Abstract
The title compound, C19H30ClNO5, was obtained by the tandem asymmetric Michael addition–elimination reaction of (5S)-3,4-dichloro-5-(l-menthyloxy)furan-2(5H)-one and l-valine in the presence of potassium hydroxide. The furanone unit is approximately planar (r.m.s. deviation = 0.0204 Å) and the six-membered cyclohexane ring adopts a chair conformation. The crystal structure is stabilized by a network of O—H⋯O and N—H⋯O hydrogen bonds.
Related literature
For biologically active 4-amino-2(5H)-furanones, see: Kimura et al. (2000 ▶); Tanoury et al., 2008 ▶). For the synthesis of the precursor, (5S)-3,4-dichloro-5-(l-menthyloxy)furan-2(5H)-one, see: Chen & Geng (1993 ▶). 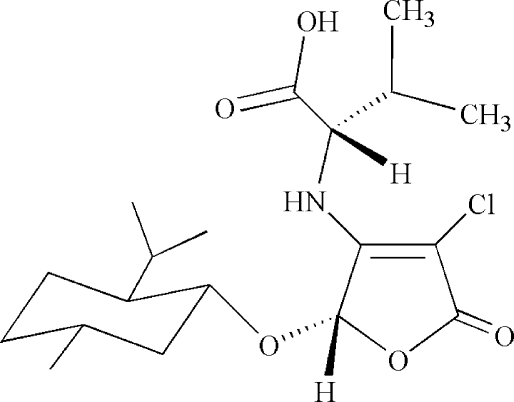
Experimental
Crystal data
C19H30ClNO5
M r = 387.89
Tetragonal,

a = 10.4540 (4) Å
c = 39.300 (3) Å
V = 4294.9 (4) Å3
Z = 8
Mo Kα radiation
μ = 0.20 mm−1
T = 293 K
0.30 × 0.23 × 0.15 mm
Data collection
Bruker APEXII CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2004 ▶) T min = 0.769, T max = 0.867 (expected range = 0.860–0.970)
22031 measured reflections
3796 independent reflections
2868 reflections with I > 2σ(I)
R int = 0.053
Refinement
R[F 2 > 2σ(F 2)] = 0.040
wR(F 2) = 0.091
S = 1.04
3796 reflections
242 parameters
H-atom parameters constrained
Δρmax = 0.18 e Å−3
Δρmin = −0.23 e Å−3
Absolute structure: Flack, (1983 ▶), 1499 Friedel pairs
Flack parameter: −0.03 (8)
Data collection: APEX2 (Bruker, 2004 ▶); cell refinement: APEX2 and SAINT (Bruker, 2004 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809022120/gk2204sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809022120/gk2204Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O2i | 0.86 | 2.25 | 3.019 (3) | 148 |
| O1—H1A⋯O3ii | 0.82 | 1.83 | 2.617 (2) | 160 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Acknowledgments
The work was supported by the National Natural Science Foundation of China (grant No. 20772035) and the Natural Science Foundation of Guangdong Province, China (grant No. 5300082).
supplementary crystallographic information
Comment
Many 4-amino-2(5H)-furanones have been patented as prodrugs or insecticides and herbicides (Kimura et al., 2000; Tanoury et al., 2008). Attracted by versatility of 4-amino-2(5H)-furanones, we synthesized the title molecule with chiral synthon 3,4-dichloro-5-(S)-(l-menthyloxy)-2(5H)-furanone and L-valine in the presence of potassium hydroxide via the tandem asymmetric Michael addition-elimination reaction. With 2(5H)-furanone moiety and polyfunctional groups (carboxyl, amino, halogeno), the title compound is expected to be a biologically active product and an excellent ligand.
The structure of the title compound is illustrated in Fig. 1. The five-membered furane ring and the six-membered cyclohexane ring are connected via C10—O2—C11 ether bond. The configuration of chiral centers is following: C4(S), C10(S), C11(R), C12(S), C17(R)). The furanone unit is approximately planar, whereas the cyclohexane ring shows a chair conformation with three substituents occupying equatorial positions. The molecules are linked by O4—H6···O3 and N1—H1···O5 hydrogen bonds forming a three-dimensional network (Table. 1 and Fig. 2).
