Abstract
The title compound, C34H47NO12, is the norditerpenoid alkaloid aconifine isolated from the leaves and tubers of Aconitum karakolicum Rapaics. It has a lycoctonine carbon skeleton and contains four six-membered rings and two five-membered rings; its geometry is similar to that observed in other lycoctonine-type diterpenoid alkaloids. There are two intramolecular O—H⋯O hydrogen bonds which close five- and seven-membered pseudo-rings, respectively. In the crystal, two intermolecular O—H⋯O hydrogen bonds cross-link the molecules into double chains along the a axis.
Related literature
For the isolation of aconifine, see: Sultankhodzhaev et al. (1973 ▶). For spectroscopic data and the chemical structure of aconifine, see: Sultankhodzhaev et al. (1980 ▶). For the neurocardiotoxic activity of aconifine, see: Dzhakhangirov et al. (1997 ▶). For the neurocardiotoxic activity of lycoctonine alkaloids, see: Dzhakhangirov et al. (1976 ▶). For general background to lycoctonine alkaloids and their structures, see: Joshi & Pelletier (1987 ▶).
Experimental
Crystal data
C34H47NO12
M r = 661.73
Orthorhombic,
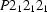
a = 12.0213 (3) Å
b = 15.4938 (6) Å
c = 17.1038 (4) Å
V = 3185.68 (16) Å3
Z = 4
Cu Kα radiation
μ = 0.87 mm−1
T = 100 K
0.40 × 0.30 × 0.25 mm
Data collection
Oxford Diffraction Xcalibur Ruby diffractometer
Absorption correction: multi-scan (CrysAlisPro; Oxford Diffraction, 2009 ▶) T min = 0.767, T max = 0.811
11111 measured reflections
6334 independent reflections
6050 reflections with I > 2σ(I)
R int = 0.024
Refinement
R[F 2 > 2σ(F 2)] = 0.037
wR(F 2) = 0.098
S = 1.06
6334 reflections
451 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.36 e Å−3
Δρmin = −0.22 e Å−3
Absolute structure: Flack (1983 ▶), 2633 Friedel pairs
Flack parameter: 0.04 (10)
Data collection: CrysAlisPro (Oxford Diffraction, 2009 ▶); cell refinement: CrysAlisPro; data reduction: CrysAlisPro; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809021436/zl2215sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809021436/zl2215Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O11—H11⋯O5 | 0.83 (3) | 2.07 (3) | 2.791 (2) | 146 (3) |
| O8—H8⋯O12 | 0.83 (3) | 2.11 (3) | 2.598 (2) | 117 (3) |
| O2—H2⋯O7i | 0.87 (3) | 2.21 (3) | 3.066 (2) | 168 (3) |
| O8—H8⋯O2ii | 0.83 (3) | 2.39 (3) | 2.928 (2) | 123 (3) |
Symmetry codes: (i) 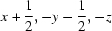 ; (ii)
; (ii) 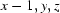 .
.
Acknowledgments
We thank the Academy of Sciences of Uzbekistan for supporting this study.
supplementary crystallographic information
Comment
The norditerpenoid alkaloid aconifine was isolated from leaves and tubers of Aconitum karakolicum Rapaics (Sultankhodzhaev et al.,1980). It exhibits neurocardiotoxic properties (Dzhakhangirov et al., 1997) similar to those of aconitine (Dzhakhangirov et al., 1976). The molecular structure of the title compound is shown in Fig. 1. Aconifine has a lycoctonine carbon skeleton; its geometry is similar to that observed in other lycoctonine type diterpenoid alkaloids (Joshi et al.,1987).
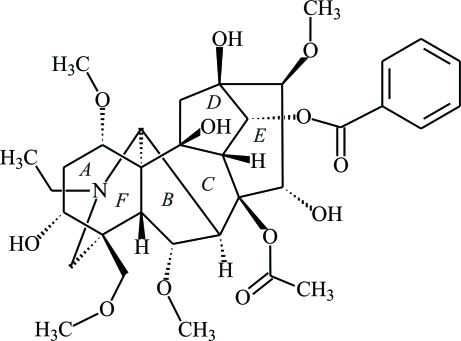
The lycoctonine carbon skeleton, contains four six-membered rings, (A, C, E and F), and two five-membered rings (B and D) (Fig. 2). Rings A and C have more or less regular chair conformations, whereas ring F shows significant distortions and ring E adopts a sofa conformation. The five-membered rings B and D have envelope conformations.
The position and orientation of the 10 oxo substituents on the carbon lycoctonine skeleton are 1α, 3α, 4β, 6α, 8β, 10β, 13β, 14α, 15α, 16β, which confirms the earlier structure assignment based on spectral data (Sultankhodzhaev et al., 1980).
