Abstract
Checkpoint kinase 2 (CHEK2) is a protein involved in arresting cell cycle in response to DNA damage. To investigate whether it plays an important role in the development of prostate cancer (PRCA) in the Ashkenazi Jewish (AJ) population, we sequenced CHEK2 in 75 AJ individuals with prostate, breast, or no cancer (n = 25 each). We identified seven coding SNPs (five are novel) that changed the amino acid sequence, resulting in R3W, E394F, Y424H, S428F, D438Y, P509S, and P509L. We determined the frequency of each variant in 76 AJ families collected by members of the International Consortium for Prostate Cancer Genetics (ICPCG) where ≥ 2 men were affected by PRCA. Only one variant, Y424H in exon 11, was identified in more than two families. Exon 11 was then screened in nine additional AJ ICPCG families (a total of 85 families). The Y424H variant occurred in nine affected cases from four different families; however it did not completely segregate with the disease. We performed bioinformatics analysis, which showed that Y424H is a non-conservative missense substitution that falls at a position that is invariant in vertebrate CHEK2 orthologs. Both SIFT and Align-GVGD predict that Y424H is a loss of function mutation, however the frequency of Y424H was not significantly different between unselected AJ cases from Montreal/Memorial Sloan Kettering Cancer Centre (MSKCC) and AJ controls from Israel/MSKCC (OR 1.18, 95%CI: 0.34–4.61, p=.99). Moreover, functional assays using S. Cerevisae revealed that the Y424H substitution did not alter function of CHEK2 protein. Although we cannot rule out a subtle influence of the CHEK2 variants on PRCA risk, these results suggest that germline CHEK2 mutations have a minor role in PRCA susceptibility in AJ men.
Keywords: checkpoint kinase 2, prostate cancer, single-nucleotide polymorphism, Ashkenazi Jewish, budding yeast
INTRODUCTION
Prostate cancer (PRCA) is a leading cause of morbidity and mortality in men. It is diagnosed in almost one-fifth of US men during their lifetime. Although many etiological factors have been implicated, genetic predisposition and age remain as the two major factors in development of PRCA [1]. Epidemiological studies suggest that up to 5% of all cases may be due to autosomal dominant genes [2; 3; 4; 5] and twin studies suggest that approximately 42% of PRCA cases diagnosed under the age of 70 years are likely due to heritable factors [6]. Men with an affected father or brother are twice as likely to develop PRCA as men with no affected relatives [7]. In addition, the relative risk of developing PRCA rises considerably as the number of cases in a family cluster increases and the average age at diagnosis in the cluster decreases [8]. A recent meta-analysis on the risk of PRCA among men with a positive family history found a 1.8–2.1-fold increased risk if a second degree relative is affected and 2.9-fold increased risk if the father or a brother is affected [9].
The CHEK2 gene, located on chromosome 22q, encodes a checkpoint kinase that acts to prevent cellular entry into mitosis in response to DNA damage, presumably to gain time needed for DNA repair [10; 11]. Activated CHEK2 phosphorylates BRCA1 and TP53 proteins, regulating tumor suppressor function of these proteins [12; 13; 14]. Mutations in CHEK2 were originally described in Li-Fraumeni syndrome and Li-Fraumeni-like families [15; 16], and the 1100delC variant was later found to be a moderate risk breast cancer susceptibility allele [17; 18; 19]. Several studies suggest that the CHEK2 locus or its variants maybe important in PRCA susceptibility. In a linkage study of 1233 PRCA families, analysis of the 269 families with at least 5 affected members identified a LOD score of 3.57 at 22q12 [20], which is near the CHEK2 locus at 22q12.1 and this has recently been further refined [21]. Additionally, four independent studies have investigated the association between CHEK2 mutations and PRCA risk with some conflicting results [22; 23; 24; 25; 26]. In this study, we investigated whether germline CHEK2 mutations play an important role in the development of PRCA in the Ashkenazi Jewish population. This ethnic group has founder mutations in other cancer-predisposing genes such as BRCA2 6174delT that have been shown to be more frequent in AJ men with prostate cancer [27] making it a good study population.
MATERIALS AND METHODS
SNP discovery and frequency estimate
In the first step in the identification of CHEK2 variants (q > 0.01), CHEK2 was sequenced in 75 AJ individuals with prostate, breast, or no cancer (n = 25 each). DNA was extracted from blood lymphocytes using standard methods. Since a portion of the CHEK2 gene (exons 10–14) shares high homology with regions on other chromosomes [28], all primers were tested by in silico PCR and BLAT (UCSC Genome Bioinformatics Website) to confirm the specificity of each primer pair. Sequencing was done in both forward and reverse directions using an ABI 3730XL DNA Sequencer (Applied Biosystems, Foster, CA). Sequences were analyzed using Chromas 2.3 (Technelysium Pty, Tewantin, Qld, Australia). Long range PCR using previously described methods [29] was used to confirm that all identified variants were located within the functional copy of CHEK2, on chromosome 22q12.
