hemoglobin <5g/dl. Severe malnutrition was defined as weight for height z score <-3 (National Centers for Health Statistics reference standards, CDC, Atlanta, USA) or kwashiorkor.17 Treatment for pneumonia and other conditions was according to current WHO guidelines.17
In order to estimate the association of respiratory viral infection with severe disease, we recruited two further sets of children resident in the census area by convenience sampling from 1st May 2007 to 30th April 2008: i) mild upper respiratory tract infection (URTI): those with symptoms including cough, runny or blocked nose, sore throat or sneezing being managed as outpatients and not meeting any criteria for pneumonia, and ii) well infants and children: those without any symptoms or signs of upper or lower respiratory infection attending for routine immunization at the hospital. Following clinical history and examination, a nasal wash specimen was taken.
Laboratory methods
Nasal wash samples were stored at −80°C and analyzed as a batch at the end of the study at the University of Pretoria, South Africa. The total viral nucleic acids were extracted from 200ul of the respiratory specimens using the Magnapure LC Total Nucleic Acid Isolation Kit (Roche, Manheim, Germany), according to the manufacturer's instructions. cDNA was synthesized using Expand Reverse Transcriptase (Roche, Mannheim, Germany). Real-time PCR reactions were performed using the LightCycler Fast Start DNA MasterPLUS HybProbe kit (Roche, Mannheim, Germany) for Adenovirus (Adeno); Parainfluenza virus (PIV) 1,2 & 3; RSV, Influenza (Flu) types A and B, human metapneumovirus (hMPV); Human bocavirus (hBoV); human coronavirus OC43 (OC43), human coronavirus Hong Kong (HKU1); human coronavirus 229E (229E) and human coronavirus NL-63 (NL63). PCR amplicons for DNA sequencing were gel purified using the Wizard® SV Gel and PCR Clean-Up System (Promega, Madison, WI, USA), according to the manufacturer’s instructions. DNA sequencing was performed with specific primers using the ABI PRISM BigDye Terminator Cycle Sequencing Reaction kit (version 3.1) on an ABI PRISM 3130 DNA sequencer (Applied Biosystems, Foster City, CA, USA), according to the manufacturer’s instructions.
Statistical Analysis
Statistical analysis was performed using STATA 9.1 (Stata Corp, TX, USA). Eligibility and classification of the clinical syndromes of pneumonia were determined from the original record of each item of history and examination on the database.
Incidence rates were estimated using data from all eligible admissions known to be resident in the census area on the day of admission and the mid-study (1st July 2007) census population estimates interpolated from the linear equation determined by regressing population size (log 10) for all enumeration rounds to the end of 2008 against the mid-date of each round.
In order to describe demographic and clinical features of cases of severe and very severe pneumonia, we compared those with a respiratory virus detected to those with no virus detected. Proportions were compared using the Chi squared and Fishers exact tests as appropriate. Continuous data were compared by t-test or the Kruskall-Wallis rank sum test.
To estimate the strength of association of detection of RSV and non-RSV viruses with severe disease, we compared admitted cases of ‘severe pneumonia’ and ‘very severe pneumonia’ to controls comprising well infants and children (excluding those with signs of URTI) using a case control design. Odds ratios were calculated by multivariable logistic regression and presented models for RSV (including those with RSV plus an additional virus) and respiratory viruses not including RSV that were i) unadjusted (Model 1) and ii) adjusted for age and season in monthly intervals (Model 2). To address a potential bias of under-representation of fatal cases, the (unadjusted) odds ratios for severe disease among all eligible admissions were modeled by imposing the proportions with RSV and other viruses detected among fatal and non-fatal sampled cases on all eligible admissions (Model 3). All statistical tests were two-sided with a significance level of 5%.
The sample size for cases was determined by previous annual admissions, which would allow the prevalence of pathogens of 5% and 20% to be estimated with precision of +/- 2% and +/-3% respectively. We aimed to recruit 100 well controls and 100 with mild URTI in order to describe specificities of 95% and 85% with a lower side precision of −6% and −8%, respectively. Power for the case-control analysis was confirmed for the actual numbers sampled using the method described by Fliess.20
Ethical Approval
The study was approved by the Kenyan National Ethical Review Committee (SSC 815) and the Oxford Tropical Ethical Review Committee (011 06). Individual written informed consent was sought from parents or guardians of all participants.
