Abstract
The nicotinamide (NA) molecules of the title compound, 2C6H6N2O·C2H3F3O, form centrosymmetric R 2 2(8) hydrogen-bonded dimers via N—H⋯O contacts. The asymmetric unit contains two molecules of NA and one trifluoroethanol molecule disordered over two sites of equal occupancy. The packing consists of alternating layers of nicotinamide dimers and disordered 2,2,2-trifluoroethanol molecules stacking in the c-axis direction. Intramolecular C—H⋯O and intermolecular N—H⋯N, O—H⋯N, C—H⋯N, C—H⋯O and C—H⋯F interactions are present.
Related literature
For nicotinamide polymorphs, see: Wright & King (1954 ▶); Miwa et al. (1999 ▶); Hino et al. (2001 ▶). For nicotinamide co-crystals and salts, see: Fleischman et al. (2003 ▶); Koman et al. (2003 ▶); Athimoolam & Natarajan (2007a
▶,b
▶); Berry et al. (2008 ▶). For graph-set motifs, see: Etter (1990 ▶). For initial identification using multi-sample foil transmission X-ray powder diffraction analysis, see: Florence et al. (2003 ▶).
Experimental
Crystal data
2C6H6N2O·C2H3F3O
M r = 344.30
Triclinic,
a = 5.0472 (3) Å
b = 11.2930 (7) Å
c = 15.0877 (10) Å
α = 107.002 (3)°
β = 96.636 (3)°
γ = 95.753 (3)°
V = 808.70 (9) Å3
Z = 2
Mo Kα radiation
μ = 0.12 mm−1
T = 123 K
0.15 × 0.10 × 0.02 mm
Data collection
Bruker APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 2002 ▶) T min = 0.903, T max = 0.998
15936 measured reflections
4008 independent reflections
3416 reflections with I > 2σ(I)
R int = 0.025
Refinement
R[F 2 > 2σ(F 2)] = 0.039
wR(F 2) = 0.114
S = 1.04
4008 reflections
260 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.39 e Å−3
Δρmin = −0.29 e Å−3
Data collection: APEX2 (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: PLATON (Spek, 2009 ▶) and Mercury (Macrae et al., 2006 ▶); software used to prepare material for publication: PLATON.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809007594/fl2234sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809007594/fl2234Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N2—H1N⋯N3i | 0.897 (16) | 2.122 (16) | 2.9994 (15) | 165.7 (15) |
| N2—H2N⋯O1ii | 0.898 (16) | 2.013 (16) | 2.9093 (13) | 176.8 (14) |
| N4—H3N⋯O2iii | 0.877 (16) | 2.047 (16) | 2.9139 (13) | 170.1 (16) |
| O3—H3O⋯N1iv | 0.84 | 1.91 | 2.7511 (15) | 178 |
| N4—H4N⋯O1v | 0.878 (17) | 2.222 (17) | 3.0867 (14) | 168.4 (15) |
| C1—H1⋯N3i | 0.95 | 2.51 | 3.4220 (16) | 161 |
| C3—H3⋯O2vi | 0.95 | 2.40 | 3.0103 (15) | 122 |
| C5—H5⋯F3Avii | 0.95 | 2.46 | 3.408 (7) | 177 |
| C7—H7⋯O1v | 0.95 | 2.36 | 3.2118 (14) | 149 |
| C11—H11⋯O3 | 0.95 | 2.50 | 3.2590 (16) | 137 |
Symmetry codes: (i) 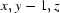 ; (ii)
; (ii) 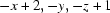 ; (iii)
; (iii) 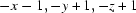 ; (iv)
; (iv) 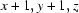 ; (v)
; (v) 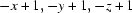 ; (vi)
; (vi) 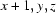 ; (vii)
; (vii) 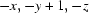 .
.
Acknowledgments
The authors thank the University of Strathclyde for funding JB, the Basic Technology programme of the UK Research Councils for project funding under the Control and Prediction of the Organic Solid State (www.cposs.org.uk), and the Glasgow Centre for Physical Organic Chemistry for access to single-crystal diffraction facilities.
supplementary crystallographic information
Comment
The crystal structure of nicotinamide (NA) was first reported in 1954 (Wright & King, 1954; Miwa et al., 1999) and a number of polymorphic forms have also been identified (Hino et al., 2001). In recent years NA has also been investigated as a pharmaceutically acceptable co-crystal former to modify the crystal structure and physico-chemical properties of drug compounds including carbamazepine (Fleischman et al., 2003), salicylic acid and both racemic and S(+)-ibuprofen (Berry et al., 2008). Crystal structures of 3,5-dinitrosalicylate (Koman et al., 2003) as well as 2R,3R-tartrate and trifluoroacetate (Athimoolam & Natarajan, 2007a and Athimoolam & Natarajan, 2007b) salts of NA have also been reported.
