Abstract
In the title compound, [Mn(C20H14N2O2)(N3)(H2O)]·0.5H2O, the MnIII ion is chelated by the N,N′,O,O′-tetradentate Schiff base ligand and further coordinated by one azide ion and one water molecule in trans positions, resulting in a distorted fac-MnN3O3 octahedral arrangement. The O atom of the uncoordinated water molecule lies on a crystallographic twofold axis. In the crystal, O—H⋯O and O—H⋯N hydrogen bonds help to establish the packing.
Related literature
For background to salicylaldehyde complexes, see: Alam et al. (2003 ▶); Zelewsky & von Knof (1999 ▶).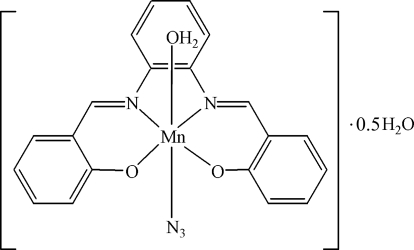
Experimental
Crystal data
[Mn(C20H14N2O2)(N3)(H2O)]·0.5H2O
M r = 438.33
Monoclinic,

a = 25.100 (10) Å
b = 11.478 (5) Å
c = 12.599 (5) Å
β = 94.175 (3)°
V = 3620 (3) Å3
Z = 8
Mo Kα radiation
μ = 0.77 mm−1
T = 293 K
0.12 × 0.10 × 0.08 mm
Data collection
Bruker APEXII CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2004 ▶) T min = 0.914, T max = 0.941
11927 measured reflections
3162 independent reflections
2371 reflections with I > 2σ(I)
R int = 0.082
Refinement
R[F 2 > 2σ(F 2)] = 0.037
wR(F 2) = 0.086
S = 1.00
3162 reflections
280 parameters
4 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.32 e Å−3
Δρmin = −0.38 e Å−3
Data collection: APEX2 (Bruker, 2004 ▶); cell refinement: SAINT-Plus (Bruker, 2004 ▶); data reduction: SAINT-Plus; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809020200/hb2977sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809020200/hb2977Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Selected bond lengths (Å).
| Mn1—O1 | 1.8636 (18) |
| Mn1—O2 | 1.8844 (18) |
| Mn1—N2 | 1.986 (2) |
| Mn1—N1 | 1.988 (2) |
| Mn1—N3 | 2.306 (2) |
| Mn1—O1W | 2.321 (2) |
Table 2. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1W—H2W⋯N3i | 0.82 (2) | 2.12 (2) | 2.937 (3) | 176 (3) |
| O1W—H1W⋯O2ii | 0.820 (11) | 2.076 (6) | 2.885 (3) | 169 (2) |
| O2W—H3W⋯N5 | 0.82 (3) | 2.18 (3) | 3.000 (3) | 173 (4) |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Acknowledgments
The authors thank the National Ministry of Science and Technology of China (grant No. 2001CB6105–07) for support.
supplementary crystallographic information
Comment
The synthesis of complexes consisting of salicylaldehyde ligand has attracted continuous research interest not only because of their appealing structural and topological novelty, but also due to their unusual optical, electronic, magnetic, and catalytic properties, as well as their potential medical application (Alam et al., 2003; Zelewsky & von Knof, 1999). In the present paper, we describe the synthesis and structural characterizations of the title compound, (I),
As shown in Fig. 1, each Mn(III) atom is chelated by Schiff base ligand via two N and two O atoms and is additionally coordinated by one azide and a water molecule, forming a distorted octahedral geometry (Table 1) in which, the Schiff base lies in the equatorial plane, and the azide and aqua ligands lie in the axial coordination sites.
With O—H···O and O—H···N hydrogen bonds (Table 2), a three-dimensional network is formed as shown in Fig. 2.