Results
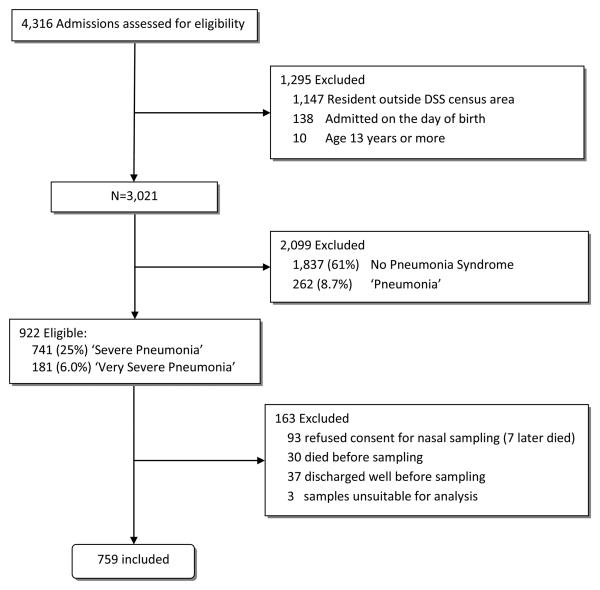
During 1st January through 31st December 2007, 922 eligible infants and children with ‘severe pneumonia’ or ‘very severe pneumonia’ were admitted and viral screening was conducted on 759 (82%) (Figure 1). Their median age was 9.0 months (IQR 3.3 to 20), 59% were male and 52 (6.9%, 95% CI 5.2 to 8.9%) had a positive HIV rapid antibody test. Children who were not sampled were more severely ill than those sampled (eTable 1). Three quarters of deaths among non-sampled admissions occurred within 24 hours of admission.
Figure 1.
Flow Diagram for Inclusion and Sampling of Infants and Children with ‘Severe’ or ‘Very Severe’ Pneumonia.
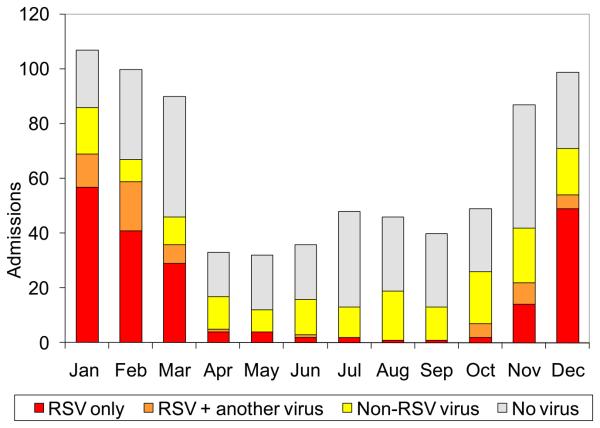
One or more respiratory viruses were detected in 425 (56%, 95% CI 52 to 60%) cases (Table 1). RSV was the most commonly detected virus, present in 260 (34%, 95% CI 31 to 38%) admissions overall, and in 192/453 (42%, 95% CI 38 to 47%) infants (eTable 2). RSV was strongly seasonal (P<0.001), detected in more than 50% of pneumonia cases in January, February and December, but in <5% from July through September (Figure 2). Other viruses showing clear seasonality were hMPV, PIV 3 and Flu A (eFigure 1).
Table 1.
Respiratory Viruses Detected and Odds Ratios for Severe Disease.
| No. (%) |
||||||
|---|---|---|---|---|---|---|
| Severe or Very Severe Pneumonia (N=759) |
Outpatient URTIa (N=96) |
Well Controls (N=57) |
||||
|
|
||||||
| No virus | 334 (44) | 54 (56) | 41 (72) | |||
|
|
||||||
| Any virus | 425 (56) | 42 (44) | 16 (28) | Odds Ratio [95% Confidence Intervals]b | ||
|
|
|
|||||
| 1 virus | 351 (46) | 36 (38) | 15 (26) |
Model 1: Unadjusted Odds Ratio |
Model 2: Adjusted Odds Ratio |
Model 3: Unadjusted Odds Ratio for All Eligible Admissions |
|
| ||||||
| 2 viruses | 66 (8.7) | 5 (5) | 1 (2) | |||
|
| ||||||
| 3 viruses | 8 (1.1) | 1 (1) | 0 (0) | |||
|
| ||||||
| RSV only | 206 (27) | 15 (16) | 2 (4) | 9.38 [2.99 to 47.3] | 6.11 [1.65 to 22.6] | 8.99 [2.87 to 45.2] |
|
| ||||||
| RSV + another virus | 54 (7.1) | 2 (2) | 1 (2) | |||
|
| ||||||
| Non-RSV virus | 165 (22) | 25 (26) | 13 (23) | 0.94 [0.48 to 1.94] | 1.27 [0.64 to 2.52] | 0.88 [0.45 to 1.83] |
Upper Respiratory Tract Infection
Regression Models for Admissions with ‘Severe’ or ‘Very Severe’ Pneumonia versus well controls, excluding outpatients with URTI (see text).
