Abstract
The title compound, C21H16N2, has been known since 1877. Although the crystal structure of 36 derivatives of lophine are known, the structure of parent compound has remained unknown until now. The three phenyl rings bonded to the imidazole core are not coplanar with the latter, with dihedral angles of 21.4 (3), 24.7 (3), and 39.0 (3)°, respectively, between the phenyl ring planes in the 2-, 4- and 5-positions of the imidazole ring. The molecules are packed in layers running perpendicular to the b axis. Although there are acceptor and donor atoms for hydrogen bonds, no such interactions are detected in the crystal in contrast to other lophine derivatives.
Related literature
For background on lophine and its derivatives, see: Fridman et al. (2008 ▶); Fridman, Kaftory & Speiser (2007 ▶); Fridman, Kaftory, Eichen & Speiser (2007 ▶); Kamidate et al. (1989 ▶); Liu et al. (2005 ▶); Nakashima (2003 ▶); Nakashima et al. (1995 ▶); Radziszewski (1877 ▶); Seethalakshmi et al. (2006 ▶); Thiruvalluvar et al. (2007 ▶); Thuer et al. (2004 ▶). For information about the Cambridge Database, see: Allen (2002 ▶). For related literature, see: Inouye & Sakaino (2000 ▶); Kaftory et al. (1998 ▶); Santos et al. (2001 ▶).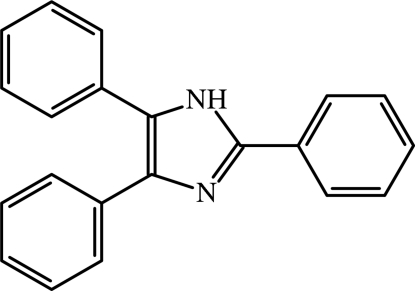
Experimental
Crystal data
C21H16N2
M r = 296.36
Orthorhombic,

a = 20.218 (4) Å
b = 7.538 (2) Å
c = 20.699 (4) Å
V = 3154.6 (12) Å3
Z = 8
Mo Kα radiation
μ = 0.07 mm−1
T = 293 K
0.50 × 0.10 × 0.05 mm
Data collection
Nonius KappaCCD diffractometer
Absorption correction: none
22192 measured reflections
2747 independent reflections
1036 reflections with I > 2σ(I)
R int = 0.143
Refinement
R[F 2 > 2σ(F 2)] = 0.088
wR(F 2) = 0.245
S = 1.09
2747 reflections
209 parameters
H-atom parameters constrained
Δρmax = 0.25 e Å−3
Δρmin = −0.30 e Å−3
Data collection: COLLECT (Nonius, 2006); cell refinement: DENZO HKL-2000 (Otwinowski & Minor, 1997 ▶); data reduction: DENZO HKL-2000; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 1999 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809006552/hb2915sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809006552/hb2915Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
Comment
Recently, heterocyclic imidazole derivatives have attracted considerable attention because of their unique optical properties (Santos et al., 1996). These compounds play a very important role in chemistry as multipurpose analytical tools (Nakashima et al., 1995 & Nakashima, 2003 & Kamidate et al., 1989). Lophine (2,4,5-triphenyl-1H-imidazole) (I) is an attractive fluorescence and chemiluminescence compound. The chemiluminescence properties of this synthetic organic compound was reported for the first time by Radziszewski (1877). He showed that lophine emits a yellow light when it reacts with oxygen in the presence of a strong base. Since then many lophine derivatives have been synthesized and studied with regards to their optical properties. Lophine was chosen as the molecule of the week (December 15, 2008) by the American Chemical Society with the following summary "Lophine (2,4,5-triphenyl-1H-imidazole) exhibits lemon yellow chemiluminescence in solution and is one of the few long-lasting chemiluminescence molecules. It forms dimmers that have piezochromic and photochromic properties. It has been proposed as an analytical reagent for trace metal ion detection." The crystal structure of 36 different lophine derivatives have been deposited at the Cambridge Crystallographic Data Center (Allen, 2002). It is interesting to note, however, that the crystal structure of the parent lophine compound was never published. We undertook a search for new lophine derivatives that will show thermo- and photochromic properties (Fridman et al., 2007, Fridman et al., 2007, Fridman et al., 2008).
