To the Editor
Nucleosomes occlude their wrapped DNA, strongly influencing the accessibility of functional DNA binding sites. This has led to interest in genome-wide mapping of nucleosome positions and in understanding the principles that govern these positions. We recently compared the positions of nucleosomes reconstituted in vitro to a map of in vivo nucleosome locations1. We found high similarity between the maps, implying that intrinsic DNA sequence preferences of nucleosomes have a major role in determining the organization of nucleosomes in vivo. A subsequent paper by Struhl and colleagues2 (henceforth Zhang et al.) used a similar approach but stated an opposite conclusion. We believe that the stated conclusion of Zhang et al.2 is inconsistent with data in both of these papers and also with previously published results and conclusions, including earlier publications by Struhl and colleagues.
Both our study1 and that of Zhang et al.2 reconstituted nucleosomes in vitro using purified histone octamers and yeast genomic DNA, then mapped the resulting nucleosomes genome-wide using micrococcal nuclease and parallel DNA sequencing. Evidence presented in these and earlier publications that proves that nucleosome sequence preferences contribute substantially to nucleosome organization in vivo includes the following. First, nucleosome-bound sequences from yeast, worm, fly, chicken and human have distinctive patterns of dinucleotide periodicities1,3–6. These patterns represent intrinsic nucleosome sequence preferences, as they appear also in nucleosomes reconstituted in vitro using only purified histones and DNA1,4,7,8. Thus, many nucleosomes in vivo occupy positions that are favored by intrinsic nucleosome sequence preferences. Second, our in vitro map1 and that of Zhang et al.2 both show strong nucleosome depletion at yeast promoters, transcription-factor binding sites and gene ends. This depletion is similar to that observed at these regions in vivo, suggesting that these in vivo patterns are largely dictated by intrinsic nucleosome sequence preferences (Fig. 1a). Third, this same conclusion was reached using an in vitro reconstitution experiment on a few genes in an earlier paper by Struhl and colleagues9. Fourth, there is a striking correspondence between in vitro and in vivo nucleosome positions over a ~10-kb region encompassing the sheep β-lactoglobulin gene10. Fifth, our computational model of intrinsic nucleosome sequence preferences was independently validated by analysis of nucleosomes reconstituted on bacteriophage λ DNA and on an 82,000–base pair (bp) DNA region from the human β-globin locus11. This experiment mapped nucleosomes with a single-molecule imaging approach that did not require the use of either micrococcal nuclease or parallel DNA sequencing. Our computational model, in turn, predicts nucleosome occupancies that are significantly correlated with nucleosome occupancy in C. elegans in vivo1. Sixth, another earlier study by Struhl and colleagues12 showed that the deletion of sequences in a yeast promoter that strongly disfavor nucleosome formation alters the in vivo nucleosome organization and consequently decreases transcription-factor accessibility and gene expression. Seventh, evolutionary changes from high to low expression of a large group of orthologous genes between two yeast species are accompanied by corresponding changes from strong to weaker DNA-encoded nucleosome depletion over their promoters, again suggesting that intrinsic nucleosome DNA sequence preferences dictate in vivo nucleosome occupancy and thereby influence gene expression13. Finally, the numbers reported in Zhang et al.2 directly show significant (20% by their count) correspondences between the in vitro and in vivo maps, again implying that the genomic DNA sequence is an important determinant of nucleosome organization in vivo.
Figure 1.