Experimental
A mixture of manganese(III) acetylacetonate (1 mmol) and N,N'-bis(2-hydroxy-5-bromobenzyl)1,2-diaminopropane (1 mmol), and dipotassium nickel tetracyanide (1 mmol) in 20 ml methanol was refluxed for several hours. The above cooled solution was filtered and the filtrate was kept in an ice box. One week later, brown blocks of (I) were obtained with a yield of 5%. Anal. Calc. for C40H34Mn2N10O7: C 54.75, H 3.88, N 15.97%; Found: C 54.71, H 3.75, N 15.82.
Refinement
All H atoms were placed in calculated positions with C—H = 0.93Å and refined as riding with Uiso(H) = 1.2Ueq(carrier). H atom on aqua were located from difference density maps and were refined with distance restraints of O–H = 0.82 (1) Å.
Figures
Fig. 1.
The molecular structure of (I), drawn with 30% probability displacement ellipsoids for the non-hydrogen atoms.
Fig. 2.
Three-dimensional network formed by hydrogen bonds (dashed lines).
Crystal data
| [Mn(C20H14N2O2)(N3)(H2O)]·0.5H2O | F(000) = 1808 |
| Mr = 438.33 | Dx = 1.612 Mg m−3 |
| Monoclinic, C2/c | Mo Kα radiation, λ = 0.71070 Å |
| Hall symbol: -C 2yc | Cell parameters from 3162 reflections |
| a = 25.10 (1) Å | θ = 3.0–25.0° |
| b = 11.478 (5) Å | µ = 0.77 mm−1 |
| c = 12.599 (5) Å | T = 293 K |
| β = 94.175 (3)° | Block, pink |
| V = 3620 (3) Å3 | 0.12 × 0.10 × 0.08 mm |
| Z = 8 |
Data collection
| Bruker APEXII CCD area-detector diffractometer | 3162 independent reflections |
| Radiation source: fine-focus sealed tube | 2371 reflections with I > 2σ(I) |
| graphite | Rint = 0.082 |
| φ and ω scans | θmax = 25.0°, θmin = 3.0° |
| Absorption correction: multi-scan (SADABS; Bruker, 2004) | h = −27→29 |
| Tmin = 0.914, Tmax = 0.941 | k = −13→13 |
| 11927 measured reflections | l = −14→13 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.037 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.086 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.00 | w = 1/[σ2(Fo2) + (0.03P)2] where P = (Fo2 + 2Fc2)/3 |
| 3162 reflections | (Δ/σ)max = 0.001 |
| 280 parameters | Δρmax = 0.32 e Å−3 |
| 4 restraints | Δρmin = −0.38 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Mn1 | 0.204685 (15) | 0.09791 (3) | 0.87813 (3) | 0.00903 (13) | |
| C1 | 0.11631 (10) | 0.2543 (2) | 0.90445 (18) | 0.0102 (6) | |
| C2 | 0.09780 (10) | 0.3688 (2) | 0.90882 (19) | 0.0124 (6) | |
| H2 | 0.1219 | 0.4301 | 0.9061 | 0.015* | |
