Abstract
There are two molecules in the asymmetric unit of the title compound, C10H9N3, with inter-ring dihedral angles of 31.1 (1) and 35.3 (1)°. The bridging C—N—C bond angles are 128.2 (1) and 129.1 (1)°. In the crystal, the two independent molecules are linked into a dimer by two N—H⋯N hydrogen bonds.
Related literature
For the structure of 4-chloro-N-(pyrimidin-2-yl)aniline, see: Maizathul Akmam et al. (2009 ▶).
Experimental
Crystal data
C10H9N3
M r = 171.20
Triclinic,
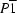
a = 8.8792 (2) Å
b = 9.9382 (2) Å
c = 10.2038 (2) Å
α = 93.186 (1)°
β = 103.665 (1)°
γ = 97.780 (1)°
V = 863.28 (3) Å3
Z = 4
Mo Kα radiation
μ = 0.08 mm−1
T = 123 K
0.35 × 0.20 × 0.10 mm
Data collection
Bruker SMART APEX diffractometer
Absorption correction: none
8238 measured reflections
3950 independent reflections
3144 reflections with I > 2σ(I)
R int = 0.020
Refinement
R[F 2 > 2σ(F 2)] = 0.039
wR(F 2) = 0.108
S = 1.03
3950 reflections
243 parameters
2 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.20 e Å−3
Δρmin = −0.23 e Å−3
Data collection: APEX2 (Bruker, 2008 ▶); cell refinement: SAINT (Bruker, 2008 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: X-SEED (Barbour, 2001 ▶); software used to prepare material for publication: publCIF (Westrip, 2009 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809007685/xu2492sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809007685/xu2492Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯N5 | 0.89 (1) | 2.10 (1) | 2.972 (1) | 164 (1) |
| N4—H4⋯N2 | 0.89 (1) | 2.15 (1) | 3.020 (1) | 165 (1) |
Acknowledgments
The authors thank the University of Malaya for supporting this study.
supplementary crystallographic information
Experimental
2-Chloropyrimidine (0.05 mol), aniline (0.05 mol) and ethanol (5 ml) were heated at 423–433 K for 3 h. The product was dissolved in water and the solution extracted with ether. The ether phase was dried over sodium sulfate; the evaporation of the solvent gave well shaped colorless crystals along with some unidentified brown material.
Refinement
Carbon-bound H-atoms were placed in calculated positions (C—H 0.95 Å) and were included in the refinement in the riding model approximation, with Uiso(H) set to 1.2Ueq(C). The amino H-atoms were located in a difference Fourier map, and were refined with a distance restraint of N–H 0.88±0.01 Å; their isotropic temperature factors were refined.
Figures
Fig. 1.
Thermal ellipsoid plot (Barbour, 2001) of the two independent molecules of C10H9N3 at the 70% probability level. Hydrogen atoms are drawn as spheres of arbitrary radius. Dashed lines denote hydrogen bonds.