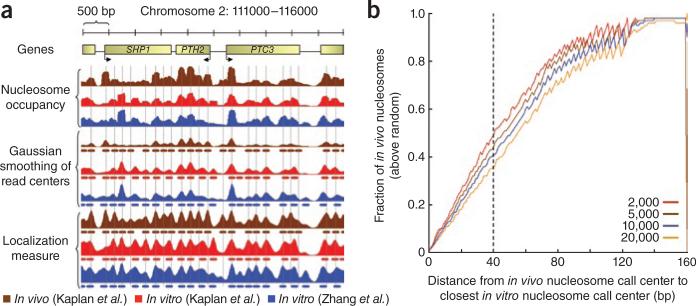
Nucleosome positions in vitro compared to those in vivo. (a) In vivo and in vitro nucleosome data on an arbitrary 5-kbp genomic region from yeast. For each of the three experimental nucleosome datasets (brown and red, in vivo and in vitro from Kaplan et al.1, respectively; blue, in vitro from Zhang et al.2) we show the nucleosome occupancy, a Gaussian smoothing (s.d. = 40 bp) of read centers (73 bp downstream from read start) and the localization measure (also smoothed with a Gaussian, s.d. = 40 bp), along with nucleosome calls made on each of the data tracks. Vertical lines, center positions of the nucleosome calls made on the Gaussian-smoothed read centers in vivo. (b) Cumulative distribution of distances between centers of nucleosome calls in vivo and in vitro, normalized by a randomized control. Nucleosomes were called in vivo and in vitro (Kaplan et al.1 datasets) by taking the track of Gaussian-smoothed read centers and iteratively selecting the position with the highest value and excluding the 147 base pairs surrounding it. We selected 50,000 nucleosomes from the in vitro data and 2,000, 5,000, 10,000 and 20,000 nucleosomes from the in vivo data. Next, we computed the cumulative distribution by calculating the fraction of in vivo nucleosome call centers that are within k base pairs of the closest in vitro call center for every k = 0...160 bp. As a control, we performed the same computation on in vivo calls that were shuffled randomly while preserving pairwise distances between neighboring nucleosomes. Finally, we subtracted the shuffled from real distribution and divided the resulting distribution by 1 minus the shuffled distribution, thus scaling the results between 0 (not improving from random) and 1 (explaining all in vivo positions). Dashed vertical line, fraction above random of in vivo nucleosome centers within 40 bp of in vitro nucleosome centers.
Thus, nucleosome occupancy measures from both studies properly capture real features of the genomically encoded nucleosome landscape in comparing the in vitro and in vivo nucleosome organizations. No other single factor has been shown quantitatively to have a greater importance for the in vivo nucleosome organization than does the genomic DNA sequence itself, through the nucleosomes’ intrinsic DNA sequence preferences.
Using a localization metric like that introduced in Zhang et al.2 (except correcting their calculation as in Struhl and colleagues’ previous work14; see below), our in vitro data account for 34–41% of the in vivo positions (Fig. 1a,b). Using a simpler Gaussian smoothing of the raw data, our in vitro map accounts for 36–49% of the in vivo nucleosome positions, and our model of the nucleosome sequence preferences, learned in cross-validation from our in vitro data, accounts for 42–57% of the in vivo nucleosome positions (Fig. 1a,b). In all cases, the exact numbers depend on the cutoffs used for calling nucleosome positions and on what is considered to be a close enough distance between nucleosomes. However, the key issue is not the exact numbers but rather that, even using positioning-based measures, both the original analysis of Zhang et al.2 and our reanalysis of both datasets show that a substantial fraction (~20–60%) of the in vivo nucleosome positions are attributable to intrinsic nucleosome sequence preferences.
Thus, while we re-emphasize that many aspects of the in vivo nucleosome organization are not explained by nucleosome sequence preferences—as we had previously noted1—many studies by many groups show that intrinsic nucleosome sequence preferences contribute substantially to nucleosome organization and chromatin function in vivo. Finally, with respect to the question of whether there exists a genomic code for nucleosome positioning, both our analysis and that of Zhang et al.2 show that the genomic DNA sequence encodes many aspects of the in vivo nucleosome organization; whether this reflects the use of a code is something we shall leave for others to debate.
There are also three specific technical points on which we disagree with the analysis in Zhang et al.2. These chiefly concern how to quantify the contribution of intrinsic nucleosome sequence preferences.