The 150 prostate cancer cases used for SNP discovery, including the 25 samples used for the preliminary frequency analysis, were obtained from an on-going recruitment effort among McGill University-affiliated hospitals described previously [30]. Twenty-five of the 150 prostate cancer cases were chosen for SNP discovery based on the presence of a family history and/or a high Gleason score (mean score = 7.4; mean age at diagnosis = 67.5 years). The 25 healthy AJ controls were recruited as part of the same study and were unrelated to the cases (mean age = 67.9 years). The 25 AJ breast cancer cases all had family history of breast cancer and had been screened for the 3 AJ founder BRCA1/2 mutations. The remaining 125 prostate cancer cases from the 150 prostate cancer cases mentioned above were genotyped to estimate the frequencies of the seven variants uncovered during SNP discovery (mean Gleason score = 5.6; mean age at diagnosis = 68.2 for 109 and 122 subjects with available information, respectively). Two hundred AJ individuals obtained from the National Laboratory for the Genetics of Israeli Populations (NLGIP) (www.tau.ac.il/medicine/NLGIP/nlgip.htm) were genotyped for all seven variants to determine reference allele frequencies in the general AJ population. The additional cohort of unselected prostate cancer cases and controls from MKSCC has been previously described [27].
A total of 76 AJ prostate cancer families with ≥ 2 affected members were included from three centers that are part of the ICPCG (John Hopkins University, University of Michigan, and Fred Hutchinson Cancer Research Center) and have been previously characterized [20; 31; 32; 33]. Study subjects and DNA samples were treated in accordance with IRB regulations in the respective institutions.
Depending on the characteristics of the variant, different methods were used for screening a larger series of cases and controls including primer specific assay (R3W), restriction digestion analysis (E394F), and SSCP (single-strand conformation polymorphism - all other variants, exons 11 and 13). All suspected positives were confirmed by direct sequencing as described above. Positive and negative controls were used in all screening tests.
Allele and genotype frequency was expressed as percentage (proportion) and analyzed by means of chi-square or Fisher’s exact test when appropriate. A two-tailed exact p value was presented unless it was less than 0.001. A p value of less than 0.05 was considered statistically significant. The statistical analysis was performed using Arcus Quickstat Biomedical Version 1.0 (Research Solutions Limited). Simultaneous screening of 25 cases and 25 controls for SNP discovery provides an 95% power to detect an allele with a frequency of 1% or more in either cases or controls from this population [34] and eliminates potential biases inherent in studying cases first and then only screening for those variants identified in cases in the control set. The frequency of these SNPs was determined in individuals from 76 AJ families collected by ICPCG members; for each family, every member on whom DNA was available was screened for all seven variants (with at least one affected member being screened in each family). In addition, these variants were screened in 125 unselected AJ PRCA cases from Montreal and 200 AJ controls from Israel. The frequency of the Y424H and S428F variants was determined by screening an additional series of 590 consecutive AJ PRCA cases and 345 controls.
CHEK2 variants, amino-acid stability, and conservation
To investigate the potential role of CHEK2 missense substitutions in silico, we prepared a CHEK2 protein multiple sequence alignment containing sequences from five mammals plus chicken, frog, zebrafish, and tunicate. Apparent missing exons or cloning artifacts in the individual sequences were repaired by exon prediction from the corresponding genomic sequence. The alignment was prepared using T-Coffee [35] and is available online at <http://agvgd.iarc.fr/alignments.php>. The fit between individual human missense substitutions and the range of variation observed at their corresponding position in the alignment was assessed using SIFT (Sorts Intolerant From Tolerant) [36] and Align-GVGD [37].
Yeast transformation and MMS treatment
Plasmids carrying mutations resulting in 1100delC, S428F, F428S, and Y424H CHEK2 variants were created using site directed mutagenesis (QuikChange XL kit, Stratagene) performed on the plasmid pMH267 (pBAD101, 2 micron LEU2 GAL-CHEK2) carrying wild type human CHEK2 kindly provided by Dr. Steven Elledge [38]. Coding regions of all plasmids were sequenced to verify their identity and confirm that no other mutations were present. Site directed mutagenesis was also performed on the S428F plasmid to re-create the wild type CHEK2 (i.e., F428S) plasmid. Yeast strain S. Cerevisae (MATa sml1::URA3 rad53::HIS3, isogenic to W303), kindly provided by Dr. R. Rothstein [39], was transformed using plasmids carrying the wild type CHEK2 or the mutants mentioned above using a combination of protocols by Elble [40] and Gietz & Woods [41]. Expression of CHEK2 by the plasmids was confirmed using two-step RT-PCR reactions. RNA was isolated from spheroblasts created by digestion of transformed yeast cultures using lyticase. First strand cDNA was synthesized using RNA digested with DNAse I to eliminate any possible contamination of RNA with transforming plasmid DNA. No amplification was observed when at least 60 nanograms of RNA digested with DNAse I was used as template in a 25 L PCR reaction using primers specific to CHEK2. To ensure viability in response to lethality due to Rad53 deletion in this yeast strain, sml1, an inhibitor of ribonucleotide reductase, whose deletion results in increased dNTP pools available for DNA replication and thus suppressing Rad53 lethality, was also deleted. Following transformation and incubation at 30°C for 4 to 6 days, three colonies from each plate were picked and inoculated into disposable cuvettes (Sigma) containing 2 mL of His-Ura-Leu- liquid media and incubated at 30°C with shaking at 200 or 250 rpm until all cultures reached a density of above 0.5 OD600 nm. Yeast cultures were then diluted down to 0.500 ± 0.001 OD600 nm and 20 μL of the diluted sample was inoculated into cuvettes containing 2 mL of fresh His-Ura-Leu- media. In addition, 20 μL of the diluted samples were inoculated into cuvettes containing the same media with 0.00125 or 0.00250% of 100% MMS (methyl methane sulphonate; v/v; Sigma). The same approach was used to inoculate 20 μL of the diluted samples into His-Ura-Leu- liquid media containing 2% glucose -instead of galactose- to suppress expression of CHEK2 from all plasmids. The cuvettes were incubated at 30°C with shaking at 200 or 250 rpm and optical density readings were recorded approximately every 5 hrs for approximately 30 hrs. Experiments were done in duplicates, repeated, and similar results were obtained. No colonies were observed when no plasmid DNA was added to the transformation reaction. Nearly zero readings were obtained when non-transformed yeast grown on His-Ura- plates and liquid media were inoculated into His-Ura-Leu- liquid media.