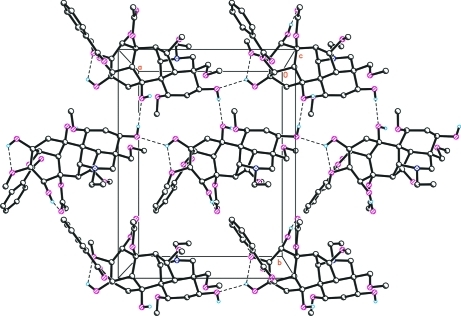
In the crystal structure of the title compound there are four acidic H atoms that can participate in H-bonds. The H8 and H11 hydroxyl hydrogen atoms take part in intramolecular H-bonds which close 5 and 7-membered pseudo-cycles, respectively. Hydroxyl hydrogen atom H7 does not take a part in any H-bonding interactions. Atoms H2 and H8 participate in intermolecular O—H···O bonds which link the molecules into infinite double chains along the a-axis (Table 1; Fig.3). The hydrogen atom H8 forms a bifurcated H-bond to both O2 and O7.
Experimental
The title compound was isolated from the chloroform fraction of the tubers of Aconitum karakolicum Rapaics by a known method (Sultankhodzhaev et al., 1973). Crystals suitable for X-ray analysis were obtained by slow evaporation of a methanol solution at room temperature (m.p. 471–473 K).
Refinement
The hydroxyl hydrogen atoms were located in a difference Fourier map and refined isotropically. The H atoms bonded to C atoms were placed geometrically (with C—H distances of 0.98 Å for CH; 0.97 Å for CH2; 0.96 Å for CH3; and 0.93 Å for Car) and included in the refinement in a riding motion approximation with Uiso=1.2Ueq(C) [Uiso=1.5Ueq(C) for methyl H atoms].
Figures
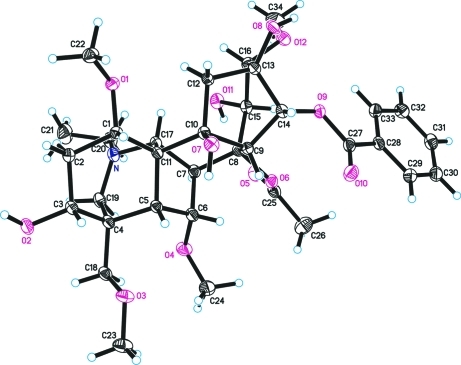
Fig. 1.
The molecular structure of aconifine, showing the atomic numbering scheme and displacement ellipsoids drawn at the 50% probability level.
Fig. 2.
Ring assignments in Aconifine
Fig. 3.
Crystal packing of aconifine, viewed down the c-axis; H-bonds shown as dashed lines. H-atoms not involved in hydrogen bonding are omitted for clarity.
Crystal data
| C34H47NO12 | Dx = 1.380 Mg m−3 |
| Mr = 661.73 | Melting point: 472(2) K |
| Orthorhombic, P212121 | Cu Kα radiation, λ = 1.54184 Å |
| Hall symbol: P 2ac 2ab | Cell parameters from 11111 reflections |
| a = 12.0213 (3) Å | θ = 3.7–75.5° |
| b = 15.4938 (6) Å | µ = 0.87 mm−1 |
| c = 17.1038 (4) Å | T = 100 K |
| V = 3185.68 (16) Å3 | Prizmatic, colourless |
| Z = 4 | 0.40 × 0.30 × 0.25 mm |
| F(000) = 1416 |
Data collection
| Oxford Diffraction Xcalibur Ruby diffractometer | 6334 independent reflections |
| Radiation source: Enhance (Cu) X-ray Source | 6050 reflections with I > 2σ(I) |
| graphite | Rint = 0.024 |
| Detector resolution: 10.2576 pixels mm-1 | θmax = 75.6°, θmin = 3.9° |
| ω scans | h = −15→9 |
| Absorption correction: multi-scan (CrysAlis PRO; Oxford Diffraction, 2009) | k = −19→19 |
| Tmin = 0.767, Tmax = 0.811 | l = −21→21 |
| 11111 measured reflections |
Refinement
| Refinement on F2 | Hydrogen site location: inferred from neighbouring sites |
| Least-squares matrix: full | H atoms treated by a mixture of independent and constrained refinement |
| R[F2 > 2σ(F2)] = 0.037 | w = 1/[σ2(Fo2) + (0.0771P)2] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.098 | (Δ/σ)max < 0.001 |
| S = 1.06 | Δρmax = 0.36 e Å−3 |
| 6334 reflections | Δρmin = −0.22 e Å−3 |
| 451 parameters | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| 0 restraints | Extinction coefficient: 0.00043 (12) |