Figure 2.
Seasonal Pattern of RSV and Other Viruses Detected Among Infants and Children Admitted with ‘Severe’ or ‘Very Severe’ Pneumonia.
Other respiratory viruses were detected in 219 (29%, 95% CI 26 to 32%)) admissions (eTable 2 & eFigure 1). These comprised: Flu A (5.8%); Flu B (0.1%); hMPV (3.0%); hBoV (2.1%); PIV 1 (2.4%); PIV 2 (1.3%); PIV 3 (3.8%); Adenovirus (3.8%); NL63 (1.3%); HKU1 (0.1%); 229E (6.7%) and OC43 (1.8%).
RSV plus one or more other respiratory virus were detected in 54 (7%) cases, representing 21% (95% CI 16 to 26%) of all RSV cases. These comprised 229E (14), Flu A (10), adenovirus (10), PIV 3 (6), hBoV (6), HMPV (3), PIV 1 (3), PIV 2 (3), NL63 (2), and OC43 (1).
The incidence of admission with ‘severe pneumonia’ or ‘very severe pneumonia’ was 4.8% per child/year in the first year of life, 1.5% per child/year in under 5s and 0.1% amongst children 5 or more years old (Table 2). The incidence of admission for any respiratory virus and with RSV was highest in the first year of life (3.0% per child/year and 2.0% per child/year, respectively).
Table 2.
Incidence of Admission with ‘Severe Pneumonia’ or ‘Very Severe Pneumonia’ Among Residents of the Kilifi DSS Census Area.
| Age Group | under 28 days | All <1y |
1 to 1.99y |
2 to 4.99y |
All <5y |
5 to 12.99y |
All < 13y |
|---|---|---|---|---|---|---|---|
| Denominator | 9,423 births | 8,837 | 44,538 | 104,505 | |||
| per 1,000 live births |
per 100,000 children/yr |
||||||
|
| |||||||
| ‘Severe’ or ‘Very Severe’ Pneumonia |
6.65 | 4,798 | 1,674 | 543 | 1,522 | 99 | 681 |
|
| |||||||
| Any respiratory virus | 3.79 | 2,993 | 871 | 213 | 862 | 36 | 380 |
|
| |||||||
| RSV | 2.46 | 2,038 | 455 | 85 | 535 | 15 | 233 |
|
| |||||||
| 229E | 0.51 | 318 | 135 | 32 | 105 | 3 | 46 |
|
| |||||||
| Influenza A | 0.31 | 244 | 97 | 32 | 82 | 15 | 39 |
|
| |||||||
| PIV3 | 0.10 | 212 | 48 | 11 | 57 | 6 | 26 |
|
| |||||||
| Adenovirus | 0.10 | 149 | 77 | 21 | 55 | 9 | 26 |
|
| |||||||
| HMPV | 0.20 | 138 | 58 | 11 | 44 | 6 | 21 |
Admissions with respiratory viruses detected had a median age of 7.5 months (IQR 2.7 to 18) and were younger than those with no virus detected (median age of 11 months (IQR 3.8 to 24), P<0.001). However, this effect was completely accounted for by RSV (Table 3). ‘Very severe pneumonia’ was less common among admissions with respiratory viruses (P<0.001). There was no association between the presence of wheeze or hypoxia and a respiratory virus being detected. Most cases with RSV did not have an admission diagnosis of bronchiolitis. Admissions with RSV were less severely ill and had fewer adverse risk factors than admissions with no virus: they were less likely to be prematurely born, shocked, severely malnourished or to die. Of the 2 deaths among children with RSV, both had congenital heart disease. Admissions with respiratory viruses other than RSV were similar in all these respects to admissions with no virus detected. Among 24 deaths, 8 (33%, 95% CI 16 to 55%) occurred in admissions with a virus detected.
Table 3.