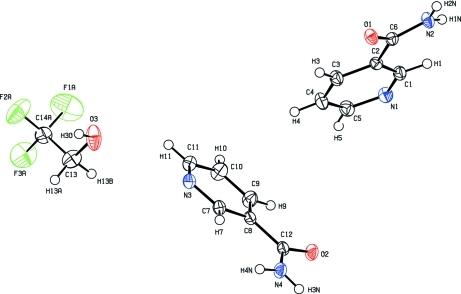
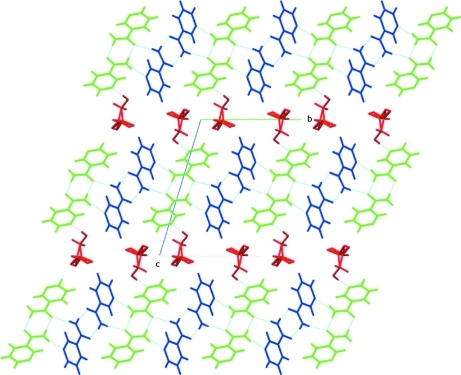
In the 2,2,2-trifluoroethanol (TFE) hemisolvate reported here, the molecules crystallize in space group P1 with two molecules of NA and one molecule of TFE in the asymetric unit (Fig. 1). Both independent NA molecules form centro-symmetric R22(8) (Etter, 1990) dimer motifs via N—H..O hydrogen bonds (Table 1). The anti-oriented hydrogen atoms on both amide groups form further contacts between adjacent non-symmetry equivalent dimers, either to the aromatic nitrogen, N3 (N2—H1N···N3) or the carbonyl oxygen atom, O1 (N4—H4N···O1).This gives rise to a two-dimensional hydrogen bonded layer of nicotinamide dimers lying parallel to the a-b plane. The remaining aromatic acceptor nitrogen atom, N1, forms an N—H···O hydrogen bond to the solvent molecule producing a structure with alternating layers of NA dimers and TFE that stack in the direction of the c-axis (Fig. 2). The structure is further stabilized by six weak C—H···O and C—H···F interactions (Table 1). The CF3 atoms of the TFE molecule are disordered over two sites with equal occupancies.
Experimental
The novel structure reported here was discovered during a study of solvate formation in organic compounds with a range of fluorinated solvents. A crystalline sample was obtained by isothermal evaporation at 263 K from a saturated TFE solution held on a Reactarray RM2 crystalliser. Identification of the novel phase was initially made using multi-sample foil transmission X-ray powder diffraction analysis (Florence et al., 2003) and a suitable single-crystal for structure determination was selected from the sample.
Refinement
The amine H atoms were located in a difference synthesis and were refined isotropically [N—H = 0.877 (16)–0.898 (16) Å]. All other H atoms were positioned geometrically at distances of 0.95, 0.99 and 0.84 Å from the parent atoms for CH, CH2 and OH groups respectively. For these atoms, a riding model was used during the refinement process. The Uiso(H) values were constrained to be 1.2 times Ueq of the carrier C atom or 1.5 times Ueq of the carrier O atom.
Figures
Fig. 1.
View of the title compound with the atom numbering scheme. Displacement ellipsoids for non-H atoms are drawn at the 50% probability level. One of the disordered site for the CF3 atoms has been omitted for clarity.
Fig. 2.
Crystal packing of the reported compound viewed down the a axis, showing the two-dimensional hydrogen-bonded network of alternating dimers, separated by layers of solvent molecules. Hydrogen bonds are shown as blue dashed lines.