Experimental
The precursor, 3,4-dichloro-5-(S)-(L-menthyloxy)-2(5H)-furanone, was prepared according to the literature procedure (Chen et al., 1993).
After the mixture of L-valine (4.5 mmol) and potassium hydroxide (5.8 mmol) was dissolved in absolute ethyl alcohol under nitrogen atmosphere, dichloromethane solution of 3,4-dichloro-5-(S)-(l-menthyloxy)-2(5H)-furanone (3.0 mmol) was added. The reaction was carried out under the stirring at room temperature for 24 h. Once the reaction was complete, the solvents were removed under reduced pressure. The residual solid was dissolved in dichloromethane, and pH of the solution was adjusted to 3–4 with 15% of aqueous HCl solution. Then the combined organic layers from extraction were concentrated under reduced pressure, and the crude product was purified by silica gel column chromatography with the gradient mixture of petroleum ether and ethyl acetate to give the product yielding (I) 0.6891 g (59.2%). Data for (I): [α]20°D = 47.616° (c 0.481, CH3CH2OH); 1H NMR (400 MHz, CDCl3, TMS): 0.832 (3H, d, J = 6.0 Hz, CH3), 0.904–0.935 (7H, m, CH, 2CH3), 0.955–1.057 (8H, m, 2CH3, CH2), 1.312–1.457 (2H, m, 2CH), 1.605–1.710 (2H, m, CH2), 2.100–2.350 (3H, m, CH2, CH), 3.505–3.609 (1H, m, CH), 4.726 (1H, s, NH), 5.116–5.138 (1H, d, J = 8.8 Hz, CH), 5.700 (1H, s, CH), 10.212 (1H, s, COOH); ESI-MS, m/z (%): Calcd for C19H31ClNO5+([M+H]+): 388.19, Found: 388.15 (100.0)
Refinement
All H atoms were positioned in calculated positions (O—H = 0.82 Å; N—H = 0.86 Å; C—H = 0.96Å - 0.98 Å) and were refined using a riding model, with Uiso(H) = 1.2 Ueq(C,N) for methylene, methine and amino H atoms and Uiso(H) = 1.5 Ueq(C,O) for methyl or hydroxyl H atoms.
Figures
Fig. 1.
Molecular structure of the title compound with displacement ellipsoids shown at the 30% probability level.
Fig. 2.
Perspective view of the crystal packing. Dashed lines represent hydrogen bonds.
Crystal data
| C19H30ClNO5 | Dx = 1.200 Mg m−3 |
| Mr = 387.89 | Mo Kα radiation, λ = 0.71073 Å |
| Tetragonal, P43212 | Cell parameters from 3405 reflections |
| Hall symbol: P 4nw 2abw | θ = 2.2–19.1° |
| a = 10.4540 (4) Å | µ = 0.20 mm−1 |
| c = 39.300 (3) Å | T = 293 K |
| V = 4294.9 (4) Å3 | Block, colourless |
| Z = 8 | 0.30 × 0.23 × 0.15 mm |
| F(000) = 1664.0 |
Data collection
| Bruker APEXII CCD area-detector diffractometer | 3796 independent reflections |
| Radiation source: fine-focus sealed tube | 2868 reflections with I > 2σ(I) |
| graphite | Rint = 0.053 |