| Primary atom site location: structure-invariant direct methods | Absolute structure: Flack (1983), 2633 Friedel pairs |
| Secondary atom site location: difference Fourier map | Flack parameter: 0.04 (10) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.66866 (9) | −0.16533 (7) | −0.10572 (6) | 0.0176 (2) | |
| O2 | 1.04961 (9) | −0.13201 (8) | −0.02826 (7) | 0.0209 (2) | |
| O3 | 0.98504 (9) | −0.08240 (8) | 0.16387 (7) | 0.0217 (2) | |
| O4 | 0.82173 (10) | 0.09161 (8) | 0.13342 (7) | 0.0232 (2) | |
| O5 | 0.55172 (11) | 0.23615 (8) | 0.08471 (7) | 0.0251 (3) | |
| O6 | 0.53676 (9) | 0.10653 (7) | 0.14475 (6) | 0.0171 (2) | |
| O7 | 0.58835 (9) | −0.18623 (7) | 0.09376 (6) | 0.0175 (2) | |
| O8 | 0.29051 (9) | −0.12191 (8) | −0.00458 (6) | 0.0183 (2) | |
| O9 | 0.33324 (9) | 0.00330 (7) | 0.12982 (6) | 0.0162 (2) | |
| O10 | 0.38215 (10) | −0.02601 (8) | 0.25398 (6) | 0.0234 (2) | |
| O11 | 0.50108 (10) | 0.13929 (8) | −0.04816 (7) | 0.0213 (2) | |
| O12 | 0.26888 (9) | 0.04325 (8) | −0.02596 (7) | 0.0210 (2) | |
| N | 0.79454 (11) | 0.02206 (9) | −0.06929 (8) | 0.0169 (3) | |
| C1 | 0.73828 (12) | −0.16204 (9) | −0.03752 (8) | 0.0145 (3) | |
| H1A | 0.7261 | −0.2152 | −0.0076 | 0.017* | |
| C2 | 0.85921 (13) | −0.16088 (10) | −0.06393 (9) | 0.0170 (3) | |
| H2A | 0.8792 | −0.2176 | −0.0834 | 0.020* | |
| H2B | 0.8673 | −0.1202 | −0.1066 | 0.020* | |
| C3 | 0.93799 (12) | −0.13616 (10) | 0.00116 (9) | 0.0163 (3) | |
| H3A | 0.9347 | −0.1807 | 0.0418 | 0.020* | |
| C4 | 0.90603 (12) | −0.04888 (9) | 0.03803 (9) | 0.0157 (3) | |
| C5 | 0.79025 (12) | −0.06011 (9) | 0.07794 (8) | 0.0143 (3) | |
| H5A | 0.7935 | −0.1039 | 0.1192 | 0.017* | |
| C6 | 0.74394 (13) | 0.02751 (10) | 0.11062 (9) | 0.0170 (3) | |
| H6A | 0.6966 | 0.0146 | 0.1558 | 0.020* | |
| C7 | 0.66856 (12) | 0.06323 (9) | 0.04426 (8) | 0.0155 (3) | |
| H7A | 0.6874 | 0.1233 | 0.0320 | 0.019* | |
| C8 | 0.54779 (12) | 0.05540 (9) | 0.07171 (8) | 0.0147 (3) | |
| C9 | 0.52897 (12) | −0.03713 (10) | 0.10283 (8) | 0.0140 (3) | |
| H9A | 0.5578 | −0.0423 | 0.1562 | 0.017* | |
| C10 | 0.58311 (13) | −0.10691 (9) | 0.04910 (8) | 0.0143 (3) | |
| C11 | 0.70101 (12) | −0.08482 (10) | 0.01411 (8) | 0.0137 (3) | |
| C12 | 0.49078 (12) | −0.12252 (10) | −0.01330 (8) | 0.0147 (3) | |
| H12A | 0.5152 | −0.1018 | −0.0640 | 0.018* | |
| H12B | 0.4746 | −0.1837 | −0.0175 | 0.018* | |
| C13 | 0.38627 (12) | −0.07295 (10) | 0.01320 (8) | 0.0156 (3) | |
| C14 | 0.40634 (12) | −0.06253 (9) | 0.10058 (8) | 0.0151 (3) | |
| H14A | 0.3941 | −0.1172 | 0.1281 | 0.018* | |
| C15 | 0.45606 (12) | 0.08664 (10) | 0.01258 (8) | 0.0157 (3) | |
| H15A | 0.4053 | 0.1236 | 0.0424 | 0.019* | |
| C16 | 0.38277 (13) | 0.01688 (10) | −0.02702 (8) | 0.0167 (3) | |
| H16A | 0.4066 | 0.0104 | −0.0815 | 0.020* | |
| C17 | 0.69192 (12) | 0.00319 (9) | −0.02677 (9) | 0.0149 (3) | |
| H17A | 0.6284 | 0.0035 | −0.0627 | 0.018* | |
| C18 | 0.99741 (13) | −0.02636 (10) | 0.09801 (9) | 0.0177 (3) | |
| H18A | 1.0703 | −0.0340 | 0.0747 | 0.021* | |