| C3 | 0.04451 (10) | 0.3928 (2) | 0.91706 (19) | 0.0156 (6) | |
| H3 | 0.0332 | 0.4699 | 0.9196 | 0.019* | |
| C4 | 0.00719 (10) | 0.3029 (2) | 0.9217 (2) | 0.0180 (6) | |
| H4 | −0.0288 | 0.3197 | 0.9260 | 0.022* | |
| C5 | 0.02444 (10) | 0.1897 (2) | 0.9198 (2) | 0.0161 (6) | |
| H5 | −0.0002 | 0.1297 | 0.9238 | 0.019* | |
| C6 | 0.07880 (10) | 0.1623 (2) | 0.91183 (18) | 0.0110 (6) | |
| C7 | 0.09346 (10) | 0.0426 (2) | 0.91314 (18) | 0.0114 (6) | |
| H7 | 0.0662 | −0.0115 | 0.9188 | 0.014* | |
| C8 | 0.15275 (10) | −0.1198 (2) | 0.91053 (18) | 0.0092 (6) | |
| C9 | 0.11626 (10) | −0.2052 (2) | 0.93648 (19) | 0.0123 (6) | |
| H9 | 0.0819 | −0.1844 | 0.9525 | 0.015* | |
| C10 | 0.13142 (10) | −0.3207 (2) | 0.93819 (18) | 0.0123 (6) | |
| H10 | 0.1071 | −0.3777 | 0.9553 | 0.015* | |
| C11 | 0.18272 (10) | −0.3529 (2) | 0.91454 (18) | 0.0124 (6) | |
| H11 | 0.1925 | −0.4311 | 0.9157 | 0.015* | |
| C12 | 0.21902 (10) | −0.2690 (2) | 0.88938 (18) | 0.0109 (6) | |
| H12 | 0.2533 | −0.2906 | 0.8736 | 0.013* | |
| C13 | 0.20450 (10) | −0.1518 (2) | 0.88751 (18) | 0.0096 (5) | |
| C14 | 0.28922 (10) | −0.0719 (2) | 0.84750 (19) | 0.0110 (6) | |
| H14 | 0.3007 | −0.1480 | 0.8385 | 0.013* | |
| C15 | 0.32741 (10) | 0.0188 (2) | 0.83746 (18) | 0.0111 (6) | |
| C16 | 0.38034 (10) | −0.0164 (2) | 0.82023 (18) | 0.0145 (6) | |
| H16 | 0.3878 | −0.0954 | 0.8143 | 0.017* | |
| C17 | 0.42059 (10) | 0.0627 (2) | 0.81212 (19) | 0.0155 (6) | |
| H17 | 0.4548 | 0.0377 | 0.7998 | 0.019* | |
| C18 | 0.40976 (10) | 0.1809 (2) | 0.82255 (18) | 0.0142 (6) | |
| H18 | 0.4371 | 0.2350 | 0.8181 | 0.017* | |
| C19 | 0.35893 (10) | 0.2184 (2) | 0.83938 (19) | 0.0136 (6) | |
| H19 | 0.3525 | 0.2978 | 0.8462 | 0.016* | |
| C20 | 0.31675 (10) | 0.1395 (2) | 0.84649 (18) | 0.0094 (6) | |
| N1 | 0.23943 (8) | −0.05699 (18) | 0.86829 (15) | 0.0098 (5) | |
| N2 | 0.14151 (8) | 0.00196 (18) | 0.90708 (15) | 0.0092 (5) | |
| N3 | 0.17896 (8) | 0.07150 (19) | 0.70029 (16) | 0.0131 (5) | |
| N4 | 0.13255 (9) | 0.05323 (19) | 0.67637 (16) | 0.0136 (5) | |
| N5 | 0.08768 (9) | 0.0341 (2) | 0.65186 (17) | 0.0234 (6) | |
| O1 | 0.16772 (7) | 0.23708 (15) | 0.89293 (13) | 0.0127 (4) | |
| O2 | 0.26797 (6) | 0.18073 (15) | 0.85790 (12) | 0.0113 (4) | |
| O1W | 0.23229 (7) | 0.08568 (17) | 1.05761 (13) | 0.0138 (4) | |
| O2W | 0.0000 | −0.0960 (3) | 0.7500 | 0.0383 (8) | |
