Abstract
Crystals of the title salt, C17H28NO3 +·C4H3O4 −, were obtained by reacting parthenolide with dimethylamine followed by conversion of the amine adduct into a water-soluble fumarate salt. Subsequent crystallization of the fumarate salt from water afforded colorless orthorhombic crystals. The amine addition is highly stereospecific yielding exclusively a single diastereomer with R-configuration at the newly formed C-11 chiral carbon. In the crystal, intermolecular O—H⋯O and N—H⋯O hydrogen bonds help to establish the packing.
Related literature
Parthenolide (PTL) is a naturally occurring sesquiterpene lactone used in the treatment of fever, migraine headaches, rheumatoid arthritis, and also as an anti-inflammatory agent (Heptinstall et al. (1988 ▶). For the potent anti-tumor and cytotoxic properties of PTL, see: Crooks et al. (2007 ▶). The absolute stereochemistry of the C-11 chiral carbon is typical of such amine adducts of parthenolide, see: Nasim et al. (2007a
▶,b
▶). For bond-length data, see: Allen et al. (1987 ▶).
Experimental
Crystal data
C17H28NO3 +·C4H3O4 −
M r = 409.47
Orthorhombic,
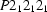
a = 6.3164 (1) Å
b = 15.1650 (2) Å
c = 22.0028 (3) Å
V = 2107.61 (5) Å3
Z = 4
Cu Kα radiation
μ = 0.80 mm−1
T = 90 K
0.26 × 0.20 × 0.10 mm
Data collection
Bruker X8 Proteum diffractometer
Absorption correction: multi-scan (SADABS in APEX2; Bruker–Nonius, 2006 ▶) T min = 0.788, T max = 0.924
16991 measured reflections
3762 independent reflections
3640 reflections with I > 2σ(I)
R int = 0.041
Refinement
R[F 2 > 2σ(F 2)] = 0.027
wR(F 2) = 0.069
S = 1.04
3762 reflections
268 parameters
H-atom parameters constrained
Δρmax = 0.20 e Å−3
Δρmin = −0.14 e Å−3
Absolute structure: Flack (1983 ▶), 1525 Friedel pairs
Flack parameter: −0.01 (4)
Data collection: APEX2 (Bruker–Nonius, 2006 ▶); cell refinement: SAINT (Bruker–Nonius, 2006 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: XP in SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXL97 and local procedures.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809021941/hg2516sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809021941/hg2516Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1N⋯O6 | 0.93 | 1.83 | 2.7563 (14) | 172 |
| O4—H4⋯O6i | 0.84 | 1.73 | 2.5544 (13) | 169 |
Symmetry code: (i) 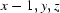 .
.
Acknowledgments
This research work was supported by the Kentucky Lung Cancer Research Program.
supplementary crystallographic information
Comment
Parthenolide (PTL) isolated from Tanacetum parthenium (commonly referred as feverfew), is a naturally occurring sesquiterpene lactone used in the treatment of fever, migraine headaches, rheumatoid arthritis, and also as an anti-inflammatory agent (Heptinstall et al., 1988). The PTL molecule and several structurally related analogs have become topics of recent interest because of their potent anti-tumor and cytotoxic properties (Crooks et al., 2007). Despite promising in vitro activity, this potent natural product has a major limitation which precludes its further development as a therapeutic agent, i.e. its poor water-solubility, thus limiting its potential as a promising clinical agent. The title compound crystallized from water as orthorombic crystals. Bond angles and bond distances within the molecule were quite regular with average normal bond lengths (Allen et al., 1987). The absolute stereochemistry of the newly formed C-11 chiral carbon was found to be R, which is typical of such amine adducts of parthenolide (Nasim et al., 2007a, 2007b). Hydrogen bonding was observed between N1—H and O6 of the carbonyl oxygen of the fumarate moiety and between O4—H and O6 (Table 1). The title compound is more water soluble and more biologically potent than the parent compound, in both in vitro and in vivo anti-leukemic activity screens. T he title compound is currently in phase 1 clinical trials.