During our investigation we have succeeded to identify crystals of neat lophine. Here we describe its crystal structure. The molecular structure of lophine depicted in Figure 1, may be described by the rotation of three phenyl rings about their bonds to the imidazole ring. The rotation angles of the phenyl rings plane are 21.4, 24.7, and 39.0° at the 2, 4 and 5 positions of the imidazole ring respectively. Although the imidazole ring bearing H-donor and H-acceptor N atoms, they do not participate in hydrogen bonding as found in five other lophine derivatives (Seethalakshmi et al., 2006, Ynouye & Sakaino, 1986, Thuer et al., 2004, Liu et al., 2005, Thiruvalluvar et al., 2007) where the lophine derivative molecules packed in chains made up of hydrogen bonded molecules of N—H···N type. All other lophine derivatives form hydrogen bonds through the N—H or the N atoms either with molecules of the same kind or with solvent molecules. The absent of strong intermolecular forces might be the reason for the lack of published crystal structure of neat lophine because it is difficult to crystallize it.
Experimental
Lophine synthesis: benzil (1 mmol), suitable benzaldehyde (1 mmol), and ammonium acetate (1.2 g) were dissolved in boiling glacial acetic acid (16 ml) and refluxed for 5–24 h. The reaction progress was monitored by TLC. After the reaction completion, the reaction mixture was poured into ice-water, washed with NaHCO3 and then washed several times with EtOAc. The combined extracts were dried over MgSO4. The purification was done by flash column chromatography. The compound was obtained in 46% yield. Colourless plates of (I) were recrystallised from DMSO.
Refinement
The quality of the crystals was poor which is also reflected in the crystal structure refinement. H atoms were clearly found in the difference Fourier maps. All H atoms were refined at idealized positions riding on the C and N atoms with C—H = 0.96 Å, and N—H = 0.86 Å, and Uiso(H) = 1.2Ueq(C or N).
Figures
Fig. 1.
Molecular structure of (I). Displacement ellipsoids are drawn at the 50% probability level.
Fig. 2.
Unit cell packing for (I).
Crystal data
| C21H16N2 | F(000) = 1248 |
| Mr = 296.36 | Dx = 1.248 Mg m−3 |
| Orthorhombic, Pbca | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ac 2ab | Cell parameters from 22192 reflections |
| a = 20.218 (4) Å | θ = 2.0–25.3° |
| b = 7.538 (2) Å | µ = 0.07 mm−1 |
| c = 20.699 (4) Å | T = 293 K |
| V = 3154.6 (12) Å3 | Plate, colorless |
| Z = 8 | 0.50 × 0.10 × 0.05 mm |
Data collection
| Nonius KappaCCD diffractometer | 1036 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.143 |