| H18B | 0.9902 | 0.0333 | 0.1143 | 0.021* | |
| C19 | 0.89793 (13) | 0.02424 (10) | −0.02277 (9) | 0.0181 (3) | |
| H19A | 0.9025 | 0.0793 | 0.0041 | 0.022* | |
| H19B | 0.9610 | 0.0203 | −0.0579 | 0.022* | |
| C20 | 0.78363 (14) | 0.09877 (11) | −0.11883 (10) | 0.0221 (3) | |
| H20A | 0.7980 | 0.1499 | −0.0878 | 0.026* | |
| H20B | 0.7080 | 0.1024 | −0.1384 | 0.026* | |
| C21 | 0.86362 (17) | 0.09632 (14) | −0.18710 (11) | 0.0345 (4) | |
| H21A | 0.8541 | 0.0433 | −0.2153 | 0.052* | |
| H21B | 0.9386 | 0.1000 | −0.1681 | 0.052* | |
| H21C | 0.8489 | 0.1442 | −0.2213 | 0.052* | |
| C22 | 0.65941 (14) | −0.24944 (10) | −0.13754 (9) | 0.0204 (3) | |
| H22A | 0.7274 | −0.2644 | −0.1636 | 0.031* | |
| H22B | 0.5991 | −0.2510 | −0.1743 | 0.031* | |
| H22C | 0.6454 | −0.2900 | −0.0963 | 0.031* | |
| C23 | 1.06760 (15) | −0.06738 (12) | 0.22176 (10) | 0.0264 (4) | |
| H23A | 1.0660 | −0.0078 | 0.2371 | 0.040* | |
| H23B | 1.1396 | −0.0811 | 0.2009 | 0.040* | |
| H23C | 1.0530 | −0.1031 | 0.2665 | 0.040* | |
| C24 | 0.84230 (16) | 0.09095 (14) | 0.21572 (11) | 0.0320 (4) | |
| H24A | 0.7750 | 0.1051 | 0.2431 | 0.048* | |
| H24B | 0.8987 | 0.1327 | 0.2280 | 0.048* | |
| H24C | 0.8670 | 0.0346 | 0.2313 | 0.048* | |
| C25 | 0.54687 (13) | 0.19314 (10) | 0.14337 (10) | 0.0206 (3) | |
| C26 | 0.55057 (18) | 0.22808 (12) | 0.22522 (11) | 0.0316 (4) | |
| H26A | 0.6043 | 0.1964 | 0.2552 | 0.047* | |
| H26B | 0.4786 | 0.2223 | 0.2489 | 0.047* | |
| H26C | 0.5711 | 0.2879 | 0.2239 | 0.047* | |
| C27 | 0.33426 (13) | 0.01855 (10) | 0.20752 (9) | 0.0170 (3) | |
| C28 | 0.26739 (12) | 0.09676 (10) | 0.22593 (9) | 0.0170 (3) | |
| C29 | 0.26246 (13) | 0.12388 (11) | 0.30387 (9) | 0.0198 (3) | |
| H29 | 0.3007 | 0.0937 | 0.3424 | 0.024* | |
| C30 | 0.19980 (14) | 0.19638 (11) | 0.32318 (9) | 0.0223 (3) | |
| H30 | 0.1962 | 0.2147 | 0.3749 | 0.027* | |
| C31 | 0.14270 (14) | 0.24149 (10) | 0.26579 (10) | 0.0227 (3) | |
| H31 | 0.1015 | 0.2902 | 0.2790 | 0.027* | |
| C32 | 0.14703 (14) | 0.21384 (11) | 0.18828 (10) | 0.0221 (3) | |
| H32 | 0.1080 | 0.2437 | 0.1500 | 0.027* | |
| C33 | 0.20932 (13) | 0.14203 (11) | 0.16821 (9) | 0.0195 (3) | |
| H33 | 0.2125 | 0.1239 | 0.1164 | 0.023* | |
| C34 | 0.24229 (16) | 0.10857 (13) | −0.08130 (11) | 0.0303 (4) | |
| H34A | 0.2836 | 0.1600 | −0.0694 | 0.045* | |
| H34B | 0.1641 | 0.1208 | −0.0790 | 0.045* | |
| H34C | 0.2613 | 0.0890 | −0.1329 | 0.045* | |
| H2 | 1.061 (2) | −0.1796 (17) | −0.0539 (15) | 0.032 (6)* | |
| H7 | 0.640 (2) | −0.1782 (18) | 0.1299 (17) | 0.040 (7)* | |
| H8 | 0.237 (2) | −0.0881 (15) | −0.0020 (13) | 0.023 (5)* | |
| H11 | 0.519 (2) | 0.1832 (19) | −0.0238 (17) | 0.041 (7)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0172 (5) | 0.0198 (5) | 0.0158 (5) | 0.0027 (4) | −0.0020 (4) | −0.0033 (4) |
| O2 | 0.0136 (5) | 0.0231 (6) | 0.0259 (5) | 0.0012 (4) | 0.0028 (4) | −0.0038 (5) |
| O3 | 0.0188 (5) | 0.0242 (6) | 0.0221 (5) | −0.0053 (5) | −0.0068 (4) | 0.0021 (5) |
| O4 | 0.0197 (6) | 0.0220 (5) | 0.0280 (6) | −0.0039 (5) | −0.0034 (4) | −0.0074 (5) |