Crystal data
| 2(C6H6N2O)·C2H3F3O | Z = 2 |
| Mr = 344.30 | F(000) = 356 |
| Triclinic, P1 | Dx = 1.414 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 5.0472 (3) Å | Cell parameters from 8696 reflections |
| b = 11.2930 (7) Å | θ = 2.9–28.2° |
| c = 15.0877 (10) Å | µ = 0.12 mm−1 |
| α = 107.002 (3)° | T = 123 K |
| β = 96.636 (3)° | Slab, colourless |
| γ = 95.753 (3)° | 0.15 × 0.10 × 0.02 mm |
| V = 808.70 (9) Å3 |
Data collection
| Bruker APEXII CCD diffractometer | 4008 independent reflections |
| Radiation source: fine-focus sealed tube | 3416 reflections with I > 2σ(I) |
| graphite | Rint = 0.025 |
| φ and ω scans | θmax = 28.3°, θmin = 1.4° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 2002) | h = −6→6 |
| Tmin = 0.903, Tmax = 0.998 | k = −15→14 |
| 15936 measured reflections | l = −20→19 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.039 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.114 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.04 | w = 1/[σ2(Fo2) + (0.0611P)2 + 0.2237P] where P = (Fo2 + 2Fc2)/3 |
| 4008 reflections | (Δ/σ)max < 0.001 |
| 260 parameters | Δρmax = 0.39 e Å−3 |
| 0 restraints | Δρmin = −0.29 e Å−3 |
Special details
| Geometry. Bond distances, angles etc. have been calculated using the rounded fractional coordinates. All su's are estimated from the variances of the (full) variance-covariance matrix. The cell e.s.d.'s are taken into account in the estimation of distances, angles and torsion angles |
| Refinement. Refinement on F2 for ALL reflections except those flagged by the user for potential systematic errors. Weighted R-factors wR and all goodnesses of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The observed criterion of F2 > σ(F2) is used only for calculating -R-factor-obs etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R-factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| O1 | 0.87250 (15) | 0.11455 (7) | 0.45065 (6) | 0.0226 (2) | |
| N1 | 0.05569 (19) | −0.02252 (9) | 0.23414 (7) | 0.0240 (3) | |
| N2 | 0.7054 (2) | −0.09004 (9) | 0.40991 (7) | 0.0225 (3) | |
| C1 | 0.2538 (2) | −0.04184 (10) | 0.29313 (8) | 0.0212 (3) | |
| C2 | 0.4782 (2) | 0.04629 (9) | 0.33748 (7) | 0.0186 (3) | |
| C3 | 0.4931 (2) | 0.16131 (10) | 0.32072 (8) | 0.0236 (3) | |
| C4 | 0.2882 (2) | 0.18307 (11) | 0.26047 (9) | 0.0274 (3) | |
| C5 | 0.0760 (2) | 0.08876 (11) | 0.21836 (8) | 0.0261 (3) | |
| C6 | 0.7000 (2) | 0.02482 (9) | 0.40354 (7) | 0.0188 (3) | |
| O2 | −0.39941 (17) | 0.44262 (7) | 0.39075 (6) | 0.0264 (3) | |
| N3 | 0.33815 (19) | 0.66946 (9) | 0.31270 (7) | 0.0236 (3) | |
| N4 | −0.2073 (2) | 0.62420 (9) | 0.49873 (7) | 0.0236 (3) | |
| C7 | 0.1753 (2) | 0.65082 (10) | 0.37294 (8) | 0.0219 (3) | |