| Detector resolution: 0 pixels mm-1 | θmax = 25.0°, θmin = 2.0° |
| φ and ω scans | h = −12→10 |
| Absorption correction: multi-scan (SADABS; Bruker, 2004) | k = −11→12 |
| Tmin = 0.769, Tmax = 0.867 | l = −46→46 |
| 22031 measured reflections |
Refinement
| Refinement on F2 | Hydrogen site location: inferred from neighbouring sites |
| Least-squares matrix: full | H-atom parameters constrained |
| R[F2 > 2σ(F2)] = 0.040 | w = 1/[σ2(Fo2) + (0.0329P)2 + 0.7758P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.091 | (Δ/σ)max = 0.001 |
| S = 1.04 | Δρmax = 0.18 e Å−3 |
| 3796 reflections | Δρmin = −0.23 e Å−3 |
| 242 parameters | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc^*^=kFc[1+0.001xFc^2^λ^3^/sin(2θ)]^-1/4^ |
| 0 restraints | Extinction coefficient: 0.0020 (3) |
| Primary atom site location: structure-invariant direct methods | Absolute structure: Flack, (1983), 1499 Friedel pairs |
| Secondary atom site location: difference Fourier map | Flack parameter: −0.03 (8) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.6886 (3) | −0.0142 (2) | −0.02302 (5) | 0.0457 (6) | |
| C2 | 0.6349 (2) | 0.0184 (2) | 0.01161 (5) | 0.0434 (6) | |
| H2 | 0.5486 | −0.0178 | 0.0132 | 0.052* | |
| C3 | 0.6252 (3) | 0.1638 (3) | 0.01705 (6) | 0.0645 (8) | |
| H3 | 0.6169 | 0.1771 | 0.0416 | 0.077* | |
| C4 | 0.5046 (4) | 0.2198 (3) | 0.00080 (9) | 0.1066 (14) | |
| H4A | 0.4929 | 0.3061 | 0.0085 | 0.160* | |
| H4B | 0.4318 | 0.1692 | 0.0072 | 0.160* | |
| H4C | 0.5135 | 0.2191 | −0.0235 | 0.160* | |
| C5 | 0.7443 (4) | 0.2337 (3) | 0.00618 (8) | 0.0901 (11) | |
| H5A | 0.8180 | 0.1926 | 0.0158 | 0.135* | |
| H5B | 0.7403 | 0.3207 | 0.0140 | 0.135* | |
| H5C | 0.7507 | 0.2325 | −0.0182 | 0.135* | |
| C6 | 0.6726 (2) | −0.0701 (2) | 0.06900 (5) | 0.0420 (6) | |
| C7 | 0.5557 (2) | −0.0781 (2) | 0.08334 (5) | 0.0478 (6) | |
| C8 | 0.5677 (3) | −0.1196 (2) | 0.11793 (5) | 0.0489 (6) | |
| C9 | 0.7698 (2) | −0.0992 (2) | 0.09642 (5) | 0.0429 (6) | |
| H9 | 0.8263 | −0.1695 | 0.0896 | 0.052* | |
| C10 | 0.9662 (2) | −0.0054 (2) | 0.11656 (5) | 0.0445 (6) | |
| H10 | 1.0136 | −0.0663 | 0.1024 | 0.053* | |
| C11 | 0.9624 (2) | −0.0546 (3) | 0.15302 (5) | 0.0529 (7) | |
| H11A | 0.9221 | −0.1381 | 0.1534 | 0.063* | |
| H11B | 0.9112 | 0.0029 | 0.1668 | 0.063* | |
| C12 | 1.0967 (3) | −0.0645 (3) | 0.16813 (6) | 0.0596 (7) | |
| H12 | 1.1447 | −0.1278 | 0.1549 | 0.072* | |
| C13 | 1.0913 (3) | −0.1106 (3) | 0.20518 (6) | 0.0868 (10) | |
| H13A | 1.0524 | −0.0458 | 0.2191 | 0.130* | |