| O5 | 0.0278 (6) | 0.0180 (5) | 0.0294 (6) | −0.0022 (5) | 0.0028 (5) | −0.0010 (5) |
| O6 | 0.0167 (5) | 0.0162 (5) | 0.0183 (5) | −0.0002 (4) | 0.0021 (4) | −0.0033 (4) |
| O7 | 0.0168 (5) | 0.0162 (5) | 0.0196 (5) | −0.0005 (4) | −0.0001 (4) | 0.0022 (4) |
| O8 | 0.0109 (5) | 0.0204 (5) | 0.0237 (5) | −0.0001 (5) | −0.0016 (4) | −0.0030 (4) |
| O9 | 0.0136 (5) | 0.0195 (5) | 0.0155 (5) | 0.0026 (4) | 0.0018 (4) | −0.0008 (4) |
| O10 | 0.0243 (6) | 0.0280 (6) | 0.0180 (5) | 0.0066 (5) | 0.0015 (4) | 0.0020 (5) |
| O11 | 0.0229 (6) | 0.0199 (5) | 0.0210 (5) | 0.0006 (5) | 0.0027 (4) | 0.0060 (5) |
| O12 | 0.0165 (5) | 0.0228 (6) | 0.0238 (5) | 0.0051 (4) | −0.0029 (4) | 0.0014 (5) |
| N | 0.0148 (6) | 0.0172 (6) | 0.0188 (6) | 0.0009 (5) | 0.0032 (5) | 0.0030 (5) |
| C1 | 0.0131 (7) | 0.0155 (6) | 0.0148 (6) | 0.0004 (5) | −0.0011 (5) | −0.0012 (5) |
| C2 | 0.0146 (7) | 0.0193 (7) | 0.0171 (6) | 0.0023 (6) | 0.0007 (5) | −0.0032 (5) |
| C3 | 0.0115 (6) | 0.0177 (7) | 0.0197 (7) | 0.0005 (5) | 0.0006 (5) | −0.0006 (6) |
| C4 | 0.0112 (6) | 0.0161 (7) | 0.0199 (6) | −0.0009 (6) | 0.0005 (5) | −0.0006 (5) |
| C5 | 0.0116 (6) | 0.0149 (6) | 0.0163 (6) | −0.0017 (5) | 0.0005 (5) | −0.0002 (5) |
| C6 | 0.0134 (7) | 0.0175 (7) | 0.0201 (7) | −0.0005 (6) | 0.0007 (5) | −0.0019 (5) |
| C7 | 0.0139 (7) | 0.0149 (6) | 0.0178 (6) | −0.0017 (5) | 0.0012 (5) | −0.0004 (5) |
| C8 | 0.0144 (6) | 0.0135 (6) | 0.0163 (6) | −0.0002 (5) | −0.0002 (5) | −0.0019 (5) |
| C9 | 0.0128 (7) | 0.0152 (6) | 0.0140 (6) | −0.0012 (5) | 0.0011 (5) | 0.0001 (5) |
| C10 | 0.0136 (7) | 0.0138 (6) | 0.0153 (6) | −0.0004 (5) | 0.0010 (5) | 0.0006 (5) |
| C11 | 0.0106 (6) | 0.0154 (6) | 0.0152 (6) | 0.0004 (5) | 0.0006 (5) | −0.0001 (5) |
| C12 | 0.0113 (6) | 0.0162 (7) | 0.0167 (6) | 0.0001 (5) | −0.0005 (5) | −0.0023 (5) |
| C13 | 0.0129 (6) | 0.0170 (7) | 0.0170 (6) | −0.0009 (6) | −0.0001 (5) | −0.0011 (5) |
| C14 | 0.0137 (7) | 0.0149 (6) | 0.0166 (6) | 0.0005 (5) | 0.0012 (5) | 0.0015 (5) |
| C15 | 0.0153 (6) | 0.0146 (6) | 0.0173 (6) | 0.0018 (6) | 0.0015 (5) | 0.0032 (5) |
| C16 | 0.0165 (7) | 0.0184 (7) | 0.0151 (6) | 0.0032 (6) | −0.0005 (5) | −0.0008 (5) |
| C17 | 0.0124 (6) | 0.0150 (6) | 0.0173 (6) | 0.0004 (5) | 0.0010 (5) | −0.0006 (5) |
| C18 | 0.0138 (6) | 0.0175 (7) | 0.0217 (7) | −0.0024 (6) | −0.0002 (5) | −0.0017 (6) |
| C19 | 0.0146 (7) | 0.0172 (7) | 0.0225 (7) | −0.0010 (6) | 0.0034 (6) | 0.0022 (6) |
| C20 | 0.0224 (8) | 0.0220 (8) | 0.0217 (7) | 0.0003 (6) | 0.0033 (6) | 0.0056 (6) |
| C21 | 0.0328 (10) | 0.0407 (11) | 0.0299 (9) | 0.0001 (8) | 0.0118 (8) | 0.0116 (8) |
| C22 | 0.0215 (7) | 0.0204 (7) | 0.0194 (7) | −0.0025 (6) | −0.0008 (6) | −0.0023 (6) |
| C23 | 0.0236 (8) | 0.0293 (9) | 0.0263 (8) | −0.0067 (7) | −0.0086 (6) | 0.0000 (7) |
| C24 | 0.0226 (8) | 0.0441 (11) | 0.0294 (9) | −0.0014 (8) | −0.0025 (6) | −0.0159 (8) |
| C25 | 0.0158 (7) | 0.0168 (7) | 0.0291 (8) | −0.0004 (6) | 0.0020 (6) | −0.0034 (6) |