RESULTS
SNP discovery and frequency estimate
Seven coding SNPs were identified (five were novel) in the prostate cancer subjects that changed the amino acid sequence, resulting in R3W, E394F, Y424H, S428F, D438Y, P509S, and P509L (Table 1). Y424H was detected in a breast cancer subject in the initial series of 75 individuals (25 prostate cancer cases, 25 breast cancer cases and 25 controls). Four of these seven variants occurred in the protein kinase domain and all were confirmed to be in the functional CHEK2 gene copy by long range PCR. To study the potential importance of these CHEK2 variants on function, bioinformatics analysis using both SIFT and Align-GVGD was performed which indicated that E394F, Y424H, S428F, and D438Y are potentially deleterious (Table 1).
TABLE 1.
Functional significance (Align-GVGD values) and frequencies of the seven CHEK2 variants in AJ subjects.
| Exon 1 7C>T R3W | Exon 10 1180G>A E394F | Exon 11 1270T>C Y424H | Exon 11 1283C>T S428F | Exon 11 1312G>T D438Y | Exon 13 1525C>T P509S | Exon 13 1526 C>T P509L | |
|---|---|---|---|---|---|---|---|
| Functional significance | |||||||
| Grantham Variation, GV | 95.81 | 0.00 | 21.61 | 46.24 | 48.73 | 164.10 | 164.10 |
| Grantham Distance, GD | 101.29 | 140.21 | 81.43 | 135.88 | 113.87 | 34.34 | 0.00 |
| Prediction | Unclassified | ED1 | ED2 | ED2 | ED2 | EN2 | EN1 |
| Frequencies in AJ PRCA | |||||||
| Families with affected male probandsa | 1/75 | 0/74 | 4/85b | 2/85 | 0/85 | 0/76 | 0/76 |
| PRCA cases from Montreal (n=150) | 0/139 | 0/136 | 0/136 | 4/141 | 1/143 | 0/141 | 0/141 |
| AJ Controls (n= 225)c | 2/225 | 0/222 | 0/221 | 5/225 | 0/225 | 0/224 | 0/224 |
| Unselected PRCA cases | 8/740 (1.08%) | 7/583 (1.2%) | |||||
| AJ Controls | 5/545 (0.92%) | 5/345 (1.5%) | |||||
n=85 for exon 11, n=76 for other exons;
Detected by SSCP while screening for S428F;
25 of the controls were from Montreal and 200 were from Israel. Abbreviations used: AJ = Ashkenazi Jewish; ED1 = enriched deleterious 1; ED2 = enriched deleterious 2; EN1 = enriched neutral 1; EN2 = enriched neutral 2; GV = Grantham Variation; GD = Grantham Deviation; MKSCC = Memorial Sloan Kettering Cancer Center; PRCA = Prostate cancer. The Align-GVGD analysis used a CHEK2 protein multiple sequence alignment containing 8 vertebrate CHEK2 sequences plus the sea urchin CHEK2 ortholog (see http://agvgd.iarc.fr/alignments.php for full description of Align-GVGD). Variant E394F has the highest potential for being disease-causing (ED1. Variants S428F, D438Y, and Y424H have a more moderate likelihood of being pathogenic (ED2).