Clinical Features of 759 Children Admitted with RSV and Other Respiratory Viruses.
| Median (IQR)a |
Median (IQR)a
P Valueb |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No virus N=334 |
Any virus N=425 |
RSV only N=206 |
RSV + another virus N=54 |
Non-RSV virus N=165 |
||||||||||
| Age (months) | 11.3 | (3.8 to 24) | 7.5 | (2.7 to 18) | <.001 | 6.1 | (2.5 to 13) | <.001 | 7.5 | (2.7 to 16) | .011 | 10 | (4.5 to 20) | .39 |
|
| ||||||||||||||
| Inpatient Stay (days) | 4 | (2 to 6) | 3 | (2 to 5) | .17 | 3 | (2 to 5) | .07 | 4 | (3 to 6) | .80 | 4 | (2 to 6) | .56 |
|
| ||||||||||||||
|
No. (%)
|
No. (%) P Valueb
|
|||||||||||||
|
No virus N=334 |
Any virus N=425 |
RSV only N=206 |
RSV + another virus N=54 |
Non-RSV virus N=165 |
||||||||||
|
| ||||||||||||||
| Very Severe Pneumonia | 73 | (22) | 53 | (13) | .001 | 22 | (11) | .001 | 8 | (15) | .24 | 23 | (14) | .035 |
|
| ||||||||||||||
| Wheeze | 50 | (15) | 59 | (14) | .67 | 30 | (15) | .90 | 8 | (15) | .98 | 2 | (13) | .50 |
|
| ||||||||||||||
| Hypoxia | 40 | (13) | 40 | (9) | .13 | 18 | (8.7) | .14 | 8 | (15) | .70 | 14 | (8.5) | .14 |
|
| ||||||||||||||
| Capillary refill ≥3 secs | 16 | (4.8) | 10 | (2.4) | .066 | 1 | (0.5) | .004 | 1 | (1.9) | .49 | 8 | (4.9) | .98 |
|
| ||||||||||||||
| Severe Anemia | 15 | (4.6) | 12 | (2.9) | .23 | 3 | (1.5) | .082 | 1 | (1.9) | .71 | 8 | (4.9) | .85 |
|
| ||||||||||||||
| History of Prematurity | 17 | (7.8) | 14 | (4.1) | .067 | 5 | (2.9) | .046 | 2 | (4.4) | .54 | 7 | (5.7) | .49 |
|
| ||||||||||||||
| Congenital Heart Disease |
12 | (3.6) | 5 | (1.2) | .05 | 2 | (1.0) | .09 | 0 | (0) | .23 | 3 | (1.8) | .40 |
|
| ||||||||||||||
| HIV | 26 | (8.0) | 26 | (6.2) | .35 | 8 | (4.0) | .07 | 4 | (7.6) | 1 | 14 | (8.6) | .82 |
|
| ||||||||||||||
| Severe Malnutrition | 26 | (7.8) | 18 | (4.2) | .038 | 5 | (2.4) | .009 | 2 | (3.7) | .40 | 11 | (6.7) | .65 |
|
| ||||||||||||||
| Bacteremia | 20 | (6.0) | 16 | (3.8) | .15 | 5 | (2.4) | .056 | 2 | (3.7) | .75 | 9 | (5.5) | .81 |
| Death | 16 | (4.8) | 8 | (1.9) | . 023 | 2 | (1.0) | .023 | 0 | (0) | .14 | 6 | (3.6) | .56 |
IQR = interquartile range
P Values in comparison with children with no virus.
Thirty six (4.7%, 95% CI 3.3 to 6.5%) admissions with ‘severe pneumonia’ or ‘very severe pneumonia’ were bacteremic, 16 (44%, 95% CI 28 to 62%) of these had a respiratory virus detected. Bacterial species were Streptococcus pneumonia (12), Escherichia coli (9), non-typhoidal Salmonella (3), Staphylococcus aureus (3), Acinetobacter sp. (3), Beta haemolytic Streptococci (3), Enterobacter sp. (2) and Haemophilus influenzae (1).
Among 57 well infants and children (median age 6.0 months, IQR 3.3 to 11) and 96 children with symptoms of mild URTI (median age 13 months, IQR 5.6 to 25), respiratory viruses were detected in 28% and 44% respectively, and were less frequent than among admitted pneumonia cases: P<0.001 and P=0.026 respectively (Table 1). These differences were due to RSV since overall, viruses other than RSV were as common in well infants and children and those with mild URTI as amongst admitted cases: P=0.85 and P=0.34 respectively (Table 1). Among those with mild URTI, viruses detected were: RSV (18%); Flu A (7%); hMPV (3%); hBoV (5%); PIV 1 (2%); Adenovirus (8%); HKU1 (2%); 229E (3%) and OC43 (2%) (eTable 3). Viruses found in well infants and children comprised: RSV (5%); hBoV (9%); OC43 (7%) and <1% for each of PIV 1, PIV 3 , Adenovirus, HKU1, and 229E. Flu A and HMPV were not detected among well infants (eTable 2).