| H1W | 0.2366 (10) | 0.1525 (7) | 1.0796 (16) | 0.023 (9)* | |
| H2W | 0.2189 (11) | 0.0413 (15) | 1.0991 (14) | 0.037 (10)* | |
| H3W | 0.0224 (11) | −0.056 (3) | 0.723 (3) | 0.064 (13)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Mn1 | 0.0082 (2) | 0.0069 (2) | 0.0122 (2) | −0.00022 (17) | 0.00244 (16) | −0.00015 (16) |
| C1 | 0.0133 (14) | 0.0136 (14) | 0.0037 (13) | 0.0011 (11) | 0.0004 (10) | 0.0013 (11) |
| C2 | 0.0163 (14) | 0.0085 (14) | 0.0124 (14) | 0.0000 (11) | 0.0008 (11) | 0.0017 (11) |
| C3 | 0.0190 (15) | 0.0117 (15) | 0.0162 (15) | 0.0046 (12) | 0.0017 (12) | 0.0029 (11) |
| C4 | 0.0105 (14) | 0.0172 (16) | 0.0271 (16) | 0.0050 (12) | 0.0059 (12) | 0.0031 (13) |
| C5 | 0.0130 (14) | 0.0117 (15) | 0.0239 (16) | −0.0027 (11) | 0.0035 (12) | 0.0033 (12) |
| C6 | 0.0149 (14) | 0.0083 (14) | 0.0101 (14) | 0.0023 (11) | 0.0031 (11) | 0.0004 (11) |
| C7 | 0.0121 (14) | 0.0118 (15) | 0.0105 (14) | −0.0045 (11) | 0.0024 (11) | −0.0004 (11) |
| C8 | 0.0129 (14) | 0.0080 (14) | 0.0064 (13) | −0.0001 (11) | −0.0005 (10) | −0.0019 (10) |
| C9 | 0.0119 (14) | 0.0114 (15) | 0.0138 (14) | −0.0013 (11) | 0.0035 (11) | −0.0016 (11) |
| C10 | 0.0159 (14) | 0.0119 (15) | 0.0092 (14) | −0.0046 (11) | 0.0014 (11) | 0.0001 (11) |
| C11 | 0.0205 (15) | 0.0067 (14) | 0.0093 (13) | 0.0021 (11) | −0.0028 (11) | 0.0002 (11) |
| C12 | 0.0129 (14) | 0.0145 (15) | 0.0052 (13) | 0.0043 (11) | 0.0005 (10) | −0.0027 (10) |
| C13 | 0.0124 (14) | 0.0120 (14) | 0.0042 (13) | −0.0028 (11) | −0.0007 (10) | −0.0015 (11) |
| C14 | 0.0142 (14) | 0.0106 (15) | 0.0082 (13) | 0.0035 (11) | 0.0011 (11) | 0.0006 (10) |
| C15 | 0.0128 (14) | 0.0144 (14) | 0.0062 (13) | −0.0007 (11) | 0.0016 (10) | 0.0010 (11) |
| C16 | 0.0170 (15) | 0.0162 (15) | 0.0104 (14) | 0.0038 (12) | 0.0021 (11) | 0.0012 (11) |
| C17 | 0.0069 (14) | 0.0278 (17) | 0.0119 (14) | 0.0028 (12) | 0.0011 (11) | 0.0015 (12) |
| C18 | 0.0107 (14) | 0.0240 (17) | 0.0077 (14) | −0.0048 (12) | −0.0008 (11) | 0.0009 (12) |
| C19 | 0.0193 (15) | 0.0119 (15) | 0.0095 (14) | −0.0023 (12) | −0.0006 (11) | −0.0023 (11) |
| C20 | 0.0082 (13) | 0.0175 (15) | 0.0026 (12) | 0.0006 (11) | 0.0005 (10) | 0.0020 (11) |
| N1 | 0.0135 (12) | 0.0086 (12) | 0.0073 (11) | 0.0005 (9) | 0.0014 (9) | 0.0004 (9) |
| N2 | 0.0118 (11) | 0.0070 (12) | 0.0088 (11) | 0.0010 (9) | 0.0018 (9) | 0.0002 (9) |