| graphite | θmax = 25.3°, θmin = 2.0° |
| φ and ω scans | h = −23→22 |
| 22192 measured reflections | k = −8→8 |
| 2747 independent reflections | l = −24→23 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.088 | H-atom parameters constrained |
| wR(F2) = 0.245 | w = 1/[σ2(Fo2) + (0.0863P)2] where P = (Fo2 + 2Fc2)/3 |
| S = 1.09 | (Δ/σ)max < 0.001 |
| 2747 reflections | Δρmax = 0.25 e Å−3 |
| 209 parameters | Δρmin = −0.30 e Å−3 |
| 0 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.0089 (18) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N1 | 0.6928 (2) | 0.1996 (5) | 0.14220 (18) | 0.0556 (11) | |
| H1 | 0.6809 | 0.1806 | 0.1029 | 0.067* | |
| N2 | 0.7563 (2) | 0.2458 (5) | 0.2252 (2) | 0.0668 (12) | |
| C1 | 0.8110 (3) | 0.1974 (7) | 0.1209 (3) | 0.0660 (14) | |
| C2 | 0.8052 (3) | 0.2208 (9) | 0.0564 (3) | 0.0928 (19) | |
| H2 | 0.7620 | 0.2437 | 0.0390 | 0.111* | |
| C3 | 0.8616 (4) | 0.2073 (9) | 0.0170 (3) | 0.108 (2) | |
| H3 | 0.8570 | 0.2267 | −0.0286 | 0.129* | |
| C4 | 0.9225 (3) | 0.1634 (8) | 0.0422 (3) | 0.0890 (19) | |
| H4 | 0.9611 | 0.1517 | 0.0155 | 0.107* | |
| C5 | 0.9267 (3) | 0.1408 (8) | 0.1067 (4) | 0.0902 (19) | |
| H5 | 0.9694 | 0.1134 | 0.1244 | 0.108* | |
| C6 | 0.8727 (3) | 0.1565 (7) | 0.1470 (3) | 0.0822 (18) | |
| H6 | 0.8778 | 0.1408 | 0.1927 | 0.099* | |
| C7 | 0.7536 (3) | 0.2141 (7) | 0.1634 (3) | 0.0767 (16) | |
| C8 | 0.6519 (3) | 0.2205 (6) | 0.1946 (3) | 0.0672 (14) | |
| C9 | 0.6911 (3) | 0.2496 (6) | 0.2465 (3) | 0.0690 (14) | |
| C10 | 0.6777 (3) | 0.2762 (7) | 0.3158 (2) | 0.0689 (15) | |
| C11 | 0.6295 (3) | 0.1845 (7) | 0.3489 (3) | 0.0771 (16) | |
| H11 | 0.6028 | 0.1011 | 0.3255 | 0.093* | |
| C12 | 0.6206 (3) | 0.2137 (9) | 0.4140 (3) | 0.0876 (18) | |
| H12 | 0.5870 | 0.1509 | 0.4375 | 0.105* | |
| C13 | 0.6578 (4) | 0.3288 (10) | 0.4475 (3) | 0.098 (2) | |
| H13 | 0.6503 | 0.3469 | 0.4928 | 0.118* | |
| C14 | 0.7082 (4) | 0.4240 (10) | 0.4148 (3) | 0.098 (2) | |
| H14 | 0.7355 | 0.5090 | 0.4367 | 0.118* | |
| C15 | 0.7179 (3) | 0.3959 (8) | 0.3504 (3) | 0.0841 (18) | |
| H15 | 0.7531 | 0.4548 | 0.3279 | 0.101* | |
| C16 | 0.5797 (3) | 0.2127 (7) | 0.1838 (2) | 0.0673 (15) | |
| C17 | 0.5352 (3) | 0.3003 (7) | 0.2245 (3) | 0.0739 (16) | |
| H17 | 0.5523 | 0.3636 | 0.2611 | 0.089* | |
| C18 | 0.4681 (3) | 0.2945 (8) | 0.2130 (3) | 0.0827 (17) | |
| H18 | 0.4383 | 0.3568 | 0.2411 | 0.099* | |
| C19 | 0.4436 (3) | 0.2035 (9) | 0.1613 (3) | 0.0944 (19) | |
| H19 | 0.3968 | 0.1983 | 0.1536 | 0.113* | |
| C20 | 0.4872 (4) | 0.1191 (8) | 0.1199 (3) | 0.095 (2) | |
| H20 | 0.4697 | 0.0557 | 0.0835 | 0.114* | |