Detection of RSV was associated with admission with severe disease compared to well controls (odds ratio 9.38 (95% CI 2.99 to 47.3), 5% in controls, Model 1, Table 1) There was no evidence of an association between viruses other than RSV and severe disease (odds ratio 0.94 (95% CI 0.48 to 1.9), 23% in controls, Model 1, Table 1). Models adjusted for age and calendar month (Model 2, Table 1) showed no evidence of lack of fit (Hosmer-Lemeshow chi squared 7.19, P=0.52) and did not qualitatively alter this result: RSV odds ratio 6.11 (95% CI 1.65 to 22.6) and non-RSV viruses 1.27 (95% CI 0.64 to 2.52). The third model including all eligible admissions, to address the potential bias of under-representation of fatal cases, did not differ significantly from the observed, unadjusted odds ratios (Model 3 & Model 1, Table 1).
Discussion
We found that in the catchment area of Kilifi District Hospital on the Kenyan coast, the incidence of admission with clinical syndromes of severe or very severe pneumonia ranged from 4.8% in the first year of life to 0.1% among those 5 years old or greater. RSV was detected in a third of cases overall and almost half of infant cases. The seasonality of severe pneumonia was almost entirely determined by RSV. No other virus was identified in more than 7% of admitted infants or children, and these cases were clinically similar to those in whom no virus was detected. Viruses other than RSV were as common among well infants and children and those with mild URTI as among those with severe disease. These findings suggest that they make only a minor contribution to the burden of severe clinical pneumonia in this setting.
In a previous study in the same enumerated population of Kilifi 2002 through 2007, RSV was detected by immunofluorescence in 15% of ‘severe pneumonia’ or ‘very severe pneumonia’ admissions in under 5′s, rising to 27% during epidemic periods. Incidence during 2006/2007 was estimated at 0.99% and 0.27% per child per year in infants and under 5′s respectively.14 In the present study, we found approximately twice the incidence. It is known that real time PCR is more sensitive than traditional diagnostics. In a study by Kuypers et al.21 RSV detected by both immunofluorescence and PCR had a mean viral load of 6.1 × 107 copies/ml. Whereas, RSV detected by PCR only had a lower mean viral load of 4.1 × 104 copies/ml. An immunofluorescence assay detected only 19% of the RSV with viral loads <106 copies/ml.
Could our findings be due to detection of persistent viral RNA or false positive laboratory results? We believe that this is unlikely. Firstly, considering our well controls, if an assumption is made that all of the RSV detected are false positives due to persistence or laboratory error, then the specificity of our test is 95% (95% CI 85 to 99%). This is similar to that reported for immunofluorescence. Such specificity would not significantly alter the interpretation of our findings among cases. This is further supported by an odds ratio suggesting that detection of RSV was strongly associated with severe disease. To compare these findings with those from traditional methods, we examined published data from 8 previous studies in developing countries conducted using immunofluorescence, serology or viral culture.7, 8, 11, 22-27 Overall, RSV was detected in 746/3,463 (21.5%) cases and 29/1,203 (2.4%) controls, giving a pooled (fixed effects) odds ratio for disease (unadjusted for age and season) of 11.1 (95% CI 7.69 to 16.0), which is close to the unadjusted odds ratio in our study.
The high prevalence of RSV in childhood admissions with severe clinical pneumonia (34%) in contrast to the prevalence in well controls (5%), provides further support that RSV vaccination may offer considerable public health benefit. While the development of a vaccine for the key target age group of early infants has focused on live virus vaccines, none has achieved requisite levels of both safety and immunogenicity.4 A combined PIV3/RSV live-attenuated vaccine (Medi-534) for use in young infants (2 months) and older children (6 to <24 months) is currently undergoing phase I/II clinical trials. Ideally, such trials should be accompanied by modelling studies of the potential consequences on effectiveness of delayed delivery to older infants28, 29 where vaccine safety and immunogenicity are demonstrably improved. 5, 30
Of equal interest to the high prevalence of cases with RSV detected is the low proportion of cases with any of the other respiratory viruses. The real time PCR assay that we used has previously been shown to be sensitive for these viruses.31 It is possible that the use of nasal washings targets a site within the upper respiratory tract is predilected by RSV over any other virus. However, to our knowledge there are no definitive studies in the literature which clearly identify selective bias in the range of respiratory viruses detectable in different parts of the upper respiratory tract using molecular diagnostics.