| C8 | −0.0429 (2) | 0.55638 (9) | 0.34852 (8) | 0.0198 (3) | |
| C9 | −0.0899 (2) | 0.47587 (11) | 0.25734 (9) | 0.0271 (3) | |
| C10 | 0.0788 (3) | 0.49317 (12) | 0.19462 (9) | 0.0303 (3) | |
| C11 | 0.2891 (2) | 0.59119 (11) | 0.22515 (8) | 0.0258 (3) | |
| C12 | −0.2296 (2) | 0.53745 (10) | 0.41530 (8) | 0.0205 (3) | |
| F1A | 0.5655 (16) | 0.6926 (4) | −0.0538 (5) | 0.0869 (18) | 0.500 |
| F1B | 0.5243 (12) | 0.6494 (4) | −0.0430 (5) | 0.0706 (13) | 0.500 |
| F2A | 0.8008 (18) | 0.8692 (5) | −0.0041 (6) | 0.0644 (16) | 0.500 |
| F2B | 0.7971 (17) | 0.8192 (6) | −0.0144 (6) | 0.0647 (19) | 0.500 |
| F3A | 0.4000 (15) | 0.8549 (5) | −0.0620 (5) | 0.0653 (16) | 0.500 |
| F3B | 0.3721 (15) | 0.8033 (4) | −0.0738 (4) | 0.0614 (12) | 0.500 |
| O3 | 0.6456 (2) | 0.78427 (9) | 0.14649 (7) | 0.0424 (3) | |
| C13 | 0.4745 (3) | 0.82394 (14) | 0.08562 (9) | 0.0345 (4) | |
| C14A | 0.5622 (6) | 0.8123 (3) | −0.0096 (2) | 0.0320 (7)* | 0.500 |
| C14B | 0.5411 (6) | 0.7718 (4) | −0.0096 (2) | 0.0322 (7)* | 0.500 |
| H1 | 0.24020 | −0.12000 | 0.30530 | 0.0250* | |
| H1N | 0.592 (3) | −0.1567 (14) | 0.3723 (11) | 0.028 (4)* | |
| H2N | 0.839 (3) | −0.0991 (14) | 0.4511 (11) | 0.030 (4)* | |
| H3 | 0.64230 | 0.22440 | 0.35030 | 0.0280* | |
| H4 | 0.29370 | 0.26120 | 0.24840 | 0.0330* | |
| H5 | −0.06240 | 0.10330 | 0.17610 | 0.0310* | |
| H3N | −0.324 (3) | 0.6138 (15) | 0.5355 (11) | 0.036 (4)* | |
| H4N | −0.094 (3) | 0.6940 (15) | 0.5172 (11) | 0.032 (4)* | |
| H7 | 0.21110 | 0.70510 | 0.43570 | 0.0260* | |
| H9 | −0.23630 | 0.40960 | 0.23810 | 0.0320* | |
| H10 | 0.05080 | 0.43890 | 0.13200 | 0.0360* | |
| H11 | 0.40360 | 0.60330 | 0.18190 | 0.0310* | |
| H3O | 0.77220 | 0.84210 | 0.17440 | 0.0640* | |
| H13A | 0.45750 | 0.91270 | 0.11620 | 0.0410* | |
| H13B | 0.29360 | 0.77480 | 0.07530 | 0.0410* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0234 (4) | 0.0156 (4) | 0.0256 (4) | −0.0015 (3) | −0.0023 (3) | 0.0052 (3) |
| N1 | 0.0222 (5) | 0.0233 (5) | 0.0252 (5) | 0.0009 (4) | 0.0002 (4) | 0.0075 (4) |
| N2 | 0.0224 (5) | 0.0162 (4) | 0.0274 (5) | −0.0010 (4) | −0.0029 (4) | 0.0081 (4) |
| C1 | 0.0214 (5) | 0.0181 (5) | 0.0244 (5) | 0.0015 (4) | 0.0030 (4) | 0.0075 (4) |
| C2 | 0.0195 (5) | 0.0167 (5) | 0.0191 (5) | 0.0031 (4) | 0.0035 (4) | 0.0044 (4) |
| C3 | 0.0243 (5) | 0.0177 (5) | 0.0283 (6) | 0.0007 (4) | 0.0023 (4) | 0.0075 (4) |
| C4 | 0.0306 (6) | 0.0211 (5) | 0.0330 (6) | 0.0033 (4) | 0.0018 (5) | 0.0133 (5) |
| C5 | 0.0251 (5) | 0.0278 (6) | 0.0274 (6) | 0.0054 (4) | 0.0009 (4) | 0.0124 (5) |
| C6 | 0.0191 (5) | 0.0167 (5) | 0.0200 (5) | 0.0013 (4) | 0.0036 (4) | 0.0051 (4) |
| O2 | 0.0278 (4) | 0.0172 (4) | 0.0324 (5) | −0.0040 (3) | 0.0079 (3) | 0.0059 (3) |
| N3 | 0.0232 (5) | 0.0181 (4) | 0.0300 (5) | 0.0001 (4) | 0.0056 (4) | 0.0084 (4) |