| C26 | 0.0426 (11) | 0.0229 (8) | 0.0293 (9) | 0.0017 (8) | 0.0006 (8) | −0.0095 (7) |
| C27 | 0.0155 (7) | 0.0184 (7) | 0.0171 (7) | −0.0026 (6) | 0.0027 (5) | 0.0007 (5) |
| C28 | 0.0131 (7) | 0.0190 (7) | 0.0190 (6) | −0.0017 (6) | 0.0030 (5) | −0.0006 (6) |
| C29 | 0.0194 (7) | 0.0213 (7) | 0.0188 (7) | 0.0001 (6) | 0.0010 (5) | −0.0006 (6) |
| C30 | 0.0205 (8) | 0.0246 (8) | 0.0216 (7) | −0.0034 (7) | 0.0036 (6) | −0.0048 (6) |
| C31 | 0.0217 (7) | 0.0179 (7) | 0.0287 (8) | 0.0011 (6) | 0.0053 (6) | −0.0042 (6) |
| C32 | 0.0207 (8) | 0.0212 (8) | 0.0245 (8) | 0.0014 (6) | −0.0016 (6) | 0.0024 (6) |
| C33 | 0.0192 (7) | 0.0215 (7) | 0.0178 (6) | −0.0017 (6) | 0.0024 (5) | −0.0001 (6) |
| C34 | 0.0267 (9) | 0.0285 (9) | 0.0358 (9) | 0.0051 (7) | −0.0108 (7) | 0.0074 (8) |
Geometric parameters (Å, °)
| O1—C22 | 1.4167 (18) | C10—C11 | 1.5762 (19) |
| O1—C1 | 1.4365 (17) | C11—C17 | 1.536 (2) |
| O2—C3 | 1.4346 (17) | C12—C13 | 1.5406 (19) |
| O2—H2 | 0.87 (3) | C12—H12A | 0.9700 |
| O3—C23 | 1.4211 (19) | C12—H12B | 0.9700 |
| O3—C18 | 1.4301 (19) | C13—C14 | 1.5225 (19) |
| O4—C6 | 1.4187 (19) | C13—C16 | 1.553 (2) |
| O4—C24 | 1.429 (2) | C14—H14A | 0.9800 |
| O5—C25 | 1.206 (2) | C15—C16 | 1.550 (2) |
| O6—C25 | 1.3476 (19) | C15—H15A | 0.9800 |
| O6—C8 | 1.4852 (17) | C16—H16A | 0.9800 |
| O7—C10 | 1.4484 (17) | C17—H17A | 0.9800 |
| O7—H7 | 0.89 (3) | C18—H18A | 0.9700 |
| O8—C13 | 1.4118 (18) | C18—H18B | 0.9700 |
| O8—H8 | 0.83 (3) | C19—H19A | 0.9700 |
| O9—C27 | 1.3500 (17) | C19—H19B | 0.9700 |
| O9—C14 | 1.4362 (18) | C20—C21 | 1.513 (2) |
| O10—C27 | 1.200 (2) | C20—H20A | 0.9700 |
| O11—C15 | 1.4275 (18) | C20—H20B | 0.9700 |
| O11—H11 | 0.83 (3) | C21—H21A | 0.9600 |
| O12—C34 | 1.422 (2) | C21—H21B | 0.9600 |
| O12—C16 | 1.4289 (18) | C21—H21C | 0.9600 |
| N—C17 | 1.4615 (18) | C22—H22A | 0.9600 |
| N—C20 | 1.466 (2) | C22—H22B | 0.9600 |
| N—C19 | 1.476 (2) | C22—H22C | 0.9600 |
| C1—C2 | 1.522 (2) | C23—H23A | 0.9600 |
| C1—C11 | 1.553 (2) | C23—H23B | 0.9600 |
| C1—H1A | 0.9800 | C23—H23C | 0.9600 |
| C2—C3 | 1.511 (2) | C24—H24A | 0.9600 |
| C2—H2A | 0.9700 | C24—H24B | 0.9600 |
| C2—H2B | 0.9700 | C24—H24C | 0.9600 |
| C3—C4 | 1.541 (2) | C25—C26 | 1.502 (2) |
| C3—H3A | 0.9800 | C26—H26A | 0.9600 |
| C4—C19 | 1.541 (2) | C26—H26B | 0.9600 |
| C4—C18 | 1.543 (2) | C26—H26C | 0.9600 |
| C4—C5 | 1.5599 (19) | C27—C28 | 1.488 (2) |
| C5—C6 | 1.570 (2) | C28—C33 | 1.398 (2) |
| C5—C11 | 1.5778 (19) | C28—C29 | 1.399 (2) |
| C5—H5A | 0.9800 | C29—C30 | 1.392 (2) |
| C6—C7 | 1.554 (2) | C29—H29 | 0.9300 |
| C6—H6A | 0.9800 | C30—C31 | 1.387 (3) |
| C7—C8 | 1.531 (2) | C30—H30 | 0.9300 |
| C7—C17 | 1.5557 (19) | C31—C32 | 1.394 (2) |
| C7—H7A | 0.9800 | C31—H31 | 0.9300 |
| C8—C9 | 1.546 (2) | C32—C33 | 1.384 (2) |
| C8—C15 | 1.573 (2) | C32—H32 | 0.9300 |
| C9—C14 | 1.526 (2) | C33—H33 | 0.9300 |
| C9—C10 | 1.561 (2) | C34—H34A | 0.9600 |
| C9—H9A | 0.9800 | C34—H34B | 0.9600 |
| C10—C12 | 1.5588 (19) | C34—H34C | 0.9600 |
| C22—O1—C1 | 112.97 (11) | O11—C15—C16 | 107.21 (12) |
| C3—O2—H2 | 107.0 (17) | O11—C15—C8 | 112.21 (12) |