Quantifying the influence of nucleosome sequence preferences on nucleosome occupancy
Our published analysis compared nucleosome occupancy per base pair between the in vitro and in vivo maps. Nucleosome occupancy is important because it governs how easily, in a thermodynamic sense, a regulatory protein will be able to access a given specific DNA target site15. Zhang et al.2 examined detailed nucleosome positioning, a different aspect of nucleosome organization which we had not examined explicitly. Thus, the analyses of Zhang et al.2 do not contradict any of our conclusions about the role of DNA sequence in directing nucleosome occupancy. Mathematically, occupancy is a simple (147-bp-wide) moving average of the raw mapping data. We reported a genome-wide correlation of 0.74 between our in vitro and in vivo maps, and Zhang et al.2 confirm this high level of similarity in occupancy, reporting a correlation per base pair of 0.69 between the two in vitro maps and 0.71 between our in vitro and in vivo maps. These correlations are even higher (0.78 and 0.74, respectively) when made on a log scale. As we emphasized earlier1, our nucleosome occupancy measure also reveals differences between the in vitro and in vivo maps, including at binding sites for certain transcription factors, and the periodic spacing of the +1 and downstream nucleosomes. In summary, regardless of the exact quantitative magnitude of the correlation in nucleosome occupancy between the in vitro and in vivo maps, both our work and that of Zhang et al.2 agree that this correlation is highly significant.
The nature and impact of experimental biases
Zhang et al.2 criticizes the occupancy measure on the grounds that it places emphasis on the actual measured occupancy values, which might be biased by, for example, possible non-uniformities in DNA amplification or sequencing efficiencies, thus overestimating the similarity between the in vitro and in vivo maps. We agree, but we note also that several other factors are likely to cause underestimation of the similarity between the maps. For example, both experiments sample the distribution of nucleosomes relatively sparsely (for example, the map of Zhang et al.2 has only 0.27 reads per base pair or 5 reads per 20-bp window used in that analysis). This means that even two replicates are expected to differ from one another, simply because of the limited number of reads. Other important differences between the in vivo and in vitro experiments include the salt concentration, histone concentration and the histones themselves (Zhang et al.2 used fly embryo histones and Kaplan et al. used chicken erythrocyte histones, to compare with the in vivo organization in yeast). All of these factors may create artificial differences between the in vivo and in vitro maps, decreasing their agreement.
As we noted above, our in vitro and in vivo nucleosome occupancy measurements were experimentally validated by us1,4,16,17, by the single-molecule imaging analysis of nucleosome positioning in previous work11 and even by Struhl and colleagues9, including using approaches that require neither micrococcal nuclease nor DNA sequencing. These studies show that several stereotypical and functionally important patterns of nucleosome occupancy are similar in vivo and in vitro and thus are caused by nucleosome sequence preferences.
Definition of nucleosome positioning and quantifying the influence of nucleosome sequence preferences on in vivo nucleosome positions
Zhang et al.2 cites their Figure 4b–d and their Supplementary Figure 3 for their estimate that intrinsic nucleosome sequence preferences account for ~20% of the in vivo nucleosome positions. As we explain next, we believe that the method used by Zhang et al.2 to derive this estimate is inappropriate (however, we also re-emphasize that even their ~20% estimate still implies that intrinsic nucleosome sequence preferences are important for many nucleosome positions). We believe the analysis of Zhang et al.2 to be inappropriate for the following reasons. Their analysis used our in vivo data to define a set of well-positioned nucleosomes, then plotted the percentage of these nucleosomes for which their in vitro data has at least k nucleosome reads within 20 bp of each nucleosome center for all values of k. This is not a direct comparison of nucleosome positions in the two maps; rather, it compares nucleosome positions in vivo to nucleosome reads (that is, occupancy) in vitro. As an example of problems inherent in this comparison, consider two well-positioned nucleosomes in vivo for which the in vitro map has 5 reads within 20 bp of the first nucleosome's center and 10 reads within 20 bp of the second nucleosome's center. The analysis of Zhang et al.2 assigns a higher positioning correspondence score to the second nucleosome because of its greater number of reads. Suppose, however, that the second nucleosome in the in vitro map is flanked also by many additional reads outside the 20-bp window, whereas the first nucleosome has no other reads nearby. In this case, the analysis of Zhang et al.2 is inconsistent with that study's own definition of positioning because, according to that definition, the first nucleosome in the in vitro map is highly positioned and the second is not, yet the second receives the higher positioning correspondence score. Moreover, in contrast to previous work by Struhl and colleagues14, the calculation in Zhang et al.2 subtracts the amount that is attributable to random chance without rescaling the remainder and thus cannot yield a result in which 100% of the in vivo nucleosome positions were explained by the in vitro data.