Crystal data
| C10H9N3 | Z = 4 |
| Mr = 171.20 | F(000) = 360 |
| Triclinic, P1 | Dx = 1.317 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 8.8792 (2) Å | Cell parameters from 3015 reflections |
| b = 9.9382 (2) Å | θ = 2.7–28.3° |
| c = 10.2038 (2) Å | µ = 0.08 mm−1 |
| α = 93.186 (1)° | T = 123 K |
| β = 103.665 (1)° | Prism, colorless |
| γ = 97.780 (1)° | 0.35 × 0.20 × 0.10 mm |
| V = 863.28 (3) Å3 |
Data collection
| Bruker SMART APEX diffractometer | 3144 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.020 |
| graphite | θmax = 27.5°, θmin = 2.1° |
| ω scans | h = −11→11 |
| 8238 measured reflections | k = −12→12 |
| 3950 independent reflections | l = −13→12 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.039 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.108 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.056P)2 + 0.1316P] where P = (Fo2 + 2Fc2)/3 |
| 3950 reflections | (Δ/σ)max = 0.001 |
| 243 parameters | Δρmax = 0.20 e Å−3 |
| 2 restraints | Δρmin = −0.22 e Å−3 |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N1 | 0.63241 (11) | 0.69760 (10) | 0.39973 (10) | 0.0229 (2) | |
| H1 | 0.5458 (13) | 0.7328 (15) | 0.4021 (15) | 0.042 (4)* | |
| N2 | 0.60532 (11) | 0.62876 (10) | 0.60461 (10) | 0.0235 (2) | |
| N3 | 0.82465 (11) | 0.58169 (10) | 0.51919 (10) | 0.0257 (2) | |
| N4 | 0.33475 (12) | 0.78540 (10) | 0.59499 (10) | 0.0256 (2) | |
| H4 | 0.4034 (15) | 0.7306 (13) | 0.5833 (14) | 0.038 (4)* | |
| N5 | 0.37902 (11) | 0.86480 (10) | 0.39948 (10) | 0.0256 (2) | |
| N6 | 0.18856 (11) | 0.94997 (10) | 0.50126 (10) | 0.0245 (2) | |
| C1 | 0.69680 (13) | 0.72230 (11) | 0.28811 (11) | 0.0212 (2) | |
| C2 | 0.59235 (14) | 0.72964 (12) | 0.16374 (12) | 0.0259 (3) | |
| H2 | 0.4827 | 0.7163 | 0.1571 | 0.031* | |
| C3 | 0.64691 (15) | 0.75616 (13) | 0.04969 (12) | 0.0305 (3) | |
| H3 | 0.5748 | 0.7621 | −0.0342 | 0.037* | |
| C4 | 0.80703 (15) | 0.77406 (13) | 0.05803 (13) | 0.0304 (3) | |
| H4A | 0.8449 | 0.7896 | −0.0203 | 0.036* | |
| C5 | 0.91044 (14) | 0.76895 (12) | 0.18192 (12) | 0.0273 (3) | |
| H5 | 1.0200 | 0.7824 | 0.1882 | 0.033* | |
| C6 | 0.85757 (13) | 0.74464 (11) | 0.29708 (12) | 0.0234 (2) | |
| H6 | 0.9304 | 0.7432 | 0.3816 | 0.028* | |
| C7 | 0.69174 (13) | 0.63447 (11) | 0.51132 (11) | 0.0208 (2) | |
| C8 | 0.66215 (14) | 0.56772 (12) | 0.71431 (12) | 0.0269 (3) | |
| H8 | 0.6055 | 0.5618 | 0.7825 | 0.032* | |
| C9 | 0.79969 (15) | 0.51238 (14) | 0.73406 (13) | 0.0321 (3) | |
| H9 | 0.8393 | 0.4706 | 0.8140 | 0.039* | |
| C10 | 0.87631 (14) | 0.52129 (13) | 0.63096 (13) | 0.0309 (3) | |
| H10 | 0.9700 | 0.4826 | 0.6402 | 0.037* | |