| C21 | 0.5545 (3) | 0.1206 (7) | 0.1311 (3) | 0.0805 (17) | |
| H21 | 0.5846 | 0.0615 | 0.1024 | 0.097* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.056 (3) | 0.067 (2) | 0.044 (2) | 0.000 (2) | −0.008 (2) | −0.0054 (19) |
| N2 | 0.065 (3) | 0.072 (3) | 0.064 (3) | −0.005 (2) | 0.003 (3) | 0.005 (2) |
| C1 | 0.069 (4) | 0.066 (3) | 0.062 (4) | 0.004 (3) | −0.003 (3) | 0.001 (3) |
| C2 | 0.077 (5) | 0.133 (5) | 0.068 (4) | 0.024 (4) | −0.010 (4) | −0.006 (4) |
| C3 | 0.120 (6) | 0.138 (6) | 0.065 (4) | 0.011 (5) | 0.008 (4) | −0.004 (4) |
| C4 | 0.076 (5) | 0.097 (4) | 0.094 (5) | 0.009 (3) | 0.005 (4) | 0.007 (4) |
| C5 | 0.074 (5) | 0.094 (4) | 0.102 (5) | 0.003 (3) | −0.003 (4) | 0.018 (4) |
| C6 | 0.085 (5) | 0.087 (4) | 0.074 (4) | −0.016 (3) | −0.011 (4) | 0.012 (3) |
| C7 | 0.094 (5) | 0.068 (3) | 0.068 (4) | −0.003 (3) | −0.002 (4) | 0.008 (3) |
| C8 | 0.074 (4) | 0.060 (3) | 0.068 (4) | 0.002 (3) | −0.005 (3) | 0.003 (3) |
| C9 | 0.082 (4) | 0.056 (3) | 0.070 (4) | −0.004 (3) | −0.002 (3) | 0.000 (3) |
| C10 | 0.066 (4) | 0.072 (3) | 0.068 (4) | −0.005 (3) | −0.006 (3) | 0.000 (3) |
| C11 | 0.070 (4) | 0.083 (4) | 0.077 (4) | −0.011 (3) | 0.000 (3) | 0.008 (3) |
| C12 | 0.069 (4) | 0.122 (5) | 0.071 (4) | 0.003 (4) | −0.003 (4) | 0.021 (4) |
| C13 | 0.081 (5) | 0.146 (6) | 0.067 (4) | 0.015 (4) | −0.006 (4) | −0.010 (4) |
| C14 | 0.102 (6) | 0.108 (5) | 0.084 (5) | −0.004 (4) | −0.012 (4) | −0.021 (4) |
| C15 | 0.086 (4) | 0.090 (4) | 0.077 (4) | −0.018 (3) | −0.001 (3) | 0.000 (3) |
| C16 | 0.081 (4) | 0.059 (3) | 0.062 (3) | −0.008 (3) | −0.004 (3) | 0.002 (3) |
| C17 | 0.086 (5) | 0.067 (3) | 0.068 (4) | 0.006 (3) | −0.010 (3) | 0.001 (3) |
| C18 | 0.075 (5) | 0.083 (4) | 0.090 (5) | 0.003 (3) | −0.006 (4) | 0.012 (4) |
| C19 | 0.075 (5) | 0.104 (5) | 0.105 (5) | −0.005 (4) | −0.012 (4) | 0.020 (4) |
| C20 | 0.106 (6) | 0.090 (4) | 0.090 (5) | −0.016 (4) | −0.027 (5) | −0.004 (4) |
| C21 | 0.086 (5) | 0.083 (4) | 0.072 (4) | −0.009 (3) | −0.010 (4) | 0.000 (3) |
Geometric parameters (Å, °)
| N1—C7 | 1.310 (7) | C10—C15 | 1.410 (7) |
| N1—C8 | 1.374 (6) | C11—C12 | 1.377 (7) |
| N1—H1 | 0.8600 | C11—H11 | 0.9600 |
| N2—C7 | 1.302 (6) | C12—C13 | 1.342 (8) |
| N2—C9 | 1.391 (6) | C12—H12 | 0.9600 |
| C1—C2 | 1.353 (7) | C13—C14 | 1.419 (9) |
| C1—C6 | 1.392 (7) | C13—H13 | 0.9600 |
| C1—C7 | 1.462 (7) | C14—C15 | 1.363 (7) |
| C2—C3 | 1.406 (8) | C14—H14 | 0.9600 |
| C2—H2 | 0.9602 | C15—H15 | 0.9599 |
| C3—C4 | 1.378 (8) | C16—C17 | 1.398 (7) |
| C3—H3 | 0.9600 | C16—C21 | 1.390 (7) |
| C4—C5 | 1.348 (7) | C17—C18 | 1.377 (7) |