| N4 | 0.0251 (5) | 0.0189 (4) | 0.0258 (5) | −0.0022 (4) | 0.0061 (4) | 0.0062 (4) |
| C7 | 0.0231 (5) | 0.0164 (5) | 0.0254 (5) | 0.0003 (4) | 0.0038 (4) | 0.0060 (4) |
| C8 | 0.0204 (5) | 0.0150 (5) | 0.0252 (5) | 0.0027 (4) | 0.0035 (4) | 0.0079 (4) |
| C9 | 0.0253 (6) | 0.0224 (5) | 0.0290 (6) | −0.0047 (4) | 0.0035 (4) | 0.0039 (4) |
| C10 | 0.0329 (6) | 0.0281 (6) | 0.0244 (6) | −0.0029 (5) | 0.0060 (5) | 0.0014 (5) |
| C11 | 0.0261 (6) | 0.0246 (5) | 0.0287 (6) | 0.0022 (4) | 0.0079 (4) | 0.0103 (5) |
| C12 | 0.0202 (5) | 0.0157 (5) | 0.0268 (5) | 0.0016 (4) | 0.0029 (4) | 0.0089 (4) |
| F1A | 0.112 (4) | 0.063 (3) | 0.058 (2) | 0.019 (3) | 0.003 (2) | −0.022 (3) |
| F1B | 0.0614 (18) | 0.055 (3) | 0.064 (2) | 0.0127 (19) | −0.0039 (15) | −0.026 (2) |
| F2A | 0.0451 (16) | 0.110 (4) | 0.048 (2) | 0.003 (3) | 0.0128 (15) | 0.040 (3) |
| F2B | 0.0340 (15) | 0.116 (5) | 0.052 (2) | 0.012 (3) | 0.0162 (13) | 0.034 (3) |
| F3A | 0.054 (2) | 0.107 (4) | 0.042 (2) | 0.018 (3) | −0.0097 (16) | 0.038 (3) |
| F3B | 0.0508 (17) | 0.097 (3) | 0.0316 (14) | 0.008 (3) | −0.0059 (11) | 0.018 (2) |
| O3 | 0.0431 (6) | 0.0369 (5) | 0.0434 (6) | −0.0155 (4) | −0.0138 (4) | 0.0212 (4) |
| C13 | 0.0278 (6) | 0.0432 (7) | 0.0291 (6) | 0.0010 (5) | 0.0013 (5) | 0.0083 (5) |
Geometric parameters (Å, °)
| F1A—C14A | 1.324 (7) | C2—C3 | 1.3906 (16) |
| F1B—C14B | 1.316 (7) | C2—C6 | 1.4978 (14) |
| F2A—C14A | 1.289 (9) | C3—C4 | 1.3858 (16) |
| F2B—C14B | 1.366 (9) | C4—C5 | 1.3796 (17) |
| F3A—C14A | 1.296 (8) | C1—H1 | 0.9500 |
| F3B—C14B | 1.365 (7) | C3—H3 | 0.9500 |
| O1—C6 | 1.2446 (13) | C4—H4 | 0.9500 |
| O2—C12 | 1.2366 (14) | C5—H5 | 0.9500 |
| O3—C13 | 1.3874 (18) | C7—C8 | 1.3882 (15) |
| O3—H3O | 0.8400 | C8—C12 | 1.5013 (15) |
| N1—C5 | 1.3421 (17) | C8—C9 | 1.3876 (17) |
| N1—C1 | 1.3383 (15) | C9—C10 | 1.3855 (18) |
| N2—C6 | 1.3311 (15) | C10—C11 | 1.3847 (19) |
| N2—H2N | 0.898 (16) | C7—H7 | 0.9500 |
| N2—H1N | 0.897 (16) | C9—H9 | 0.9500 |
| N3—C7 | 1.3401 (15) | C10—H10 | 0.9500 |
| N3—C11 | 1.3361 (15) | C11—H11 | 0.9500 |
| N4—C12 | 1.3345 (15) | C13—C14B | 1.477 (3) |
| N4—H3N | 0.877 (16) | C13—C14A | 1.525 (3) |
| N4—H4N | 0.878 (17) | C13—H13A | 0.9900 |
| C1—C2 | 1.3894 (15) | C13—H13B | 0.9900 |
| F1A···O3 | 2.859 (7) | C4···O2ii | 3.1510 (15) |
| F1A···C10i | 3.342 (7) | C5···C3vi | 3.5363 (15) |
| F1A···F2A | 2.096 (9) | C6···N1ii | 3.2396 (14) |
| F1A···F3A | 2.119 (9) | C6···C1ii | 3.4524 (15) |
| F1B···F3B | 2.118 (8) | C7···O2ii | 3.3847 (14) |
| F1B···O3 | 2.780 (7) | C7···C12ii | 3.4460 (15) |
| F2A···F3A | 2.080 (12) | C7···O1v | 3.2118 (14) |
| F2A···O3 | 2.869 (8) | C9···C11vi | 3.5486 (15) |
| F2B···O3 | 2.750 (8) | C10···F1Ai | 3.342 (7) |