| C11 | 0.27870 (12) | 0.77031 (12) | 0.71236 (11) | 0.0219 (2) | |
| C12 | 0.28250 (13) | 0.64404 (12) | 0.76560 (12) | 0.0245 (3) | |
| H12 | 0.3171 | 0.5728 | 0.7199 | 0.029* | |
| C13 | 0.23654 (14) | 0.62121 (13) | 0.88390 (12) | 0.0281 (3) | |
| H13 | 0.2399 | 0.5349 | 0.9191 | 0.034* | |
| C14 | 0.18554 (15) | 0.72450 (14) | 0.95093 (13) | 0.0322 (3) | |
| H14 | 0.1532 | 0.7093 | 1.0319 | 0.039* | |
| C15 | 0.18214 (16) | 0.84974 (14) | 0.89877 (13) | 0.0336 (3) | |
| H15 | 0.1470 | 0.9204 | 0.9447 | 0.040* | |
| C16 | 0.22895 (14) | 0.87466 (12) | 0.78070 (12) | 0.0274 (3) | |
| H16 | 0.2271 | 0.9617 | 0.7469 | 0.033* | |
| C17 | 0.29837 (13) | 0.87077 (11) | 0.49672 (11) | 0.0220 (2) | |
| C18 | 0.34955 (14) | 0.95120 (12) | 0.30445 (12) | 0.0277 (3) | |
| H18 | 0.4052 | 0.9520 | 0.2356 | 0.033* | |
| C19 | 0.24158 (14) | 1.03965 (12) | 0.30177 (12) | 0.0281 (3) | |
| H19 | 0.2233 | 1.1016 | 0.2342 | 0.034* | |
| C20 | 0.16165 (14) | 1.03296 (12) | 0.40278 (12) | 0.0271 (3) | |
| H20 | 0.0840 | 1.0901 | 0.4020 | 0.033* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0200 (5) | 0.0288 (5) | 0.0229 (5) | 0.0082 (4) | 0.0084 (4) | 0.0053 (4) |
| N2 | 0.0250 (5) | 0.0245 (5) | 0.0232 (5) | 0.0057 (4) | 0.0090 (4) | 0.0032 (4) |
| N3 | 0.0237 (5) | 0.0303 (5) | 0.0251 (5) | 0.0093 (4) | 0.0068 (4) | 0.0043 (4) |
| N4 | 0.0280 (5) | 0.0276 (5) | 0.0273 (5) | 0.0120 (4) | 0.0134 (4) | 0.0068 (4) |
| N5 | 0.0250 (5) | 0.0288 (5) | 0.0260 (5) | 0.0065 (4) | 0.0102 (4) | 0.0052 (4) |
| N6 | 0.0223 (5) | 0.0262 (5) | 0.0260 (5) | 0.0066 (4) | 0.0062 (4) | 0.0031 (4) |
| C1 | 0.0246 (5) | 0.0192 (5) | 0.0215 (5) | 0.0043 (4) | 0.0086 (4) | 0.0019 (4) |
| C2 | 0.0239 (6) | 0.0279 (6) | 0.0260 (6) | 0.0052 (5) | 0.0055 (5) | 0.0043 (5) |
| C3 | 0.0364 (7) | 0.0325 (7) | 0.0221 (6) | 0.0062 (5) | 0.0054 (5) | 0.0046 (5) |
| C4 | 0.0393 (7) | 0.0294 (6) | 0.0262 (6) | 0.0035 (5) | 0.0159 (5) | 0.0041 (5) |
| C5 | 0.0273 (6) | 0.0255 (6) | 0.0319 (7) | 0.0025 (5) | 0.0140 (5) | 0.0014 (5) |
| C6 | 0.0230 (5) | 0.0241 (6) | 0.0232 (6) | 0.0032 (4) | 0.0063 (4) | 0.0007 (4) |
| C7 | 0.0208 (5) | 0.0207 (5) | 0.0206 (5) | 0.0022 (4) | 0.0055 (4) | −0.0003 (4) |
| C8 | 0.0319 (6) | 0.0291 (6) | 0.0220 (6) | 0.0050 (5) | 0.0107 (5) | 0.0036 (5) |
| C9 | 0.0345 (7) | 0.0394 (7) | 0.0246 (6) | 0.0114 (6) | 0.0061 (5) | 0.0114 (5) |
| C10 | 0.0263 (6) | 0.0364 (7) | 0.0317 (7) | 0.0122 (5) | 0.0055 (5) | 0.0073 (5) |
| C11 | 0.0182 (5) | 0.0257 (6) | 0.0228 (6) | 0.0043 (4) | 0.0060 (4) | 0.0035 (4) |
| C12 | 0.0245 (6) | 0.0243 (6) | 0.0259 (6) | 0.0054 (4) | 0.0080 (5) | 0.0010 (5) |
| C13 | 0.0299 (6) | 0.0269 (6) | 0.0294 (6) | 0.0042 (5) | 0.0096 (5) | 0.0075 (5) |