| C4—H4 | 0.9600 | C17—H17 | 0.9602 |
| C5—C6 | 1.378 (7) | C18—C19 | 1.364 (8) |
| C5—H5 | 0.9600 | C18—H18 | 0.9599 |
| C6—H6 | 0.9602 | C19—C20 | 1.384 (9) |
| C8—C9 | 1.353 (7) | C19—H19 | 0.9600 |
| C8—C16 | 1.477 (7) | C20—C21 | 1.380 (8) |
| C9—C10 | 1.474 (7) | C20—H20 | 0.9601 |
| C10—C11 | 1.377 (7) | C21—H21 | 0.9599 |
| C7—N1—C8 | 107.0 (4) | C12—C11—C10 | 119.9 (6) |
| C7—N1—H1 | 126.5 | C12—C11—H11 | 121.6 |
| C8—N1—H1 | 126.5 | C10—C11—H11 | 118.5 |
| C7—N2—C9 | 106.0 (5) | C13—C12—C11 | 122.4 (6) |
| C2—C1—C6 | 119.3 (6) | C13—C12—H12 | 117.0 |
| C2—C1—C7 | 120.9 (6) | C11—C12—H12 | 120.6 |
| C6—C1—C7 | 119.8 (5) | C12—C13—C14 | 118.9 (6) |
| C1—C2—C3 | 119.5 (6) | C12—C13—H13 | 120.6 |
| C1—C2—H2 | 118.3 | C14—C13—H13 | 120.6 |
| C3—C2—H2 | 122.2 | C15—C14—C13 | 119.4 (6) |
| C4—C3—C2 | 121.5 (6) | C15—C14—H14 | 118.9 |
| C4—C3—H3 | 119.7 | C13—C14—H14 | 121.6 |
| C2—C3—H3 | 118.8 | C14—C15—C10 | 120.9 (6) |
| C5—C4—C3 | 117.5 (6) | C14—C15—H15 | 120.6 |
| C5—C4—H4 | 120.5 | C10—C15—H15 | 118.5 |
| C3—C4—H4 | 122.0 | C17—C16—C21 | 118.2 (5) |
| C4—C5—C6 | 122.5 (6) | C17—C16—C8 | 121.7 (5) |
| C4—C5—H5 | 117.4 | C21—C16—C8 | 120.1 (5) |
| C6—C5—H5 | 120.0 | C18—C17—C16 | 121.0 (5) |
| C5—C6—C1 | 119.6 (6) | C18—C17—H17 | 120.3 |
| C5—C6—H6 | 120.0 | C16—C17—H17 | 118.7 |
| C1—C6—H6 | 120.4 | C19—C18—C17 | 120.5 (6) |
| N1—C7—N2 | 112.5 (6) | C19—C18—H18 | 119.5 |
| N1—C7—C1 | 122.4 (5) | C17—C18—H18 | 120.0 |
| N2—C7—C1 | 125.0 (6) | C18—C19—C20 | 119.1 (6) |
| C9—C8—N1 | 107.0 (5) | C18—C19—H19 | 120.6 |
| C9—C8—C16 | 134.8 (6) | C20—C19—H19 | 120.4 |
| N1—C8—C16 | 118.1 (5) | C21—C20—C19 | 121.4 (6) |
| C8—C9—N2 | 107.5 (5) | C21—C20—H20 | 119.9 |
| C8—C9—C10 | 133.5 (6) | C19—C20—H20 | 118.7 |
| N2—C9—C10 | 119.0 (5) | C20—C21—C16 | 119.8 (6) |
| C11—C10—C15 | 118.5 (5) | C20—C21—H21 | 121.3 |
| C11—C10—C9 | 123.1 (5) | C16—C21—H21 | 118.9 |
| C15—C10—C9 | 118.4 (5) | ||
| C6—C1—C2—C3 | −1.4 (9) | C8—C9—C10—C11 | 38.5 (8) |
| C7—C1—C2—C3 | 178.7 (6) | N2—C9—C10—C11 | −139.1 (5) |
| C1—C2—C3—C4 | 2.7 (10) | C8—C9—C10—C15 | −144.2 (6) |
| C2—C3—C4—C5 | −2.6 (10) | N2—C9—C10—C15 | 38.2 (7) |
| C3—C4—C5—C6 | 1.3 (9) | C15—C10—C11—C12 | 1.4 (9) |
| C4—C5—C6—C1 | −0.1 (9) | C9—C10—C11—C12 | 178.7 (5) |
| C2—C1—C6—C5 | 0.1 (8) | C10—C11—C12—C13 | −0.5 (10) |
| C7—C1—C6—C5 | 180.0 (5) | C11—C12—C13—C14 | 0.0 (10) |
| C8—N1—C7—N2 | −1.1 (6) | C12—C13—C14—C15 | −0.3 (10) |
| C8—N1—C7—C1 | 179.4 (5) | C13—C14—C15—C10 | 1.2 (10) |
| C9—N2—C7—N1 | 1.0 (6) | C11—C10—C15—C14 | −1.8 (9) |
| C9—N2—C7—C1 | −179.5 (5) | C9—C10—C15—C14 | −179.2 (6) |
| C2—C1—C7—N1 | 20.7 (8) | C9—C8—C16—C17 | 24.5 (9) |