| N3 | 0.0097 (12) | 0.0190 (14) | 0.0108 (12) | −0.0020 (9) | 0.0013 (9) | 0.0014 (9) |
| N4 | 0.0198 (14) | 0.0138 (13) | 0.0078 (12) | 0.0027 (10) | 0.0047 (10) | 0.0002 (9) |
| N5 | 0.0122 (13) | 0.0395 (17) | 0.0183 (13) | 0.0007 (12) | −0.0002 (10) | 0.0000 (11) |
| O1 | 0.0099 (9) | 0.0074 (10) | 0.0213 (10) | −0.0001 (7) | 0.0038 (7) | −0.0005 (8) |
| O2 | 0.0114 (9) | 0.0097 (10) | 0.0133 (10) | −0.0007 (8) | 0.0037 (7) | 0.0011 (8) |
| O1W | 0.0192 (11) | 0.0090 (11) | 0.0133 (10) | −0.0039 (8) | 0.0027 (8) | −0.0004 (9) |
| O2W | 0.025 (2) | 0.028 (2) | 0.063 (2) | 0.000 | 0.0130 (18) | 0.000 |
Geometric parameters (Å, °)
| Mn1—O1 | 1.8636 (18) | C10—H10 | 0.9300 |
| Mn1—O2 | 1.8844 (18) | C11—C12 | 1.379 (3) |
| Mn1—N2 | 1.986 (2) | C11—H11 | 0.9300 |
| Mn1—N1 | 1.988 (2) | C12—C13 | 1.393 (3) |
| Mn1—N3 | 2.306 (2) | C12—H12 | 0.9300 |
| Mn1—O1W | 2.321 (2) | C13—N1 | 1.430 (3) |
| C1—O1 | 1.324 (3) | C14—N1 | 1.307 (3) |
| C1—C2 | 1.397 (3) | C14—C15 | 1.427 (3) |
| C1—C6 | 1.422 (3) | C14—H14 | 0.9300 |
| C2—C3 | 1.377 (3) | C15—C20 | 1.418 (4) |
| C2—H2 | 0.9300 | C15—C16 | 1.420 (3) |
| C3—C4 | 1.398 (4) | C16—C17 | 1.367 (4) |
| C3—H3 | 0.9300 | C16—H16 | 0.9300 |
| C4—C5 | 1.371 (4) | C17—C18 | 1.392 (4) |
| C4—H4 | 0.9300 | C17—H17 | 0.9300 |
| C5—C6 | 1.411 (3) | C18—C19 | 1.378 (3) |
| C5—H5 | 0.9300 | C18—H18 | 0.9300 |
| C6—C7 | 1.422 (4) | C19—C20 | 1.401 (3) |
| C7—N2 | 1.301 (3) | C19—H19 | 0.9300 |
| C7—H7 | 0.9300 | C20—O2 | 1.330 (3) |
| C8—C9 | 1.397 (3) | N3—N4 | 1.200 (3) |
| C8—C13 | 1.401 (3) | N4—N5 | 1.167 (3) |
| C8—N2 | 1.425 (3) | O1W—H1W | 0.820 (11) |
| C9—C10 | 1.379 (4) | O1W—H2W | 0.82 (2) |
| C9—H9 | 0.9300 | O2W—H3W | 0.82 (3) |
| C10—C11 | 1.393 (3) | ||
| O1—Mn1—O2 | 90.68 (8) | C11—C10—H10 | 119.7 |
| O1—Mn1—N2 | 92.69 (8) | C12—C11—C10 | 120.0 (2) |
| O2—Mn1—N2 | 175.37 (8) | C12—C11—H11 | 120.0 |
| O1—Mn1—N1 | 175.33 (8) | C10—C11—H11 | 120.0 |
| O2—Mn1—N1 | 93.71 (8) | C11—C12—C13 | 120.1 (2) |
| N2—Mn1—N1 | 82.83 (9) | C11—C12—H12 | 119.9 |
| O1—Mn1—N3 | 95.95 (8) | C13—C12—H12 | 119.9 |
| O2—Mn1—N3 | 96.55 (7) | C12—C13—C8 | 119.7 (2) |
| N2—Mn1—N3 | 86.26 (8) | C12—C13—N1 | 125.1 (2) |
| N1—Mn1—N3 | 85.12 (8) | C8—C13—N1 | 115.1 (2) |
| O1—Mn1—O1W | 94.00 (7) | N1—C14—C15 | 125.5 (2) |
| O2—Mn1—O1W | 88.14 (7) | N1—C14—H14 | 117.2 |