| F2B···F3B | 2.196 (11) | C11···C9ii | 3.5486 (15) |
| F3A···F1A | 2.119 (9) | C11···O3 | 3.2590 (16) |
| F3A···F2A | 2.080 (12) | C12···C7vi | 3.4460 (15) |
| F3B···F2B | 2.196 (11) | C12···C12xii | 3.5927 (16) |
| F3B···F1B | 2.118 (8) | C12···N3vi | 3.2557 (15) |
| F1A···H10i | 2.7300 | C12···N4xii | 3.3776 (15) |
| F1B···H10i | 2.7800 | C13···C1xi | 3.4220 (18) |
| F2A···H3O | 2.8200 | C13···N1viii | 3.4382 (18) |
| F2B···H13Bii | 2.8700 | C1···H13Ax | 2.9000 |
| F2B···H3O | 2.8000 | C1···H1N | 2.627 (16) |
| F3A···H5iii | 2.4600 | C1···H3Oix | 2.8000 |
| F3B···H5iii | 2.5800 | C3···H3Nxii | 3.086 (16) |
| F3B···H9iii | 2.8600 | C5···H3Oix | 2.8900 |
| O1···N2iv | 2.9093 (13) | C6···H2Niv | 2.878 (16) |
| O1···N4v | 3.0867 (14) | C7···H4N | 2.649 (16) |
| O1···C7v | 3.2118 (14) | C7···H1xi | 3.0500 |
| O2···C7vi | 3.3847 (14) | C7···H1Nxi | 2.872 (16) |
| O2···C3vi | 3.0103 (15) | C12···H3Nvii | 2.984 (16) |
| O2···N4vii | 2.9139 (13) | H1···N3x | 2.5100 |
| O2···C4vi | 3.1510 (15) | H1···C7x | 3.0500 |
| O3···N1viii | 2.7511 (15) | H1···N2 | 2.6100 |
| O3···F1A | 2.859 (7) | H1···H1N | 2.0800 |
| O3···F2A | 2.869 (8) | H1N···N3x | 2.122 (16) |
| O3···C11 | 3.2590 (16) | H1N···C1 | 2.627 (16) |
| O3···F2B | 2.750 (8) | H1N···H1 | 2.0800 |
| O3···F1B | 2.780 (7) | H1N···C7x | 2.872 (16) |
| O1···H7v | 2.3600 | H2N···H2Niv | 2.57 (2) |
| O1···H2Niv | 2.013 (16) | H2N···O1iv | 2.013 (16) |
| O1···H4Nv | 2.222 (17) | H2N···C6iv | 2.878 (16) |
| O1···H3 | 2.4800 | H3···O1 | 2.4800 |
| O2···H3Nvii | 2.047 (16) | H3···O2ii | 2.4000 |
| O2···H4vi | 2.6900 | H3N···C12vii | 2.984 (16) |
| O2···H3vi | 2.4000 | H3N···C3xii | 3.086 (16) |
| O2···H9 | 2.4700 | H3N···O2vii | 2.047 (16) |
| O3···H11 | 2.5000 | H3O···C1viii | 2.8000 |
| N1···C6vi | 3.2396 (14) | H3O···C5viii | 2.8900 |
| N1···C13ix | 3.4382 (18) | H3O···N1viii | 1.9100 |
| N1···O3ix | 2.7510 (15) | H3O···F2A | 2.8200 |
| N2···O1iv | 2.9093 (13) | H3O···F2B | 2.8000 |
| N2···N3x | 2.9994 (15) | H4···O2ii | 2.6900 |
| N3···C1xi | 3.4220 (16) | H4N···O1v | 2.222 (17) |
| N3···C12ii | 3.2557 (15) | H4N···C7 | 2.649 (16) |
| N3···N2xi | 2.9994 (15) | H4N···H7 | 2.0900 |
| N4···C12xii | 3.3776 (15) | H5···F3Biii | 2.5800 |
| N4···O1v | 3.0867 (14) | H5···F3Aiii | 2.4600 |
| N4···O2vii | 2.9139 (13) | H7···N4 | 2.6000 |
| N1···H13Ax | 2.8600 | H7···H4N | 2.0900 |
| N1···H3Oix | 1.9100 | H7···O1v | 2.3600 |
| N2···H1 | 2.6100 | H9···O2 | 2.4700 |
| N3···H1Nxi | 2.122 (16) | H9···F3Biii | 2.8600 |
| N3···H1xi | 2.5100 | H10···F1Ai | 2.7300 |
| N4···H7 | 2.6000 | H10···F1Bi | 2.7800 |
| C1···N3x | 3.4220 (16) | H11···O3 | 2.5000 |
| C1···C13x | 3.4220 (18) | H13A···N1xi | 2.8600 |
| C1···C6vi | 3.4524 (15) | H13A···C1xi | 2.9000 |
| C3···O2ii | 3.0103 (15) | H13B···F2Bvi | 2.8700 |