We screened all seven CHEK2 variants in a series of 125 unselected AJ PRCA cases in 200 AJ controls, and in individuals from 76 AJ PRCA families. In the series of unselected cases and controls, variant S428F, which was previously associated with an increased risk of breast cancer in AJ women [42], was not found at a significantly increased frequency (2.8% in cases versus 2.2% in controls, p=.74). Of the CHEK2 variants identified in this study, only Y424H in exon 11 was observed in more than one of the 76 ICPCG-PRCA families studied. Exon 11, which includes both Y424H and S248F, was screened in 9 additional ICPCG families (for a total of 85 families with at least one affected genotyped). The S248F variant was found in 2 men from unique families. However, it was not present in the other affected male in the family in either case. The Y424H variant occurred in a total of 9 affected men in 4 different families. In one family, all 3 affected men had the Y424H variant and in a second family 4 of the 5 affected men carried the Y424H variant. For the other 2 families, only 1 affected out of 2 or 3 affected men had this variant. Thus, Y424H did not fully co-segregate with the PRCA phenotype and the variant was also observed in five unaffected individuals from the families. In order to estimate the frequency of Y424H and S428F in the AJ population, additional unselected AJ cases and controls were screened for the variant but no significant differences in frequency were seen (table 1). Immunohistochemical staining of CHEK2 in prostate cancer tissue samples from the Montreal cohort did not reveal any difference in intensity with normal tissue and there was no correlation with gleason score (data not shown).
Yeast complementation assay and MMS treatment
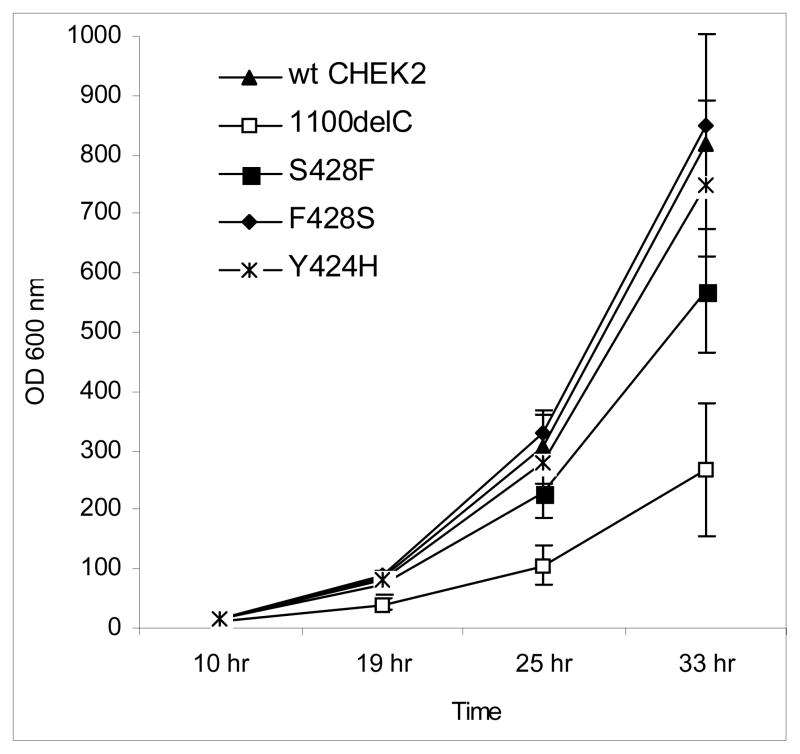
We performed functional assays in the budding yeast S. Cerevisae to investigate whether the Y424H or S428F substitution in CHEK2 protein would affect its function in growth of yeast cells. Transformation of Rad53-null sml1-null yeast strain with wild type human CHEK2 resulted in exponential growth (Figure 1), indicating that expression of wild type human CHEK2 suppressed lethality of the Rad53 deletion in yeast, presumably because of its functions in cell cycle checkpoints as previously reported [38; 42]. Yeast cells carrying the 1100delC mutant grew at a significantly lower rate than those carrying the wild type CHEK2. These results indicate that the 1100delC mutant fails to fully replace functions of Rad53 in the cell cycle, probably because the reduced kinase checkpoint activity in this mutant results in increased apoptosis. Transformation with the S428F variant also decreased cell proliferation, although the effect was weaker than that observed with 1100delC (Figure 1). Normal growth rate was restored in yeast transformed with the F428S plasmid, created using S428F to re-create the wild type CHEK2. Introduction of the Y424H mutation did not significantly affect growth rate. Cells transformed with all variants grew at rates similar to the wild type when their expression of CHEK2 was suppressed by replacing the 2% galactose in liquid media with 2% glucose, indicating that differences in growth rates observed in Figure 1 were due to expression of mutant or wild type CHEK2 from these plasmids (data not shown).
FIGURE 1.
Transformation with plasmids carrying wild type (wt) human CHEK2, as well as the Y424H variant, represses the normally reduced viability due to Rad53 deletion in the Rad53-null sml1-null yeast S. Cerevisae. Yeast carrying the CHEK2*1100delC variant grows less than that carrying the wild type, while yeast cells carrying the CHEK2*S428F variant grow more than cells carrying 1100delC but less than those carrying the wild type. Yeast carrying F428S, a mutant created from the S428F plasmid to re-create a wild type CHEK2 plasmid, grows as well as yeast carrying genuine wild type CHEK2. Time = time of measurement after inoculation of known number of yeast cells into fresh medium.
We determined the cytotoxic and genotoxic concentrations of MMS in our yeast strain. Homologous recombination-deficient cells are sensitive to MMS as it induces stalls in replication forks [43]. Cells carrying the 1100delC variant showed greater viability but overall slower growth rates over a 27-hr period as compared to S428F and Y424H in response to MMS treatment at a concentration of 0.00125%. These results are in agreement with previously published data showing that yeast transformed with the Rad53Ha allele, which has a reduced Rad53 kinase activity (and thus could be comparable to our 1100delC variant) was hypersensitive to hydroxyurea but hyposensitive to MMS treatment [44].