| N2—Mn1—O1W | 88.47 (7) | C15—C14—H14 | 117.2 |
| N1—Mn1—O1W | 84.58 (7) | C20—C15—C16 | 118.3 (2) |
| N3—Mn1—O1W | 168.94 (7) | C20—C15—C14 | 125.0 (2) |
| O1—C1—C2 | 118.3 (2) | C16—C15—C14 | 116.6 (2) |
| O1—C1—C6 | 123.5 (2) | C17—C16—C15 | 121.8 (3) |
| C2—C1—C6 | 118.2 (2) | C17—C16—H16 | 119.1 |
| C3—C2—C1 | 121.3 (2) | C15—C16—H16 | 119.1 |
| C3—C2—H2 | 119.4 | C16—C17—C18 | 119.3 (2) |
| C1—C2—H2 | 119.4 | C16—C17—H17 | 120.3 |
| C2—C3—C4 | 120.9 (3) | C18—C17—H17 | 120.3 |
| C2—C3—H3 | 119.6 | C19—C18—C17 | 120.6 (2) |
| C4—C3—H3 | 119.6 | C19—C18—H18 | 119.7 |
| C5—C4—C3 | 119.1 (2) | C17—C18—H18 | 119.7 |
| C5—C4—H4 | 120.5 | C18—C19—C20 | 121.3 (3) |
| C3—C4—H4 | 120.5 | C18—C19—H19 | 119.3 |
| C4—C5—C6 | 121.4 (2) | C20—C19—H19 | 119.3 |
| C4—C5—H5 | 119.3 | O2—C20—C19 | 118.9 (2) |
| C6—C5—H5 | 119.3 | O2—C20—C15 | 122.5 (2) |
| C5—C6—C1 | 119.2 (2) | C19—C20—C15 | 118.6 (2) |
| C5—C6—C7 | 117.7 (2) | C14—N1—C13 | 122.8 (2) |
| C1—C6—C7 | 123.1 (2) | C14—N1—Mn1 | 124.04 (18) |
| N2—C7—C6 | 125.9 (2) | C13—N1—Mn1 | 113.16 (16) |
| N2—C7—H7 | 117.1 | C7—N2—C8 | 122.2 (2) |
| C6—C7—H7 | 117.1 | C7—N2—Mn1 | 124.61 (18) |
| C9—C8—C13 | 119.9 (2) | C8—N2—Mn1 | 112.96 (15) |
| C9—C8—N2 | 124.3 (2) | N4—N3—Mn1 | 117.69 (16) |
| C13—C8—N2 | 115.8 (2) | N5—N4—N3 | 178.8 (3) |
| C10—C9—C8 | 119.6 (2) | C1—O1—Mn1 | 129.44 (16) |
| C10—C9—H9 | 120.2 | C20—O2—Mn1 | 128.82 (16) |
| C8—C9—H9 | 120.2 | Mn1—O1W—H1W | 107.2 (17) |
| C9—C10—C11 | 120.7 (2) | Mn1—O1W—H2W | 123.5 (18) |
| C9—C10—H10 | 119.7 | H1W—O1W—H2W | 114.6 (19) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1W—H2W···N3i | 0.82 (2) | 2.12 (2) | 2.937 (3) | 176 (3) |
| O1W—H1W···O2ii | 0.82 (1) | 2.08 (1) | 2.885 (3) | 169 (2) |
| O2W—H3W···N5 | 0.82 (3) | 2.18 (3) | 3.000 (3) | 173 (4) |
Symmetry codes: (i) x, −y, z+1/2; (ii) −x+1/2, −y+1/2, −z+2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HB2977).
References
- Alam, M. A., Nethaji, M. & Ray, M. (2003). Angew. Chem. Int. Ed.42, 1940–1942. [DOI] [PubMed]
- Bruker (2004). APEX2, SAINT-Plus and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Zelewsky, A. & von Knof, U. (1999). Angew. Chem. Int. Ed.38, 302–322. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809020200/hb2977sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809020200/hb2977Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