To obtain a more direct comparison of nucleosome positions between the in vitro and in vivo maps, we used two measures (see below) of positioning to separately assign discrete nucleosome positions in both maps. From these discrete positions, we then calculated the fraction of the positioned nucleosomes in vivo that are explainable by the positions adopted by nucleosomes in vitro—that is, the fraction of the positioned nucleosomes in vivo that is attributable to intrinsic nucleosome sequence preferences. One measure is essentially that used by Zhang et al.2; we refer to it as ‘localization’. It defines the positioning at every base pair i as the number of nucleosome reads that fall within a 40-bp region centered on i, divided by the number of reads within 160 bp of i and then smoothed with a Gaussian. This is identical to the measure used by Zhang et al.2 except that we smooth the results and use a 40-bp window instead of one of 20 bp. This is done to better accommodate the sparseness of the data (summarized above: the map of Zhang et al.2 has only ~5 reads per 20-bp window) and the limited accuracy with which nucleosome centers are known (the distribution of nucleosome lengths that result from micrococcal nuclease digestion is much greater than 40 bp wide6,17,18). We also corrected their calculation to allow for the full possible range of answers (as in ref. 14). Despite these improvements to the Zhang et al.2 metric, we consider it problematic because we find that different results are obtained by slight variations of its parameters. We therefore also introduced a second measure, based on simple Gaussian smoothing of the raw nucleosome read data. As intended in Zhang et al.2, both measures assign favorable scores to highly positioned nucleosomes, regardless of whether those nucleosomes are very abundant in the cell population, or very rare. The results using these two metrics and our original occupancy metric are included in Figure 1a,b.
In summary, although significant differences do exist between the in vitro and in vivo nucleosome maps, as we had previously noted, the existing literature and comparisons using both our data and those of Zhang et al.2 all show that the genome explicitly encodes many aspects of the in vivo nucleosome organization through the nucleosomes’ intrinsic DNA sequence preferences.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Kaplan N, et al. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Y, et al. Nat. Struct. Mol. Biol. 2009;16:847–852. doi: 10.1038/nsmb.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Satchwell SC, Drew HR, Travers AA. J. Mol. Biol. 1986;191:659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- 4.Segal E, et al. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson SM, Tan FJ, McCullough HL, Riordan DP, Fire AZ. Genome Res. 2006;16:1505–1516. doi: 10.1101/gr.5560806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Albert I, et al. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- 7.Lowary PT, Widom J. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 8.Widlund HR, et al. J. Mol. Biol. 1997;267:807–817. doi: 10.1006/jmbi.1997.0916. [DOI] [PubMed] [Google Scholar]
- 9.Sekinger EA, Moqtaderi Z, Struhl K. Mol. Cell. 2005;18:735–748. doi: 10.1016/j.molcel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 10.Fraser RM, Keszenman-Pereyra D, Simmen MW, Allan J. J. Mol. Biol. 2009;390:292–305. doi: 10.1016/j.jmb.2009.04.079. [DOI] [PubMed] [Google Scholar]
- 11.Visnapuu ML, Greene EC. Nat. Struct. Mol. Biol. 2009;16:1056–1062. doi: 10.1038/nsmb.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iyer V, Struhl K. EMBO J. 1995;14:2570–2579. doi: 10.1002/j.1460-2075.1995.tb07255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Field Y, et al. Nat. Genet. 2009;41:438–445. doi: 10.1038/ng.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peckham HE, et al. Genome Res. 2007;17:1170–1177. doi: 10.1101/gr.6101007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Segal E, Widom J. Nat. Rev. Genet. 2009;10:443–456. doi: 10.1038/nrg2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thastrom A, Bingham LM, Widom J. J. Mol. Biol. 2004;338:695–709. doi: 10.1016/j.jmb.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 17.Field Y, et al. PLoS Comput. Biol. 2008;4:e1000216. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valouev A, et al. Genome Res. 2008;18:1051–1063. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]