| C3···C5ii | 3.5363 (15) | ||
| C13—O3—H3O | 109.00 | C9—C10—C11 | 118.64 (12) |
| C1—N1—C5 | 117.62 (10) | N3—C11—C10 | 122.95 (11) |
| C6—N2—H2N | 116.2 (11) | N4—C12—C8 | 118.69 (10) |
| C6—N2—H1N | 122.9 (10) | O2—C12—N4 | 122.47 (10) |
| H1N—N2—H2N | 120.7 (15) | O2—C12—C8 | 118.84 (10) |
| C7—N3—C11 | 117.80 (10) | C8—C7—H7 | 118.00 |
| H3N—N4—H4N | 117.7 (15) | N3—C7—H7 | 118.00 |
| C12—N4—H4N | 124.6 (10) | C8—C9—H9 | 120.00 |
| C12—N4—H3N | 117.4 (11) | C10—C9—H9 | 120.00 |
| N1—C1—C2 | 123.48 (11) | C9—C10—H10 | 121.00 |
| C1—C2—C6 | 123.54 (10) | C11—C10—H10 | 121.00 |
| C3—C2—C6 | 118.67 (9) | C10—C11—H11 | 119.00 |
| C1—C2—C3 | 117.78 (10) | N3—C11—H11 | 119.00 |
| C2—C3—C4 | 119.37 (10) | O3—C13—C14B | 107.67 (18) |
| C3—C4—C5 | 118.54 (12) | O3—C13—C14A | 115.28 (16) |
| N1—C5—C4 | 123.19 (11) | F1A—C14A—F3A | 108.0 (5) |
| O1—C6—C2 | 119.14 (9) | F1A—C14A—F2A | 106.7 (5) |
| N2—C6—C2 | 118.66 (9) | F1A—C14A—C13 | 108.7 (4) |
| O1—C6—N2 | 122.20 (10) | F2A—C14A—F3A | 107.2 (5) |
| C2—C1—H1 | 118.00 | F2A—C14A—C13 | 113.6 (4) |
| N1—C1—H1 | 118.00 | F3A—C14A—C13 | 112.5 (4) |
| C2—C3—H3 | 120.00 | F1B—C14B—F2B | 106.3 (5) |
| C4—C3—H3 | 120.00 | F1B—C14B—F3B | 104.3 (4) |
| C3—C4—H4 | 121.00 | F1B—C14B—C13 | 117.2 (4) |
| C5—C4—H4 | 121.00 | F2B—C14B—F3B | 107.0 (5) |
| N1—C5—H5 | 118.00 | F2B—C14B—C13 | 110.6 (4) |
| C4—C5—H5 | 118.00 | F3B—C14B—C13 | 110.7 (4) |
| N3—C7—C8 | 123.44 (11) | O3—C13—H13A | 108.00 |
| C9—C8—C12 | 118.67 (10) | O3—C13—H13B | 108.00 |
| C7—C8—C9 | 117.85 (10) | C14A—C13—H13A | 108.00 |
| C7—C8—C12 | 123.48 (10) | C14B—C13—H13B | 98.00 |
| C8—C9—C10 | 119.30 (11) | ||
| C5—N1—C1—C2 | −0.71 (16) | N3—C7—C8—C9 | 1.42 (17) |
| C1—N1—C5—C4 | −0.62 (17) | C9—C8—C12—N4 | −171.07 (11) |
| C11—N3—C7—C8 | −1.22 (17) | C7—C8—C12—N4 | 8.74 (16) |
| C7—N3—C11—C10 | 0.22 (18) | C9—C8—C12—O2 | 8.43 (16) |
| N1—C1—C2—C6 | 180.00 (13) | C7—C8—C12—O2 | −171.77 (11) |
| N1—C1—C2—C3 | 1.37 (16) | C7—C8—C9—C10 | −0.60 (17) |
| C1—C2—C6—O1 | −169.45 (10) | C12—C8—C9—C10 | 179.21 (11) |
| C3—C2—C6—N2 | −170.96 (10) | C8—C9—C10—C11 | −0.30 (19) |
| C3—C2—C6—O1 | 9.14 (15) | C9—C10—C11—N3 | 0.5 (2) |
| C1—C2—C6—N2 | 10.45 (15) | O3—C13—C14B—F1B | 58.1 (4) |
| C6—C2—C3—C4 | −179.39 (10) | O3—C13—C14B—F2B | −64.0 (5) |
| C1—C2—C3—C4 | −0.72 (16) | O3—C13—C14B—F3B | 177.6 (3) |
| C2—C3—C4—C5 | −0.49 (17) | O3—C13—C14A—F1A | 62.9 (4) |
| C3—C4—C5—N1 | 1.21 (18) | O3—C13—C14A—F2A | −55.7 (4) |
| N3—C7—C8—C12 | −178.39 (11) | O3—C13—C14A—F3A | −177.7 (4) |
Symmetry codes: (i) −x+1, −y+1, −z; (ii) x+1, y, z; (iii) −x, −y+1, −z; (iv) −x+2, −y, −z+1; (v) −x+1, −y+1, −z+1; (vi) x−1, y, z; (vii) −x−1, −y+1, −z+1; (viii) x+1, y+1, z; (ix) x−1, y−1, z; (x) x, y−1, z; (xi) x, y+1, z; (xii) −x, −y+1, −z+1.