| C14 | 0.0363 (7) | 0.0406 (7) | 0.0259 (6) | 0.0105 (6) | 0.0160 (5) | 0.0087 (5) |
| C15 | 0.0417 (7) | 0.0368 (7) | 0.0295 (7) | 0.0172 (6) | 0.0166 (6) | 0.0036 (5) |
| C16 | 0.0328 (6) | 0.0253 (6) | 0.0275 (6) | 0.0097 (5) | 0.0104 (5) | 0.0048 (5) |
| C17 | 0.0199 (5) | 0.0228 (6) | 0.0232 (6) | 0.0025 (4) | 0.0060 (4) | 0.0017 (4) |
| C18 | 0.0291 (6) | 0.0300 (6) | 0.0254 (6) | 0.0028 (5) | 0.0102 (5) | 0.0049 (5) |
| C19 | 0.0329 (6) | 0.0254 (6) | 0.0257 (6) | 0.0047 (5) | 0.0054 (5) | 0.0074 (5) |
| C20 | 0.0268 (6) | 0.0257 (6) | 0.0287 (6) | 0.0082 (5) | 0.0042 (5) | 0.0019 (5) |
Geometric parameters (Å, °)
| N1—C7 | 1.3613 (14) | C5—H5 | 0.9500 |
| N1—C1 | 1.4090 (14) | C6—H6 | 0.9500 |
| N1—H1 | 0.892 (9) | C8—C9 | 1.3829 (17) |
| N2—C8 | 1.3294 (15) | C8—H8 | 0.9500 |
| N2—C7 | 1.3555 (14) | C9—C10 | 1.3819 (17) |
| N3—C10 | 1.3322 (15) | C9—H9 | 0.9500 |
| N3—C7 | 1.3423 (14) | C10—H10 | 0.9500 |
| N4—C17 | 1.3584 (15) | C11—C16 | 1.3950 (16) |
| N4—C11 | 1.4093 (14) | C11—C12 | 1.3963 (16) |
| N4—H4 | 0.894 (8) | C12—C13 | 1.3831 (16) |
| N5—C18 | 1.3339 (15) | C12—H12 | 0.9500 |
| N5—C17 | 1.3570 (14) | C13—C14 | 1.3859 (17) |
| N6—C20 | 1.3338 (15) | C13—H13 | 0.9500 |
| N6—C17 | 1.3407 (14) | C14—C15 | 1.3818 (18) |
| C1—C6 | 1.3949 (15) | C14—H14 | 0.9500 |
| C1—C2 | 1.3958 (16) | C15—C16 | 1.3883 (17) |
| C2—C3 | 1.3873 (16) | C15—H15 | 0.9500 |
| C2—H2 | 0.9500 | C16—H16 | 0.9500 |
| C3—C4 | 1.3903 (18) | C18—C19 | 1.3824 (17) |
| C3—H3 | 0.9500 | C18—H18 | 0.9500 |
| C4—C5 | 1.3839 (18) | C19—C20 | 1.3824 (17) |
| C4—H4A | 0.9500 | C19—H19 | 0.9500 |
| C5—C6 | 1.3860 (16) | C20—H20 | 0.9500 |
| C7—N1—C1 | 128.19 (9) | C10—C9—H9 | 121.9 |
| C7—N1—H1 | 114.6 (10) | C8—C9—H9 | 121.9 |
| C1—N1—H1 | 117.0 (10) | N3—C10—C9 | 122.94 (11) |
| C8—N2—C7 | 115.61 (10) | N3—C10—H10 | 118.5 |
| C10—N3—C7 | 116.06 (10) | C9—C10—H10 | 118.5 |
| C17—N4—C11 | 129.13 (10) | C16—C11—C12 | 119.15 (10) |
| C17—N4—H4 | 115.5 (9) | C16—C11—N4 | 124.17 (10) |
| C11—N4—H4 | 115.4 (9) | C12—C11—N4 | 116.58 (10) |
| C18—N5—C17 | 115.71 (10) | C13—C12—C11 | 120.91 (11) |
| C20—N6—C17 | 115.97 (10) | C13—C12—H12 | 119.5 |
| C6—C1—C2 | 119.10 (10) | C11—C12—H12 | 119.5 |
| C6—C1—N1 | 123.56 (10) | C12—C13—C14 | 119.88 (11) |
| C2—C1—N1 | 117.30 (10) | C12—C13—H13 | 120.1 |
| C3—C2—C1 | 120.70 (11) | C14—C13—H13 | 120.1 |
| C3—C2—H2 | 119.7 | C15—C14—C13 | 119.35 (11) |
| C1—C2—H2 | 119.7 | C15—C14—H14 | 120.3 |
| C2—C3—C4 | 120.04 (11) | C13—C14—H14 | 120.3 |
| C2—C3—H3 | 120.0 | C14—C15—C16 | 121.49 (11) |
| C4—C3—H3 | 120.0 | C14—C15—H15 | 119.3 |
| C5—C4—C3 | 119.13 (11) | C16—C15—H15 | 119.3 |