| H13B | 1.1764 | −0.1271 | 0.2132 | 0.130* | |
| H13C | 1.0416 | −0.1877 | 0.2065 | 0.130* | |
| C14 | 1.1649 (3) | 0.0631 (3) | 0.16501 (7) | 0.0753 (9) | |
| H14A | 1.1233 | 0.1250 | 0.1797 | 0.090* | |
| H14B | 1.2525 | 0.0534 | 0.1727 | 0.090* | |
| C15 | 1.1654 (3) | 0.1133 (3) | 0.12899 (7) | 0.0723 (9) | |
| H15A | 1.2151 | 0.0562 | 0.1147 | 0.087* | |
| H15B | 1.2062 | 0.1966 | 0.1286 | 0.087* | |
| C16 | 1.0302 (2) | 0.1249 (2) | 0.11446 (6) | 0.0522 (7) | |
| H16 | 0.9828 | 0.1814 | 0.1299 | 0.063* | |
| C17 | 1.0229 (3) | 0.1862 (3) | 0.07910 (7) | 0.0698 (8) | |
| H17 | 0.9327 | 0.1846 | 0.0723 | 0.084* | |
| C18 | 1.0627 (4) | 0.3269 (3) | 0.08046 (11) | 0.1295 (16) | |
| H18A | 1.0213 | 0.3678 | 0.0993 | 0.194* | |
| H18B | 1.0380 | 0.3685 | 0.0597 | 0.194* | |
| H18C | 1.1538 | 0.3325 | 0.0832 | 0.194* | |
| C19 | 1.0969 (4) | 0.1145 (4) | 0.05192 (7) | 0.1001 (12) | |
| H19A | 1.1866 | 0.1177 | 0.0571 | 0.150* | |
| H19B | 1.0818 | 0.1534 | 0.0302 | 0.150* | |
| H19C | 1.0692 | 0.0270 | 0.0514 | 0.150* | |
| Cl1 | 0.40649 (7) | −0.05720 (10) | 0.066293 (16) | 0.0818 (3) | |
| N1 | 0.71379 (18) | −0.0411 (2) | 0.03775 (4) | 0.0463 (5) | |
| H1 | 0.7919 | −0.0586 | 0.0327 | 0.056* | |
| O1 | 0.60255 (18) | 0.0096 (2) | −0.04637 (4) | 0.0835 (7) | |
| H1A | 0.6322 | −0.0068 | −0.0652 | 0.125* | |
| O2 | 0.79397 (18) | −0.05167 (19) | −0.02856 (4) | 0.0633 (5) | |
| O3 | 0.48370 (18) | −0.14530 (18) | 0.13838 (4) | 0.0625 (5) | |
| O4 | 0.69263 (17) | −0.13404 (16) | 0.12582 (3) | 0.0515 (5) | |
| O5 | 0.83805 (15) | 0.01070 (15) | 0.10286 (4) | 0.0472 (4) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0580 (17) | 0.0518 (16) | 0.0274 (11) | 0.0057 (12) | −0.0053 (11) | 0.0020 (11) |
| C2 | 0.0501 (15) | 0.0544 (16) | 0.0259 (10) | 0.0075 (12) | −0.0023 (10) | 0.0030 (10) |
| C3 | 0.095 (2) | 0.0608 (18) | 0.0382 (13) | 0.0207 (17) | 0.0001 (14) | −0.0027 (13) |
| C4 | 0.135 (4) | 0.086 (3) | 0.099 (2) | 0.056 (2) | −0.020 (2) | 0.002 (2) |
| C5 | 0.132 (3) | 0.067 (2) | 0.072 (2) | −0.014 (2) | 0.006 (2) | 0.0021 (17) |
| C6 | 0.0515 (15) | 0.0469 (15) | 0.0276 (11) | 0.0046 (11) | −0.0012 (11) | −0.0033 (10) |
| C7 | 0.0480 (15) | 0.0655 (17) | 0.0301 (11) | 0.0041 (13) | 0.0008 (11) | 0.0020 (11) |
| C8 | 0.0632 (19) | 0.0531 (16) | 0.0303 (11) | −0.0084 (13) | 0.0000 (12) | −0.0034 (11) |
| C9 | 0.0542 (15) | 0.0481 (15) | 0.0265 (11) | −0.0019 (12) | −0.0016 (10) | 0.0020 (11) |
| C10 | 0.0485 (15) | 0.0488 (15) | 0.0361 (12) | 0.0063 (12) | −0.0051 (11) | −0.0040 (11) |