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H1N···N3x | 0.897 (16) | 2.122 (16) | 2.9994 (15) | 165.7 (15) |
| N2—H2N···O1iv | 0.898 (16) | 2.013 (16) | 2.9093 (13) | 176.8 (14) |
| N4—H3N···O2vii | 0.877 (16) | 2.047 (16) | 2.9139 (13) | 170.1 (16) |
| O3—H3O···N1viii | 0.84 | 1.91 | 2.7511 (15) | 178 |
| N4—H4N···O1v | 0.878 (17) | 2.222 (17) | 3.0867 (14) | 168.4 (15) |
| C1—H1···N3x | 0.95 | 2.51 | 3.4220 (16) | 161 |
| C3—H3···O2ii | 0.95 | 2.40 | 3.0103 (15) | 122 |
| C5—H5···F3Aiii | 0.95 | 2.46 | 3.408 (7) | 177 |
| C7—H7···O1v | 0.95 | 2.36 | 3.2118 (14) | 149 |
| C11—H11···O3 | 0.95 | 2.50 | 3.2590 (16) | 137 |
Symmetry codes: (x) x, y−1, z; (iv) −x+2, −y, −z+1; (vii) −x−1, −y+1, −z+1; (viii) x+1, y+1, z; (v) −x+1, −y+1, −z+1; (ii) x+1, y, z; (iii) −x, −y+1, −z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: FL2234).
References
- Athimoolam, S. & Natarajan, S. (2007a). Acta Cryst. E63, o1811–o1813.
- Athimoolam, S. & Natarajan, S. (2007b). Acta Cryst. E63, o2430–o2432.
- Berry, D. J., Seaton, C. C., Clegg, W., Harrington, R. W., Coles, S. J., Horton, P. N., Hurthouse, M. B., Storey, R., Jones, W., Friščić, T. & Blagden, N. (2008). Cryst. Growth Des.8, 1697–1712.
- Bruker (2007). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Etter, M. C. (1990). Acc. Chem. Res 23, 120–126.
- Fleischman, S. G., Kuduva, S. S., McMahon, J. A., Moulton, B., Bailey Walsh, R. D., Rodríguez-Hornedo, N. & Zaworotko, M. J. (2003). Cryst. Growth Des 3, 909–919.
- Florence, A. J., Baumgartner, B., Weston, C., Shankland, N., Kennedy, A. R., Shankland, K. & David, W. I. F. (2003). J. Pharm. Sci.92, 1930–1938. [DOI] [PubMed]
- Hino, T., Ford, J. L. & Powell, M. W. (2001). Thermochimica Acta, 374, 85–92.
- Koman, M., Martiška, L., Valigura, D. & Glowiak, T. (2003). Acta Cryst. E59, o441–o442.
- Macrae, C. F., Edgington, P. R., McCabe, P., Pidcock, E., Shields, G. P., Taylor, R., Towler, M. & van de Streek, J. (2006). J. Appl. Cryst.39, 453–457.
- Miwa, Y., Mizuno, T., Tsuchida, K., Taga, T. & Iwata, Y. (1999). Acta Cryst. B55, 78–84. [DOI] [PubMed]
- Sheldrick, G. M. (2002). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Wright, W. B. & King, G. S. D. (1954). Acta Cryst.7, 283–288.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809007594/fl2234sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809007594/fl2234Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report