Influenza and parainfluenza 3 have been the most frequently detected viruses, other than RSV, in studies employing traditional diagnostics in sub-Saharan Africa. We found these, 229E and hMPV, were the most commonly detected non-RSV viruses in cases. However, these viruses appear to be common in the community causing URTI, and some are present in well controls. In a non-epidemic period, we estimate that up to approximately 6% of severe disease is associated with influenza type A. There appears to be little potential to prevent severe disease through vaccination against PIV 3.
Strengths of our study include systematic sampling of well-characterized hospitalized children from a well established, enumerated population base and detection of a comprehensive set of respiratory viruses. Our study has several limitations. In common with similar studies, our sampling missed most deaths because nasal washing cannot be conducted in the sickest children at admission. However, our sensitivity analysis suggests this in unlikely to significantly alter the main findings. We did not screen for rhinovirus and there was a lack of x-ray data. Our data reflect a single site and findings in other ecological settings may differ. We applied WHO guidelines carefully and there was good availability of clinical staff, oxygen and drugs. Case fatality may differ where resources are more constrained. Our well controls were limited in number, since we did not achieve our target sample size due to factors outside our control, and they were not matched to exactly the same calendar year. Small numbers resulted in wide confidence intervals in some subgroups. However, power was sufficient (>99%) for our case-control analysis of RSV.
It is likely that not all cases of severe pneumonia in the community were admitted to hospital, thus our incidence estimates are therefore the minimum. There is a demonstrable decay with increased distance from the hospital in the incidences of all childhood admissions and of admissions with detectable RSV.14 A study spanning only one year and from a single location cannot expect to account for likely variation in the occurrence of respiratory viruses year-to-year or between low resource settings.
In summary, our study of the occurrence of respiratory viruses in children admitted with clinical syndromes of ‘severe pneumonia’ or ‘very severe pneumonia’ to a rural district hospital in coastal Kenya, has identified over 50% of cases with a detectable virus, of which RSV was clearly predominant. We estimate that the effective prevention of RSV-associated severe pneumonia in this population might reduce all-cause clinical ‘severe pneumonia’ or ‘very severe pneumonia’ admissions to the District hospital by a third. This contrasts with no evidence to suggest a marked effect on such admissions would arise from the prevention of any other respiratory virus, with the possible exception of influenza type A. Further molecular based studies of respiratory virus etiology of severe pneumonia over longer periods and in multiple settings in sub-Saharan Africa are needed.
Supplementary Material
Acknowledgements
The authors are grateful to the staff and patients of the pediatric wards and outpatient department of Kilifi District Hospital and to the clinical, ICT and laboratory staff of the KEMRI/Wellcome Trust Research Programme who contributed to this study. We thank Dr Norbert Peshu M.P.H., Director of the Centre for Geographic Medicine Research – coast and Dr Iqbal Khandwalla M.Med., the Medical Superintendent of Kilifi District Hospital for their oversight and making facilities available for this research. No compensation was received for participation in this research. This paper is published with permission of the director of KEMRI.
The study was conceived and designed by JAB, JAGS and DJN. JAB, PM, MN and SK collected the clinical data, nasal wash samples and provided patient care. JA, AB, RL, TK, PAC and MV developed and undertook laboratory analyses. Analysis was conducted by JAB, JAGS and DJN. JAB wrote the manuscript and all authors contributed to the final version. JAB had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
The study was funded by a project grant (081186) from the Wellcome Trust (UK). JAB, JAGS and DJN are supported by fellowships from the Wellcome Trust (UK). The funder had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript.
Footnotes
All authors declare no conflict of interest.