| C6—C1—C7—N1 | −159.2 (5) | N1—C8—C16—C17 | −153.3 (5) |
| C2—C1—C7—N2 | −158.7 (6) | C9—C8—C16—C21 | −157.2 (5) |
| C6—C1—C7—N2 | 21.4 (8) | N1—C8—C16—C21 | 25.0 (7) |
| C7—N1—C8—C9 | 0.7 (5) | C21—C16—C17—C18 | 0.5 (8) |
| C7—N1—C8—C16 | 179.1 (4) | C8—C16—C17—C18 | 178.8 (4) |
| N1—C8—C9—N2 | −0.1 (5) | C16—C17—C18—C19 | −0.3 (8) |
| C16—C8—C9—N2 | −178.1 (5) | C17—C18—C19—C20 | −0.9 (9) |
| N1—C8—C9—C10 | −177.9 (5) | C18—C19—C20—C21 | 2.0 (10) |
| C16—C8—C9—C10 | 4.1 (10) | C19—C20—C21—C16 | −1.9 (9) |
| C7—N2—C9—C8 | −0.5 (5) | C17—C16—C21—C20 | 0.6 (8) |
| C7—N2—C9—C10 | 177.6 (4) | C8—C16—C21—C20 | −177.7 (5) |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HB2915).
References
- Allen, F. H. (2002). Acta Cryst. B58, 380–388. [DOI] [PubMed]
- Farrugia, L. J. (1999). J. Appl. Cryst.32, 837–838.
- Fridman, N., Kaftory, M., Eichen, Y. & Speiser, S. (2007). J. Photochem. Photobiol. A, 188, 25-33.
- Fridman, N., Kaftory, M., Eichen, Y. & Speiser, S. (2008). J. Mol. Struct.917, 101-109.
- Fridman, N., Kaftory, M. & Speiser, S. (2007). Sens. Actuators, B126, 107-115.
- Inouye, Y. & Sakaino, Y. (2000). Acta Cryst. C56, 884–887. [DOI] [PubMed]
- Kaftory, M., Taycher, H. & Botoshansky, M. (1998). J. Chem. Soc. Perkin Trans. 2, pp. 407-412.
- Kamidate, T., Yamaguchi, K., Segawa, T. & Watanabe, H. (1989). Anal. Sci 5, 429-33.
- Liu, X.-F., Zhong, Z.-P. & Xu, Z.-L. (2005). Acta Cryst. E61, o1976–o1977.
- Nakashima, K. (2003). Biomed. Chromatogr 17, 83-95. [DOI] [PubMed]
- Nakashima, K., Yamasaki, H., Kuroda, N. & Akiyama, S. (1995). Anal. Chim. Acta, 303, 103-107.
- Nonius (2000). COLLECT Nonius BV, Delft, The Netherlands.
- Otwinowski, Z. & Minor, W. (1997). Methods in Enzymology, Vol. 276, Macromolecular Crystallography, Part A, edited by C. W. Carter Jr & R. M. Sweet, pp. 307-326. New York: Academic Press.
- Radziszewski, B. (1877). Chem. Ber 10, 70-75.
- Santos, J., Mintz, E. A., Zehnder, O., Bosshard, C., Bu, X. R. & Gunter, P. (2001). Tetrahedron Lett 42, 805-808.
- Seethalakshmi, T., Puratchikody, A., Lynch, D. E., Kaliannan, P. & Thamotharan, S. (2006). Acta Cryst. E62, o2803–o2804.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Thiruvalluvar, A., Balamurugan, S., Puratchikody, A. & Nallu, M. (2007). Acta Cryst. E63, o1650–o1652.
- Thuer, W., Gompper, R. & Polborn, K. (2004). Private communication (deposition CCDC 259545). CCDC, Cambridge, England.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809006552/hb2915sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809006552/hb2915Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