DISCUSSION
The results presented here suggest that although the CHEK2 Y424H variant occurs at a highly conserved position, it does not seem to play a significant role in predisposition to PRCA. While the results do not exclude the possibility that this is a modest-risk predisposition allele in the AJ population, functional studies in yeast indicate that Y424H does not have a significant deleterious effect.
Previous studies of CHEK2 mutations in men with PRCA from the US, Finland, Poland, and Sweden have found conflicting results. Dong et al. reported a total of 28 (4.8%) germline CHEK2 mutations (16 of which were unique) among 578 sporadic PRCA cases collected at the Mayo clinic compared to 6/423 (1.4%) unaffected men. Additional screening for CHEK2 mutations in 149 families with familial PRCA prostate cancer (a minimum of three men over at least two generations) revealed 11 mutations (5 unique) in nine families, but the frequency of CHEK2 mutations in the familial group was not significantly different from that in the control group [24].
In a Finnish study of 120 patients with hereditary PRCA, 537 unselected PRCA cases, and 480 controls, the 1100delC variant was at an increased frequency among the patients with hereditary PRCA compared to controls (OR 8.24; 95% confidence intervals, CI 1.49–45.54; p=0.02) [25]. The I157T missense variant also had significantly higher frequency among hereditary PRCA patients in Finland (OR 2.12; 95% C.I. 1.06–4.27; p=0.04). However, only small associations were found between patients with unselected PRCA cases and the variants 1100delC (odds ratio, OR 3.14; 95% confidence intervals, CI 0.65–15.16; p=.15) and I127T (odds ratio, OR 1.48; 95% confidence intervals, CI 0.89–2.46; p=.13). Cybulski et al. studied three common Polish CHEK2 mutations, 1100delC, IVS2+1G>A, and I157T, in 98 familial PRCA cases, 690 unselected cases, and 1921 controls [22; 23]. They reported an OR of 2.2 (p=.04) for unselected PRCA cases and the protein-truncating alleles (1100delC and IVS2+1G>A) and an OR of 1.7 (p=.002) for the I157T variant [22]. In the 98 familial cases (2.2 cases per pedigree), IVS2+1G>A had an OR of 12.1 (C.I. 2.8–51.4, p=.0002), 1100delC had an OR of 4.9 (C.I. 0.5–44.6 p=.11), I157T had an OR of 3.8 (C.I. 2.0–7.4, p=.00002), and at least one of the three variants was identified in 20.4% of familial cases [23]. In contrast, in a recent study where the CHEK2*1100delC variant was screened in 419 men diagnosed with PRCA in southern Sweden (145 sporadic cases and 274 familial/hereditary cases) the CHEK2*1100delC variant was found in 1.2% of the cases (sporadic: 0.7%; familial: 1.6%; hereditary: 1.4%) and in 1.0% of the controls, indicating that CHEK2*1100delC is not a clinically important high-risk variant for hereditary PRCA susceptibility in this population [26]. By comparison, in the study presented here we found one 1100delC variant in 131 cases (0.76%) in the Montreal series compared to an established population frequency of 0.3% in Ashkenazi Jewish controls [45]. However, the numbers of cases analyzed (131) is too small to draw any strong conclusions and it would be interesting to study larger numbers to investigate this further.
In summary, the most compelling CHEK2 variant identified in this study was Y424H. The complete conservation among eight vertebrate sequences plus sea urchin of Y at position 424 suggests that changing this amino acid may have biologically significant consequences, but the genetic and functional assay data presented here do not provide sufficient support for this view. Overall, based on the results of this study, we conclude that germline CHEK2 variants play a minor role, if any, in PRCA susceptibility in the AJ population and conservation alone is not a sufficient reason to suspect that an amino acid substitution in a protein may be pathogenic. Some ultra-conserved regions in human genome with no apparent functions have been reported [46], supporting this conclusion.