References
- 1.Cutts FT, Zaman SM, Enwere G, et al. Efficacy of nine-valent pneumococcal conjugate vaccine against pneumonia and invasive pneumococcal disease in The Gambia: randomised, double-blind, placebo-controlled trial. Lancet. 2005 Mar 26;365(9465):1139–1146. doi: 10.1016/S0140-6736(05)71876-6. Apr 1. [DOI] [PubMed] [Google Scholar]
- 2.Mulholland K, Hilton S, Adegbola R, et al. Randomised trial of Haemophilus influenzae type-b tetanus protein conjugate vaccine [corrected] for prevention of pneumonia and meningitis in Gambian infants. Lancet. 1997 Apr 26;349(9060):1191–1197. doi: 10.1016/s0140-6736(96)09267-7. [DOI] [PubMed] [Google Scholar]
- 3.Scott JA, English M. What are the implications for childhood pneumonia of successfully introducing Hib and pneumococcal vaccines in developing countries? PLoS Med. 2008 Apr 22;5(4):e86. doi: 10.1371/journal.pmed.0050086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karron RA, Wright PF, Belshe RB, et al. Identification of a recombinant live attenuated respiratory syncytial virus vaccine candidate that is highly attenuated in infants. J Infect Dis. 2005 Apr 1;191(7):1093–1104. doi: 10.1086/427813. [DOI] [PubMed] [Google Scholar]
- 5.Wright PF, Karron RA, Belshe RB, et al. The absence of enhanced disease with wild type respiratory syncytial virus infection occurring after receipt of live, attenuated, respiratory syncytial virus vaccines. Vaccine. 2007 Oct 16;25(42):7372–7378. doi: 10.1016/j.vaccine.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmidt AC, McAuliffe JM, Murphy BR, Collins PL. Recombinant bovine/human parainfluenza virus type 3 (B/HPIV3) expressing the respiratory syncytial virus (RSV) G and F proteins can be used to achieve simultaneous mucosal immunization against RSV and HPIV3. J Virol. 2001;75(10):4594–4603. doi: 10.1128/JVI.75.10.4594-4603.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sobeslavsky O, Sebikari SR, Harland PS, Skrtic N, Fayinka OA, Soneji AD. The viral etiology of acute respiratory infections in children in Uganda. Bull World Health Organ. 1977;55(5):625–631. [PMC free article] [PubMed] [Google Scholar]
- 8.Forgie IM, O'Neill KP, Lloyd-Evans N, et al. Etiology of acute lower respiratory tract infections in Gambian children: I. Acute lower respiratory tract infections in infants presenting at the hospital. Pediatr Infect Dis J. 1991 Jan;10(1):33–41. doi: 10.1097/00006454-199101000-00008. [DOI] [PubMed] [Google Scholar]
- 9.Adegbola RA, Falade AG, Sam BE, et al. The etiology of pneumonia in malnourished and well-nourished Gambian children. Pediatr Infect Dis J. 1994 Nov;13(11):975–982. doi: 10.1097/00006454-199411000-00008. [DOI] [PubMed] [Google Scholar]
- 10.Johnson BR, Osinusi K, Aderele WI, Tomori O. Viral pathogens of acute lower respiratory infections in pre-school Nigerian children and clinical implications of multiple microbial identifications. West Afr J Med. 1993 Jan-Mar;12(1):11–20. [PubMed] [Google Scholar]
- 11.Mulholland EK, Ogunlesi OO, Adegbola RA, et al. Etiology of serious infections in young Gambian infants. Pediatr Infect Dis J. 1999 Oct;18(10 Suppl):S35–41. doi: 10.1097/00006454-199910001-00007. [DOI] [PubMed] [Google Scholar]
- 12.Mahony JB. Detection of respiratory viruses by molecular methods. Clin Microbiol Rev. 2008 Oct;21(4):716–747. doi: 10.1128/CMR.00037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stempel HE, Martin ET, Kuypers J, Englund JA, Zerr DM. Multiple viral respiratory pathogens in children with bronchiolitis. Acta Paediatr. 2009 Jan;98(1):123–126. doi: 10.1111/j.1651-2227.2008.01023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nokes DJ, Ngama M, Bett A, et al. Incidence and severity of respiratory syncytial virus pneumonia in rural Kenyan children identified through hospital surveillance. Clin Infect Dis. 2009 Nov 1;49(9):1341–1349. doi: 10.1086/606055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ndiritu M, Cowgill KD, Ismail A, et al. Immunization coverage and risk factors for failure to immunize within the Expanded Programme on Immunization in Kenya after introduction of new Haemophilus influenzae type b and hepatitis b virus antigens. BMC Public Health. 2006;6:132. doi: 10.1186/1471-2458-6-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cowgill KD, Ndiritu M, Nyiro J, et al. Effectiveness of Haemophilus influenzae type b Conjugate vaccine introduction into routine childhood immunization in Kenya. Jama. 2006 Aug 9;296(6):671–678. doi: 10.1001/jama.296.6.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.WHO . Pocket book of hospital care for children - guidelines for the management of common illnesses with limited resources. WHO; Geneva: 2005. [PubMed] [Google Scholar]