| C23—O3—C18 | 112.14 (12) | C16—C15—C8 | 117.71 (12) |
| C6—O4—C24 | 112.32 (14) | O11—C15—H15A | 106.3 |
| C25—O6—C8 | 120.55 (12) | C16—C15—H15A | 106.3 |
| C10—O7—H7 | 106.2 (18) | C8—C15—H15A | 106.3 |
| C13—O8—H8 | 106.3 (16) | O12—C16—C15 | 109.86 (12) |
| C27—O9—C14 | 117.48 (12) | O12—C16—C13 | 106.06 (12) |
| C15—O11—H11 | 102 (2) | C15—C16—C13 | 114.58 (12) |
| C34—O12—C16 | 114.23 (13) | O12—C16—H16A | 108.7 |
| C17—N—C20 | 111.98 (12) | C15—C16—H16A | 108.7 |
| C17—N—C19 | 116.56 (12) | C13—C16—H16A | 108.7 |
| C20—N—C19 | 111.61 (12) | N—C17—C11 | 110.11 (11) |
| O1—C1—C2 | 108.40 (11) | N—C17—C7 | 114.92 (12) |
| O1—C1—C11 | 108.72 (11) | C11—C17—C7 | 100.84 (11) |
| C2—C1—C11 | 115.78 (12) | N—C17—H17A | 110.2 |
| O1—C1—H1A | 107.9 | C11—C17—H17A | 110.2 |
| C2—C1—H1A | 107.9 | C7—C17—H17A | 110.2 |
| C11—C1—H1A | 107.9 | O3—C18—C4 | 108.21 (12) |
| C3—C2—C1 | 112.51 (12) | O3—C18—H18A | 110.1 |
| C3—C2—H2A | 109.1 | C4—C18—H18A | 110.1 |
| C1—C2—H2A | 109.1 | O3—C18—H18B | 110.1 |
| C3—C2—H2B | 109.1 | C4—C18—H18B | 110.1 |
| C1—C2—H2B | 109.1 | H18A—C18—H18B | 108.4 |
| H2A—C2—H2B | 107.8 | N—C19—C4 | 113.58 (12) |
| O2—C3—C2 | 109.83 (12) | N—C19—H19A | 108.8 |
| O2—C3—C4 | 109.72 (12) | C4—C19—H19A | 108.8 |
| C2—C3—C4 | 111.57 (12) | N—C19—H19B | 108.8 |
| O2—C3—H3A | 108.6 | C4—C19—H19B | 108.8 |
| C2—C3—H3A | 108.6 | H19A—C19—H19B | 107.7 |
| C4—C3—H3A | 108.6 | N—C20—C21 | 111.65 (15) |
| C3—C4—C19 | 112.64 (12) | N—C20—H20A | 109.3 |
| C3—C4—C18 | 107.04 (12) | C21—C20—H20A | 109.3 |
| C19—C4—C18 | 109.11 (12) | N—C20—H20B | 109.3 |
| C3—C4—C5 | 107.68 (11) | C21—C20—H20B | 109.3 |
| C19—C4—C5 | 108.72 (12) | H20A—C20—H20B | 108.0 |
| C18—C4—C5 | 111.68 (12) | C20—C21—H21A | 109.5 |
| C4—C5—C6 | 112.07 (12) | C20—C21—H21B | 109.5 |
| C4—C5—C11 | 109.32 (11) | H21A—C21—H21B | 109.5 |
| C6—C5—C11 | 102.42 (11) | C20—C21—H21C | 109.5 |
| C4—C5—H5A | 110.9 | H21A—C21—H21C | 109.5 |
| C6—C5—H5A | 110.9 | H21B—C21—H21C | 109.5 |
| C11—C5—H5A | 110.9 | O1—C22—H22A | 109.5 |
| O4—C6—C7 | 109.63 (12) | O1—C22—H22B | 109.5 |
| O4—C6—C5 | 117.99 (12) | H22A—C22—H22B | 109.5 |
| C7—C6—C5 | 104.75 (12) | O1—C22—H22C | 109.5 |
| O4—C6—H6A | 108.0 | H22A—C22—H22C | 109.5 |
| C7—C6—H6A | 108.0 | H22B—C22—H22C | 109.5 |
| C5—C6—H6A | 108.0 | O3—C23—H23A | 109.5 |
| C8—C7—C6 | 107.51 (11) | O3—C23—H23B | 109.5 |
| C8—C7—C17 | 111.31 (12) | H23A—C23—H23B | 109.5 |
| C6—C7—C17 | 104.61 (12) | O3—C23—H23C | 109.5 |
| C8—C7—H7A | 111.1 | H23A—C23—H23C | 109.5 |
| C6—C7—H7A | 111.1 | H23B—C23—H23C | 109.5 |
| C17—C7—H7A | 111.1 | O4—C24—H24A | 109.5 |
| O6—C8—C7 | 107.49 (11) | O4—C24—H24B | 109.5 |
| O6—C8—C9 | 101.06 (11) | H24A—C24—H24B | 109.5 |
| C7—C8—C9 | 108.54 (12) | O4—C24—H24C | 109.5 |
| O6—C8—C15 | 108.32 (11) | H24A—C24—H24C | 109.5 |
| C7—C8—C15 | 116.32 (12) | H24B—C24—H24C | 109.5 |
| C9—C8—C15 | 113.84 (12) | O5—C25—O6 | 124.70 (15) |
| C14—C9—C8 | 111.83 (12) | O5—C25—C26 | 125.10 (15) |
| C14—C9—C10 | 102.08 (11) | O6—C25—C26 | 110.20 (14) |