Acknowledgments
We acknowledge support from the National Institutes of Health, on behalf of ICPCG, (grant recipient and grant number in parentheses) (WBI, U01CA89600; KAC, CA79596; and PSN, CA78836), the Koodish Fellowship and the Evan Frankel Foundation (TK), the National Human Genome Research Institute and National Institutes of Health (EAO), the U.S. National Cancer Institute (JLS, CA080122), the Prostate Cancer Foundation and the Fred Hutchinson Cancer Research Center (JLS), the Department of Defense (DMK, W81XWH-04-1-0083), the Canadian Genetic Disease Network (WDF, DAMD17-00-1-0033), the Montreal Center for Experimental Therapeutics in cancer (AY), and the Intramural Program of the National Human Genome Research Institute. We are also grateful for the financial contribution of the Fonds de Recherche en Santé du Québec via their continuing support to the Research Institute of the McGill University Health Center. The authors gratefully acknowledge technical support from Drs. A. Al-Moustafa, S. Elledge, R. Rothstein, B. Bhullar, B. Andreson, and U. Stohaj in performing yeast functional assays.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Jacobs EJ, Rodriguez C, Mondul AM, Connell CJ, Henley SJ, Calle EE, Thun MJ. A large cohort study of aspirin and other nonsteroidal anti-inflammatory drugs and prostate cancer incidence. J Natl Cancer Inst. 2005;97:975–80. doi: 10.1093/jnci/dji173. [DOI] [PubMed] [Google Scholar]
- Carter BS, Beaty TH, Steinberg GD, Childs B, Walsh PC. Mendelian inheritance of familial prostate cancer. Proc Natl Acad Sci U S A. 1992;89:3367–71. doi: 10.1073/pnas.89.8.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronberg H, Damber L, Damber JE. Familial prostate cancer in Sweden. A nationwide register cohort study. Cancer. 1996;77:138–43. doi: 10.1002/(SICI)1097-0142(19960101)77:1<138::AID-CNCR23>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Cui J, Staples MP, Hopper JL, English DR, McCredie MR, Giles GG. Segregation analyses of 1,476 population-based Australian families affected by prostate cancer. Am J Hum Genet. 2001;68:1207–18. doi: 10.1086/320114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matikaine MP, Pukkala E, Schleutker J, Tammela TL, Koivisto P, Sankila R, Kallioniemi OP. Relatives of prostate cancer patients have an increased risk of prostate and stomach cancers: a population-based, cancer registry study in Finland. Cancer Causes Control. 2001;12:223–30. doi: 10.1023/a:1011283123610. [DOI] [PubMed] [Google Scholar]
- Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- Steinberg GD, Carter BS, Beaty TH, Childs B, Walsh PC. Family history and the risk of prostate cancer. Prostate. 1990;17:337–47. doi: 10.1002/pros.2990170409. [DOI] [PubMed] [Google Scholar]
- Simard J, Dumont M, Labuda D, Sinnett D, Meloche C, El-Alfy M, Berger L, Lees E, Labrie F, Tavtigian SV. Prostate cancer susceptibility genes: lessons learned and challenges posed. Endocr Relat Cancer. 2003;10:225–59. doi: 10.1677/erc.0.0100225. [DOI] [PubMed] [Google Scholar]
- Bruner DW, Moore D, Parlanti A, Dorgan J, Engstrom P. Relative risk of prostate cancer for men with affected relatives: systematic review and meta-analysis. Int J Cancer. 2003;107:797–803. doi: 10.1002/ijc.11466. [DOI] [PubMed] [Google Scholar]
- Chaturvedi P, Eng WK, Zhu Y, Mattern MR, Mishra R, Hurle MR, Zhang X, Annan RS, Lu Q, Faucette LF, Scott GF, Li X, Carr SA, Johnson RK, Winkler JD, Zhou BB. Mammalian Chk2 is a downstream effector of the ATM-dependent DNA damage checkpoint pathway. Oncogene. 1999;18:4047–54. doi: 10.1038/sj.onc.1202925. [DOI] [PubMed] [Google Scholar]
- Falck J, Mailand N, Syljuasen RG, Bartek J, Lukas J. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature. 2001;410:842–7. doi: 10.1038/35071124. [DOI] [PubMed] [Google Scholar]
- Chehab NH, Malikzay A, Appel M, Halazonetis TD. Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes Dev. 2000;14:278–88. [PMC free article] [PubMed] [Google Scholar]
- Shieh SY, Ahn J, Tamai K, Taya Y, Prives C. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 2000;14:289–300. [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Collins KM, Brown AL, Lee CH, Chung JH. hCds1-mediated phosphorylation of BRCA1 regulates the DNA damage response. Nature. 2000;404:201–4. doi: 10.1038/35004614. [DOI] [PubMed] [Google Scholar]
- Bell DW, Varley JM, Szydlo TE, Kang DH, Wahrer DC, Shannon KE, Lubratovich M, Verselis SJ, Isselbacher KJ, Fraumeni JF, Birch JM, Li FP, Garber JE, Haber DA. Heterozygous germ line hCHK2 mutations in Li-Fraumeni syndrome. Science. 1999;286:2528–31. doi: 10.1126/science.286.5449.2528. [DOI] [PubMed] [Google Scholar]