| C5—C4—H4A | 120.4 | C15—C16—C11 | 119.20 (11) |
| C3—C4—H4A | 120.4 | C15—C16—H16 | 120.4 |
| C4—C5—C6 | 121.39 (11) | C11—C16—H16 | 120.4 |
| C4—C5—H5 | 119.3 | N6—C17—N5 | 125.94 (11) |
| C6—C5—H5 | 119.3 | N6—C17—N4 | 119.51 (10) |
| C5—C6—C1 | 119.59 (11) | N5—C17—N4 | 114.55 (10) |
| C5—C6—H6 | 120.2 | N5—C18—C19 | 122.92 (11) |
| C1—C6—H6 | 120.2 | N5—C18—H18 | 118.5 |
| N3—C7—N2 | 125.92 (10) | C19—C18—H18 | 118.5 |
| N3—C7—N1 | 119.10 (10) | C18—C19—C20 | 116.34 (11) |
| N2—C7—N1 | 114.96 (10) | C18—C19—H19 | 121.8 |
| N2—C8—C9 | 123.22 (11) | C20—C19—H19 | 121.8 |
| N2—C8—H8 | 118.4 | N6—C20—C19 | 123.03 (11) |
| C9—C8—H8 | 118.4 | N6—C20—H20 | 118.5 |
| C10—C9—C8 | 116.22 (11) | C19—C20—H20 | 118.5 |
| C7—N1—C1—C6 | −31.08 (18) | C17—N4—C11—C16 | −28.92 (18) |
| C7—N1—C1—C2 | 151.47 (11) | C17—N4—C11—C12 | 154.69 (11) |
| C6—C1—C2—C3 | 1.23 (17) | C16—C11—C12—C13 | 0.54 (17) |
| N1—C1—C2—C3 | 178.80 (10) | N4—C11—C12—C13 | 177.12 (10) |
| C1—C2—C3—C4 | 0.81 (18) | C11—C12—C13—C14 | 0.15 (18) |
| C2—C3—C4—C5 | −1.85 (18) | C12—C13—C14—C15 | −0.37 (19) |
| C3—C4—C5—C6 | 0.85 (18) | C13—C14—C15—C16 | −0.1 (2) |
| C4—C5—C6—C1 | 1.19 (17) | C14—C15—C16—C11 | 0.8 (2) |
| C2—C1—C6—C5 | −2.21 (17) | C12—C11—C16—C15 | −1.00 (18) |
| N1—C1—C6—C5 | −179.62 (10) | N4—C11—C16—C15 | −177.30 (11) |
| C10—N3—C7—N2 | −1.59 (17) | C20—N6—C17—N5 | −2.37 (17) |
| C10—N3—C7—N1 | −179.97 (10) | C20—N6—C17—N4 | 178.70 (10) |
| C8—N2—C7—N3 | 1.75 (16) | C18—N5—C17—N6 | 3.36 (17) |
| C8—N2—C7—N1 | −179.81 (10) | C18—N5—C17—N4 | −177.67 (10) |
| C1—N1—C7—N3 | −3.22 (17) | C11—N4—C17—N6 | −3.96 (18) |
| C1—N1—C7—N2 | 178.23 (10) | C11—N4—C17—N5 | 176.99 (11) |
| C7—N2—C8—C9 | −0.28 (17) | C17—N5—C18—C19 | −1.45 (17) |
| N2—C8—C9—C10 | −1.14 (19) | N5—C18—C19—C20 | −1.09 (18) |
| C7—N3—C10—C9 | −0.06 (18) | C17—N6—C20—C19 | −0.57 (17) |
| C8—C9—C10—N3 | 1.3 (2) | C18—C19—C20—N6 | 2.18 (18) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···N5 | 0.89 (1) | 2.10 (1) | 2.972 (1) | 164 (1) |
| N4—H4···N2 | 0.89 (1) | 2.15 (1) | 3.020 (1) | 165 (1) |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: XU2492).
References
- Barbour, L. J. (2001). J. Supramol. Chem.1, 189–191.
- Bruker (2008). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Maizathul Akmam, A. B., Abdullah, Z. & Ng, S. W. (2009). Acta Cryst. E65, o94. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Westrip, S. P. (2009). publCIF In preparation.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809007685/xu2492sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809007685/xu2492Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report