| C11 | 0.0620 (17) | 0.0601 (17) | 0.0365 (12) | 0.0011 (14) | −0.0035 (12) | −0.0009 (12) |
| C12 | 0.0670 (18) | 0.070 (2) | 0.0418 (13) | 0.0108 (15) | −0.0119 (13) | −0.0125 (13) |
| C13 | 0.106 (3) | 0.108 (3) | 0.0464 (16) | 0.017 (2) | −0.0248 (17) | −0.0055 (17) |
| C14 | 0.070 (2) | 0.092 (3) | 0.0641 (18) | −0.0008 (18) | −0.0196 (15) | −0.0204 (17) |
| C15 | 0.064 (2) | 0.073 (2) | 0.079 (2) | −0.0123 (17) | −0.0053 (16) | −0.0122 (17) |
| C16 | 0.0554 (17) | 0.0466 (16) | 0.0545 (15) | 0.0019 (12) | 0.0008 (13) | −0.0082 (12) |
| C17 | 0.074 (2) | 0.0622 (19) | 0.0737 (19) | −0.0058 (16) | 0.0041 (16) | 0.0165 (15) |
| C18 | 0.153 (4) | 0.071 (3) | 0.164 (4) | −0.030 (3) | 0.003 (3) | 0.035 (3) |
| C19 | 0.119 (3) | 0.123 (3) | 0.0585 (18) | 0.011 (3) | 0.014 (2) | 0.019 (2) |
| Cl1 | 0.0488 (4) | 0.1483 (8) | 0.0483 (4) | 0.0136 (5) | 0.0026 (3) | 0.0132 (4) |
| N1 | 0.0460 (12) | 0.0683 (14) | 0.0247 (9) | 0.0099 (10) | 0.0023 (8) | 0.0061 (9) |
| O1 | 0.0698 (13) | 0.151 (2) | 0.0296 (8) | 0.0369 (14) | −0.0086 (9) | −0.0045 (11) |
| O2 | 0.0639 (13) | 0.0890 (15) | 0.0369 (9) | 0.0293 (11) | 0.0062 (8) | 0.0049 (9) |
| O3 | 0.0717 (13) | 0.0841 (14) | 0.0317 (8) | −0.0182 (10) | 0.0114 (9) | 0.0017 (8) |
| O4 | 0.0601 (12) | 0.0674 (12) | 0.0270 (8) | −0.0094 (9) | −0.0030 (8) | 0.0084 (7) |
| O5 | 0.0530 (11) | 0.0463 (11) | 0.0422 (9) | 0.0000 (8) | −0.0070 (8) | 0.0009 (8) |
Geometric parameters (Å, °)
| C1—O2 | 1.189 (3) | C11—C12 | 1.528 (3) |
| C1—O1 | 1.309 (3) | C11—H11A | 0.9700 |
| C1—C2 | 1.511 (3) | C11—H11B | 0.9700 |
| C2—N1 | 1.456 (3) | C12—C14 | 1.517 (4) |
| C2—C3 | 1.538 (3) | C12—C13 | 1.534 (3) |
| C2—H2 | 0.9800 | C12—H12 | 0.9800 |
| C3—C5 | 1.506 (4) | C13—H13A | 0.9600 |
| C3—C4 | 1.530 (4) | C13—H13B | 0.9600 |
| C3—H3 | 0.9800 | C13—H13C | 0.9600 |
| C4—H4A | 0.9600 | C14—C15 | 1.510 (4) |
| C4—H4B | 0.9600 | C14—H14A | 0.9700 |
| C4—H4C | 0.9600 | C14—H14B | 0.9700 |
| C5—H5A | 0.9600 | C15—C16 | 1.529 (4) |
| C5—H5B | 0.9600 | C15—H15A | 0.9700 |
| C5—H5C | 0.9600 | C15—H15B | 0.9700 |
| C6—N1 | 1.336 (3) | C16—C17 | 1.532 (3) |
| C6—C7 | 1.348 (3) | C16—H16 | 0.9800 |
| C6—C9 | 1.511 (3) | C17—C19 | 1.518 (4) |
| C7—C8 | 1.432 (3) | C17—C18 | 1.529 (4) |
| C7—Cl1 | 1.712 (2) | C17—H17 | 0.9800 |
| C8—O3 | 1.220 (3) | C18—H18A | 0.9600 |
| C8—O4 | 1.351 (3) | C18—H18B | 0.9600 |
| C9—O5 | 1.376 (3) | C18—H18C | 0.9600 |
| C9—O4 | 1.455 (3) | C19—H19A | 0.9600 |
| C9—H9 | 0.9800 | C19—H19B | 0.9600 |