- Vahteristo P, Tamminen A, Karvinen P, Eerola H, Eklund C, Aaltonen LA, Blomqvist C, Aittomaki K, Nevanlinna H. p53, CHK2, and CHK1 genes in Finnish families with Li-Fraumeni syndrome: further evidence of CHK2 in inherited cancer predisposition. Cancer Res. 2001;61:5718–22. [PubMed] [Google Scholar]
- Vahteristo P, Bartkova J, Eerola H, Syrjakoski K, Ojala S, Kilpivaara O, Tamminen A, Kononen J, Aittomaki K, Heikkila P, Holli K, Blomqvist C, Bartek J, Kallioniemi OP, Nevanlinna H. A CHEK2 genetic variant contributing to a substantial fraction of familial breast cancer. Am J Hum Genet. 2002;71:432–8. doi: 10.1086/341943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijers-Heijboer H, van den Ouweland A, Klijn J, Wasielewski M, de Snoo A, Oldenburg R, Hollestelle A, Houben M, Crepin E, van Veghel-Plandsoen M, Elstrodt F, van Duijn C, Bartels C, Meijers C, Schutte M, McGuffog L, Thompson D, Easton D, Sodha N, Seal S, Barfoot R, Mangion J, Chang-Claude J, Eccles D, Eeles R, Evans DG, Houlston R, Murday V, Narod S, Peretz T, Peto J, Phelan C, Zhang HX, Szabo C, Devilee P, Goldgar D, Futreal PA, Nathanson KL, Weber B, Rahman N, Stratton MR. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet. 2002;31:55–9. doi: 10.1038/ng879. [DOI] [PubMed] [Google Scholar]
- Oldenburg RA, Kroeze-Jansema K, Kraan J, Morreau H, Klijn JG, Hoogerbrugge N, Ligtenberg MJ, van Asperen CJ, Vasen HF, Meijers C, Meijers-Heijboer H, de Bock TH, Cornelisse CJ, Devilee P. The CHEK2*1100delC variant acts as a breast cancer risk modifier in non-BRCA1/BRCA2 multiple-case families. Cancer Res. 2003;63:8153–7. [PubMed] [Google Scholar]
- Xu J, Gillanders EM, Isaacs SD, Chang BL, Wiley KE, Zheng SL, Jones M, Gildea D, Riedesel E, Albertus J, Freas-Lutz D, Markey C, Meyers DA, Walsh PC, Trent JM, Isaacs WB. Genome-wide scan for prostate cancer susceptibility genes in the Johns Hopkins hereditary prostate cancer families. Prostate. 2003;57:320–5. doi: 10.1002/pros.10306. [DOI] [PubMed] [Google Scholar]
- Camp NJ, Cannon-Albright LA, Farnham JM, Baffoe-Bonnie AB, George A, Powell I, Bailey-Wilson JE, Carpten JD, Giles GG, Hopper JL, Severi G, English DR, Foulkes WD, Maehle L, Moller P, Eeles R, Easton D, Badzioch MD, Whittemore AS, Oakley-Girvan I, Hsieh CL, Dimitrov L, Xu J, Stanford JL, Johanneson B, Deutsch K, McIntosh L, Ostrander EA, Wiley KE, Isaacs SD, Walsh PC, Thibodeau SN, McDonnell SK, Hebbring S, Schaid DJ, Lange EM, Cooney KA, Tammela TL, Schleutker J, Paiss T, Maier C, Gronberg H, Wiklund F, Emanuelsson M, Isaacs WB. Compelling evidence for a prostate cancer gene at 22q12.3 by the International Consortium for Prostate Cancer Genetics. Hum Mol Genet. 2007;16:1271–8. doi: 10.1093/hmg/ddm075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cybulski C, Gorski B, Huzarski T, Masojc B, Mierzejewski M, Debniak T, Teodorczyk U, Byrski T, Gronwald J, Matyjasik J, Zlowocka E, Lenner M, Grabowska E, Nej K, Castaneda J, Medrek K, Szymanska A, Szymanska J, Kurzawski G, Suchy J, Oszurek O, Witek A, Narod SA, Lubinski J. CHEK2 is a multiorgan cancer susceptibility gene. Am J Hum Genet. 2004;75:1131–5. doi: 10.1086/426403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cybulski C, Huzarski T, Gorski B, Masojc B, Mierzejewski M, Debniak T, Gliniewicz B, Matyjasik J, Zlowocka E, Kurzawski G, Sikorski A, Posmyk M, Szwiec M, Czajka R, Narod SA, Lubinski J. A novel founder CHEK2 mutation is associated with increased prostate cancer risk. Cancer Res. 2004;64:2677–9. doi: 10.1158/0008-5472.can-04-0341. [DOI] [PubMed] [Google Scholar]
- Dong X, Wang L, Taniguchi K, Wang X, Cunningham JM, McDonnell SK, Qian C, Marks AF, Slager SL, Peterson BJ, Smith DI, Cheville JC, Blute ML, Jacobsen SJ, Schaid DJ, Tindall DJ, Thibodeau SN, Liu W. Mutations in CHEK2 associated with prostate cancer risk. Am J Hum Genet. 2003;72:270–80. doi: 10.1086/346094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppala EH, Ikonen T, Mononen N, Autio V, Rokman A, Matikainen MP, Tammela TL, Schleutker J. CHEK2 variants associate with hereditary prostate cancer. Br J Cancer. 2003;89:1966–70. doi: 10.1038/sj.bjc.6601425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagenius M, Borg A, Johansson L, Giwercman A, Bratt O. CHEK2*1100delC is not an important high-risk gene in families with hereditary prostate cancer in southern Sweden. Scand J Urol Nephrol. 2006;40:23–5. doi: 10.1080/00365590500368518. [DOI] [PubMed] [Google Scholar]