- 18.Berkley JA, Lowe BS, Mwangi I, et al. Bacteremia among children admitted to a rural hospital in Kenya. N Engl J Med. 2005 Jan 6;352(1):39–47. doi: 10.1056/NEJMoa040275. [DOI] [PubMed] [Google Scholar]
- 19.Nokes DJ, Okiro EA, Ngama M, et al. Respiratory syncytial virus epidemiology in a birth cohort from Kilifi district, Kenya: infection during the first year of life. J Infect Dis. 2004 Nov 15;190(10):1828–1832. doi: 10.1086/425040. [DOI] [PubMed] [Google Scholar]
- 20.Fliess JL. Statistical Methods for Rates and Proportions 3rd Ed. 3rd ed. Wiley; New York: 1981. [Google Scholar]
- 21.Kuypers J, Wright N, Morrow R. Evaluation of quantitative and type-specific real-time RT-PCR assays for detection of respiratory syncytial virus in respiratory specimens from children. J Clin Virol. 2004 Oct;31(2):123–129. doi: 10.1016/j.jcv.2004.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vieira SE, Stewien KE, Queiroz DA, et al. Clinical patterns and seasonal trends in respiratory syncytial virus hospitalizations in Sao Paulo, Brazil. Rev Inst Med Trop Sao Paulo. 2001 May-Jun;43(3):125–131. doi: 10.1590/s0036-46652001000300002. [DOI] [PubMed] [Google Scholar]
- 23.Forgie IM, O'Neill KP, Lloyd-Evans N, et al. Etiology of acute lower respiratory tract infections in Gambian children: II. Acute lower respiratory tract infection in children ages one to nine years presenting at the hospital. Pediatr Infect Dis J. 1991 Jan;10(1):42–47. doi: 10.1097/00006454-199101000-00009. [DOI] [PubMed] [Google Scholar]
- 24.Loscertales MP, Roca A, Ventura PJ, et al. Epidemiology and clinical presentation of respiratory syncytial virus infection in a rural area of southern Mozambique. Pediatr Infect Dis J. 2002 Feb;21(2):148–155. doi: 10.1097/00006454-200202000-00013. [DOI] [PubMed] [Google Scholar]
- 25.Albargish KA, Hasony HJ. Respiratory syncytial virus infection among young children with acute respiratory tract infection in Iraq. East Mediterr Health J. 1999 Sep;5(5):941–948. [PubMed] [Google Scholar]
- 26.Dagan R, Landau D, Haikin H, Tal A. Hospitalization of Jewish and Bedouin infants in southern Israel for bronchiolitis caused by respiratory syncytial virus. Pediatr Infect Dis J. 1993 May;12(5):381–386. doi: 10.1097/00006454-199305000-00006. [DOI] [PubMed] [Google Scholar]
- 27.Phillips PA, Lehmann D, Spooner V, et al. Viruses associated with acute lower respiratory tract infections in children from the eastern highlands of Papua New Guinea (1983-1985) Southeast Asian J Trop Med Public Health. 1990 Sep;21(3):373–382. [PubMed] [Google Scholar]
- 28.Nokes DJ, Okiro EA, Ngama M, et al. Respiratory syncytial virus infection and disease in infants and young children observed from birth in Kilifi District, Kenya. Clin Infect Dis. 2008 Jan 1;46(1):50–57. doi: 10.1086/524019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nokes JD, Cane PA. New strategies for control of respiratory syncytial virus infection. Curr Opin Infect Dis. 2008 Dec;21(6):639–643. doi: 10.1097/QCO.0b013e3283184245. [DOI] [PubMed] [Google Scholar]
- 30.Wright PF, Karron RA, Belshe RB, et al. Evaluation of a live, cold-passaged, temperature-sensitive, respiratory syncytial virus vaccine candidate in infancy. J Infect Dis. 2000 Nov;182(5):1331–1342. doi: 10.1086/315859. [DOI] [PubMed] [Google Scholar]
- 31.Lassauniére R, Kresfelder T, Venter M. A Novel Multiplex Real-Time RT-PCR Assay with FRET Hybridization Probes for the Detection and Quantification of 13 Traditional and Newly Identified Respiratory Viruses. J Virological Methods. 2010 doi: 10.1016/j.jviromet.2010.02.005. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.