| C10—O5 | 1.454 (3) | C19—H19C | 0.9600 |
| C10—C16 | 1.520 (3) | N1—H1 | 0.8600 |
| C10—C11 | 1.523 (3) | O1—H1A | 0.8200 |
| C10—H10 | 0.9800 | ||
| O2—C1—O1 | 124.8 (2) | C14—C12—C11 | 109.9 (2) |
| O2—C1—C2 | 125.7 (2) | C14—C12—C13 | 111.7 (2) |
| O1—C1—C2 | 109.5 (2) | C11—C12—C13 | 110.9 (2) |
| N1—C2—C1 | 109.21 (18) | C14—C12—H12 | 108.1 |
| N1—C2—C3 | 111.14 (19) | C11—C12—H12 | 108.1 |
| C1—C2—C3 | 111.9 (2) | C13—C12—H12 | 108.1 |
| N1—C2—H2 | 108.2 | C12—C13—H13A | 109.5 |
| C1—C2—H2 | 108.2 | C12—C13—H13B | 109.5 |
| C3—C2—H2 | 108.2 | H13A—C13—H13B | 109.5 |
| C5—C3—C4 | 112.2 (3) | C12—C13—H13C | 109.5 |
| C5—C3—C2 | 112.7 (2) | H13A—C13—H13C | 109.5 |
| C4—C3—C2 | 112.0 (2) | H13B—C13—H13C | 109.5 |
| C5—C3—H3 | 106.5 | C15—C14—C12 | 112.5 (2) |
| C4—C3—H3 | 106.5 | C15—C14—H14A | 109.1 |
| C2—C3—H3 | 106.5 | C12—C14—H14A | 109.1 |
| C3—C4—H4A | 109.5 | C15—C14—H14B | 109.1 |
| C3—C4—H4B | 109.5 | C12—C14—H14B | 109.1 |
| H4A—C4—H4B | 109.5 | H14A—C14—H14B | 107.8 |
| C3—C4—H4C | 109.5 | C14—C15—C16 | 112.0 (2) |
| H4A—C4—H4C | 109.5 | C14—C15—H15A | 109.2 |
| H4B—C4—H4C | 109.5 | C16—C15—H15A | 109.2 |
| C3—C5—H5A | 109.5 | C14—C15—H15B | 109.2 |
| C3—C5—H5B | 109.5 | C16—C15—H15B | 109.2 |
| H5A—C5—H5B | 109.5 | H15A—C15—H15B | 107.9 |
| C3—C5—H5C | 109.5 | C10—C16—C15 | 108.4 (2) |
| H5A—C5—H5C | 109.5 | C10—C16—C17 | 113.7 (2) |
| H5B—C5—H5C | 109.5 | C15—C16—C17 | 114.7 (2) |
| N1—C6—C7 | 133.7 (2) | C10—C16—H16 | 106.5 |
| N1—C6—C9 | 119.0 (2) | C15—C16—H16 | 106.5 |
| C7—C6—C9 | 107.34 (18) | C17—C16—H16 | 106.5 |
| C6—C7—C8 | 109.7 (2) | C19—C17—C18 | 111.2 (3) |
| C6—C7—Cl1 | 130.86 (17) | C19—C17—C16 | 114.0 (2) |
| C8—C7—Cl1 | 119.30 (18) | C18—C17—C16 | 110.9 (3) |
| O3—C8—O4 | 121.3 (2) | C19—C17—H17 | 106.8 |
| O3—C8—C7 | 129.0 (2) | C18—C17—H17 | 106.8 |
| O4—C8—C7 | 109.6 (2) | C16—C17—H17 | 106.8 |
| O5—C9—O4 | 110.51 (17) | C17—C18—H18A | 109.5 |
| O5—C9—C6 | 108.15 (19) | C17—C18—H18B | 109.5 |
| O4—C9—C6 | 104.14 (18) | H18A—C18—H18B | 109.5 |
| O5—C9—H9 | 111.3 | C17—C18—H18C | 109.5 |
| O4—C9—H9 | 111.3 | H18A—C18—H18C | 109.5 |
| C6—C9—H9 | 111.3 | H18B—C18—H18C | 109.5 |
| O5—C10—C16 | 106.35 (18) | C17—C19—H19A | 109.5 |
| O5—C10—C11 | 111.29 (19) | C17—C19—H19B | 109.5 |
| C16—C10—C11 | 111.45 (19) | H19A—C19—H19B | 109.5 |
| O5—C10—H10 | 109.2 | C17—C19—H19C | 109.5 |
| C16—C10—H10 | 109.2 | H19A—C19—H19C | 109.5 |
| C11—C10—H10 | 109.2 | H19B—C19—H19C | 109.5 |
| C10—C11—C12 | 111.4 (2) | C6—N1—C2 | 124.25 (19) |
| C10—C11—H11A | 109.4 | C6—N1—H1 | 117.9 |