- Kirchhoff T, Kauff ND, Mitra N, Nafa K, Huang H, Palmer C, Gulati T, Wadsworth E, Donat S, Robson ME, Ellis NA, Offit K. BRCA mutations and risk of prostate cancer in Ashkenazi Jews. Clin Cancer Res. 2004;10:2918–21. doi: 10.1158/1078-0432.ccr-03-0604. [DOI] [PubMed] [Google Scholar]
- Sodha N, Williams R, Mangion J, Bullock SL, Yuille MR, Eeles RA. Screening hCHK2 for mutations. Science. 2000;289:359. doi: 10.1126/science.289.5478.359a. [DOI] [PubMed] [Google Scholar]
- Sodha N, Houlston RS, Williams R, Yuille MA, Mangion J, Eeles RA. A robust method for detecting CHK2/RAD53 mutations in genomic DNA. Hum Mutat. 2002;19:173–7. doi: 10.1002/humu.10031. [DOI] [PubMed] [Google Scholar]
- Hamel N, Kotar K, Foulkes WD. Founder mutations in BRCA1/2 are not frequent in Canadian Ashkenazi Jewish men with prostate cancer. BMC Med Genet. 2003;4:7. doi: 10.1186/1471-2350-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange EM, Gillanders EM, Davis CC, Brown WM, Campbell JK, Jones M, Gildea D, Riedesel E, Albertus J, Freas-Lutz D, Markey C, Giri V, Dimmer JB, Montie JE, Trent JM, Cooney KA. Genome-wide scan for prostate cancer susceptibility genes using families from the University of Michigan prostate cancer genetics project finds evidence for linkage on chromosome 17 near BRCA1. Prostate. 2003;57:326–34. doi: 10.1002/pros.10307. [DOI] [PubMed] [Google Scholar]
- Janer M, Friedrichsen DM, Stanford JL, Badzioch MD, Kolb S, Deutsch K, Peters MA, Goode EL, Welti R, DeFrance HB, Iwasaki L, Li S, Hood L, Ostrander EA, Jarvik GP. Genomic scan of 254 hereditary prostate cancer families. Prostate. 2003;57:309–19. doi: 10.1002/pros.10305. [DOI] [PubMed] [Google Scholar]
- Friedrichsen DM, Stanford JL, Isaacs SD, Janer M, Chang BL, Deutsch K, Gillanders E, Kolb S, Wiley KE, Badzioch MD, Zheng SL, Walsh PC, Jarvik GP, Hood L, Trent JM, Isaacs WB, Ostrander EA, Xu J. Identification of a prostate cancer susceptibility locus on chromosome 7q11–21 in Jewish families. Proc Natl Acad Sci U S A. 2004;101:1939–44. doi: 10.1073/pnas.0308336100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Nickerson DA. Variation is the spice of life. Nat Genet. 2001;27:234–6. doi: 10.1038/85776. [DOI] [PubMed] [Google Scholar]
- Poirot O, Suhre K, Abergel C, O’Toole E, Notredame C. 3DCoffee@igs: a web server for combining sequences and structures into a multiple sequence alignment. Nucleic Acids Res. 2004;32:W37–40. doi: 10.1093/nar/gkh382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavtigian SV, Deffenbaugh AM, Yin L, Judkins T, Scholl T, Samollow PB, de Silva D, Zharkikh A, Thomas A. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J Med Genet. 2006;43:295–305. doi: 10.1136/jmg.2005.033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Huang M, Elledge SJ. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science. 1998;282:1893–7. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- Zhao X, Chabes A, Domkin V, Thelander L, Rothstein R. The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. Embo J. 2001;20:3544–53. doi: 10.1093/emboj/20.13.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elble R. A simple and efficient procedure for transformation of yeasts. Biotechniques. 1992;13:18–20. [PubMed] [Google Scholar]
- Gietz RD, Woods RA. Yeast transformation by the LiAc/SS Carrier DNA/PEG method. Methods Mol Biol. 2006;313:107–20. doi: 10.1385/1-59259-958-3:107. [DOI] [PubMed] [Google Scholar]
- Shaag A, Walsh T, Renbaum P, Kirchhoff T, Nafa K, Shiovitz S, Mandell JB, Welcsh P, Lee MK, Ellis N, Offit K, Levy-Lahad E, King MC. Functional and genomic approaches reveal an ancient CHEK2 allele associated with breast cancer in the Ashkenazi Jewish population. Hum Mol Genet. 2005 doi: 10.1093/hmg/ddi052. [DOI] [PubMed] [Google Scholar]
- Lundin C, North M, Erixon K, Walters K, Jenssen D, Goldman AS, Helleday T. Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res. 2005;33:3799–811. doi: 10.1093/nar/gki681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordon-Preciado V, Ufano S, Bueno A. Limiting amounts of budding yeast Rad53 S-phase checkpoint activity results in increased resistance to DNA alkylation damage. Nucleic Acids Res. 2006;34:5852–62. doi: 10.1093/nar/gkl741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offit K, Pierce H, Kirchhoff T, Kolachana P, Rapaport B, Gregersen P, Johnson S, Yossepowitch O, Huang H, Satagopan J, Robson M, Scheuer L, Nafa K, Ellis N. Frequency of CHEK2*1100delC in New York breast cancer cases and controls. BMC Med Genet. 2003;4:1. doi: 10.1186/1471-2350-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–5. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]