| C8—C9—C10 | 112.24 (11) | C25—C26—H26A | 109.5 |
| C14—C9—H9A | 110.1 | C25—C26—H26B | 109.5 |
| C8—C9—H9A | 110.1 | H26A—C26—H26B | 109.5 |
| C10—C9—H9A | 110.1 | C25—C26—H26C | 109.5 |
| O7—C10—C12 | 105.10 (11) | H26A—C26—H26C | 109.5 |
| O7—C10—C9 | 107.18 (11) | H26B—C26—H26C | 109.5 |
| C12—C10—C9 | 102.33 (11) | O10—C27—O9 | 123.76 (14) |
| O7—C10—C11 | 110.21 (12) | O10—C27—C28 | 126.00 (14) |
| C12—C10—C11 | 114.45 (11) | O9—C27—C28 | 110.24 (13) |
| C9—C10—C11 | 116.62 (12) | C33—C28—C29 | 120.07 (14) |
| C17—C11—C1 | 116.46 (12) | C33—C28—C27 | 121.94 (14) |
| C17—C11—C10 | 107.55 (11) | C29—C28—C27 | 117.98 (14) |
| C1—C11—C10 | 107.94 (12) | C30—C29—C28 | 119.43 (15) |
| C17—C11—C5 | 98.50 (11) | C30—C29—H29 | 120.3 |
| C1—C11—C5 | 112.60 (12) | C28—C29—H29 | 120.3 |
| C10—C11—C5 | 113.66 (11) | C31—C30—C29 | 120.43 (15) |
| C13—C12—C10 | 107.57 (11) | C31—C30—H30 | 119.8 |
| C13—C12—H12A | 110.2 | C29—C30—H30 | 119.8 |
| C10—C12—H12A | 110.2 | C30—C31—C32 | 119.99 (15) |
| C13—C12—H12B | 110.2 | C30—C31—H31 | 120.0 |
| C10—C12—H12B | 110.2 | C32—C31—H31 | 120.0 |
| H12A—C12—H12B | 108.5 | C33—C32—C31 | 120.20 (15) |
| O8—C13—C14 | 113.43 (12) | C33—C32—H32 | 119.9 |
| O8—C13—C12 | 109.50 (12) | C31—C32—H32 | 119.9 |
| C14—C13—C12 | 102.26 (11) | C32—C33—C28 | 119.88 (14) |
| O8—C13—C16 | 111.35 (12) | C32—C33—H33 | 120.1 |
| C14—C13—C16 | 110.12 (12) | C28—C33—H33 | 120.1 |
| C12—C13—C16 | 109.79 (12) | O12—C34—H34A | 109.5 |
| O9—C14—C13 | 108.67 (11) | O12—C34—H34B | 109.5 |
| O9—C14—C9 | 113.52 (12) | H34A—C34—H34B | 109.5 |
| C13—C14—C9 | 101.84 (11) | O12—C34—H34C | 109.5 |
| O9—C14—H14A | 110.8 | H34A—C34—H34C | 109.5 |
| C13—C14—H14A | 110.8 | H34B—C34—H34C | 109.5 |
| C9—C14—H14A | 110.8 |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O11—H11···O5 | 0.83 (3) | 2.07 (3) | 2.791 (2) | 146 (3) |
| O8—H8···O12 | 0.83 (3) | 2.11 (3) | 2.598 (2) | 117 (3) |
| O2—H2···O7i | 0.87 (3) | 2.21 (3) | 3.066 (2) | 168 (3) |
| O8—H8···O2ii | 0.83 (3) | 2.39 (3) | 2.928 (2) | 123 (3) |
Symmetry codes: (i) x+1/2, −y−1/2, −z; (ii) x−1, y, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: ZL2215).
References
- Dzhakhangirov, F. N. (1976). Dokl. Akad. Nauk UzSSR, pp. 32–33.
- Dzhakhangirov, F. N., Sultankhodzhaev, M. N., Tashkhodjaev, B. & Salimov, B. T. (1997). Khim. Prir. Soedin. pp. 254–270.
- Flack, H. D. (1983). Acta Cryst. A39, 876–881.
- Joshi, B. S. & Pelletier, S. W. (1987). Heterocycles, 26, 2503–2518.
- Oxford Diffraction (2009). CrysAlisPro Oxford Diffraction Ltd, Yarnton, England.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sultankhodzhaev, M. N., Beshitaishvili, L. V., Yagudaev, M. R., Yunusov, M. S. & Yunusov, S. Yu. (1980). Khim. Prir. Soedin. pp. 665–672.
- Sultankhodzhaev, M. N., Yunusov, M. S. & Yunusov, S. Yu. (1973). Khim. Prir. Soedin. pp. 127–129.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809021436/zl2215sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809021436/zl2215Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report