| C12—C11—H11A | 109.4 | C2—N1—H1 | 117.9 |
| C10—C11—H11B | 109.4 | C1—O1—H1A | 109.5 |
| C12—C11—H11B | 109.4 | C8—O4—C9 | 109.00 (16) |
| H11A—C11—H11B | 108.0 | C9—O5—C10 | 116.73 (18) |
| O2—C1—C2—N1 | −18.5 (4) | C13—C12—C14—C15 | −177.1 (3) |
| O1—C1—C2—N1 | 163.8 (2) | C12—C14—C15—C16 | 56.1 (3) |
| O2—C1—C2—C3 | 105.0 (3) | O5—C10—C16—C15 | 179.41 (19) |
| O1—C1—C2—C3 | −72.8 (3) | C11—C10—C16—C15 | 58.0 (3) |
| N1—C2—C3—C5 | 76.6 (3) | O5—C10—C16—C17 | −51.8 (3) |
| C1—C2—C3—C5 | −45.8 (3) | C11—C10—C16—C17 | −173.2 (2) |
| N1—C2—C3—C4 | −155.9 (2) | C14—C15—C16—C10 | −56.8 (3) |
| C1—C2—C3—C4 | 81.8 (3) | C14—C15—C16—C17 | 175.0 (2) |
| N1—C6—C7—C8 | 177.3 (3) | C10—C16—C17—C19 | −65.7 (3) |
| C9—C6—C7—C8 | −4.1 (3) | C15—C16—C17—C19 | 59.8 (3) |
| N1—C6—C7—Cl1 | 1.9 (4) | C10—C16—C17—C18 | 167.9 (3) |
| C9—C6—C7—Cl1 | −179.5 (2) | C15—C16—C17—C18 | −66.5 (4) |
| C6—C7—C8—O3 | −175.2 (3) | C7—C6—N1—C2 | 14.1 (4) |
| Cl1—C7—C8—O3 | 0.9 (4) | C9—C6—N1—C2 | −164.4 (2) |
| C6—C7—C8—O4 | 1.9 (3) | C1—C2—N1—C6 | −155.7 (2) |
| Cl1—C7—C8—O4 | 177.94 (17) | C3—C2—N1—C6 | 80.5 (3) |
| N1—C6—C9—O5 | 66.0 (3) | O3—C8—O4—C9 | 178.6 (2) |
| C7—C6—C9—O5 | −112.9 (2) | C7—C8—O4—C9 | 1.3 (3) |
| N1—C6—C9—O4 | −176.5 (2) | O5—C9—O4—C8 | 112.3 (2) |
| C7—C6—C9—O4 | 4.7 (3) | C6—C9—O4—C8 | −3.6 (2) |
| O5—C10—C11—C12 | −176.8 (2) | O4—C9—O5—C10 | 92.0 (2) |
| C16—C10—C11—C12 | −58.3 (3) | C6—C9—O5—C10 | −154.55 (17) |
| C10—C11—C12—C14 | 54.4 (3) | C16—C10—O5—C9 | 168.55 (17) |
| C10—C11—C12—C13 | 178.4 (2) | C11—C10—O5—C9 | −69.9 (2) |
| C11—C12—C14—C15 | −53.6 (3) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O2i | 0.86 | 2.25 | 3.019 (3) | 148 |
| O1—H1A···O3ii | 0.82 | 1.83 | 2.617 (2) | 160 |
Symmetry codes: (i) y+1, x−1, −z; (ii) −y+1/2, x−1/2, z−1/4.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: GK2204).
References
- Bruker (2004). APEX2, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Chen, Q. H. & Geng, Z. (1993). Acta Chim. Sin.51, 622–624.
- Flack, H. D. (1983). Acta Cryst. A39, 876–881.
- Kimura, Y., Mizuno, T., Kawano, T., Okada, K. & Shimad, A. (2000). Phytochemistry, 53, 829–831. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Tanoury, G. J., Chen, M. Z., Dong, Y., Forslund, R. E. & Magdziak, D. (2008). Org. Lett.10, 185–188. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809022120/gk2204sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809022120/gk2204Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