Experimental
The title compound (Systematic name: 13-(N,N-dimethyl)-amino-4 α,5 β- epoxy-4,10-dimethyl-6 α-hydroxy-12-oicacid-γ-lactone-germacra-1(10)-ene monofumarate) was synthesized by dissolving PTL (25 mg, 0.1 mmol) in 8 ml of methanol followed by addition of dimethylamine (0.15 mmol, 2.0M solution in methanol). The mixture stirred under ambient conditions for 6 hrs. The crude product was subjected to flash silica gel column chromatography to afford the pure dimethylamino analog free base. The free base was then converted to the fumarate salt by dissolving it in diethyl ether followed by addition of one equivalent of fumaric acid. The white solid that precipitated out of the diethyl ether solution was filtered, washed with diethyl ether and then dried under vacuum. Crystallization of the obtained white solid from water afforded colorless crystals that were suitable for X-ray analysis. 1H-NMR (D20, 300 MHz):δ 6.67 2H, s, (HOOC-CH)2–), 5.24 (1H, dd, J=2.1, 12.0 Hz, 1-CH), 4.30 (1H, t, J=9.0 Hz, 6-CH), 1.88–3.60 (13H, m, 2-CH2, 3-CH2, 8-CH2, 9-CH2, 13-CH2, 5-CH, 7-CH, 11-CH), 2.98 (6H, s, N-(CH3)2), 1.70 (3H, s, 14-CH3), 1.34 (3H, s, 15-CH3) p.p.m. 13C-NMR (DMSO-d6, 75 MHz): δ 179.8 ((HOOC-CH)2–), 173.8 (12-C=O), 138.2 ((HOOC-CH)2–),137.2 (10-C), 127.3 (1-C), 85.8 (6-C), 69.2 (N-(CH3)2), 67.1 (5-C), 58.3 (4-C), 49.6 (7-C), 44.9 (11-C), 42.5 (9-C), 38.1 (3-C), 30.9 (8-C), 26.1(2-C), 18.7 (15-C), 18.6 (14-C)p.p.m. Elemental analysis: calc. for C21H31NO7: C 61.60%, H 7.63%, N 3.42%. Found: C 61.81%, H 7.64%, N 3.47%.
Refinement
H atoms were found in difference Fourier maps and subsequently placed in idealized positions with constrained distances of 0.98 Å (RCH3), 0.99 Å (R2CH2), 1.00 Å (R3CH), 0.95 Å (Csp2H), 0.84 Å (O—H), 0.93 Å (N—H), and with Uiso(H) values set to either 1.2Ueq or 1.5Ueq (RCH3, OH) of the attached atom.
Figures
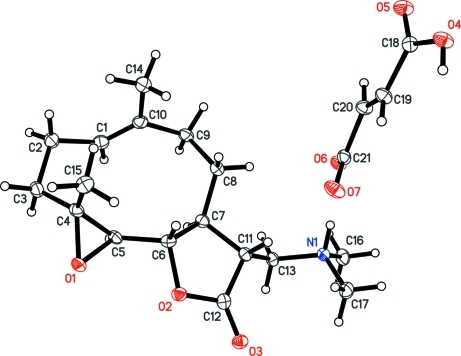
Fig. 1.
A view of the molecule (I), showing the molecule and the atom numbering scheme. Displacement ellipsoids are drawn at the 50% probability level.
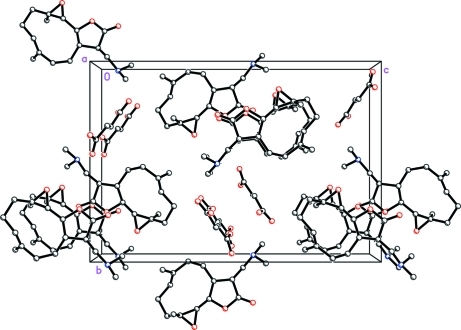
Fig. 2.
Crystal packing of (I) viewed along the a axis. H atoms have been omitted for clarity.
Crystal data
| C17H28NO3+·C4H3O4− | F(000) = 880 |
| Mr = 409.47 | Dx = 1.290 Mg m−3 |
| Orthorhombic, P212121 | Cu Kα radiation, λ = 1.54178 Å |
| Hall symbol: P 2ac 2ab | Cell parameters from 5373 reflections |
| a = 6.3164 (1) Å | θ = 3.5–68.0° |
| b = 15.1650 (2) Å | µ = 0.80 mm−1 |
| c = 22.0028 (3) Å | T = 90 K |
| V = 2107.61 (5) Å3 | Block, colourless |
| Z = 4 | 0.26 × 0.20 × 0.10 mm |
Data collection
| Bruker X8 Proteum diffractometer | 3762 independent reflections |
| Radiation source: fine-focus rotating anode | 3640 reflections with I > 2σ(I) |
| graded multilayer optics | Rint = 0.041 |
| Detector resolution: 5.6 pixels mm-1 | θmax = 68.0°, θmin = 3.5° |
| φ and ω scans | h = −7→7 |
| Absorption correction: multi-scan (SADABS in APEX2; Bruker–Nonius, 2006) | k = −15→18 |
| Tmin = 0.788, Tmax = 0.924 | l = −26→26 |
| 16991 measured reflections |
Refinement
| Refinement on F2 | Hydrogen site location: inferred from neighbouring sites |
| Least-squares matrix: full | H-atom parameters constrained |
| R[F2 > 2σ(F2)] = 0.027 | w = 1/[σ2(Fo2) + (0.0359P)2 + 0.3552P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.069 | (Δ/σ)max = 0.001 |
| S = 1.04 | Δρmax = 0.20 e Å−3 |
| 3762 reflections | Δρmin = −0.14 e Å−3 |
| 268 parameters | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| 0 restraints | Extinction coefficient: 0.00093 (19) |
| Primary atom site location: structure-invariant direct methods | Absolute structure: Flack (1983), 1525 Friedel pairs |
| Secondary atom site location: difference Fourier map | Flack parameter: −0.01 (4) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger.Flack x(u) obtained by Parsons quotient method, as implemented in XPREP. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N1 | 0.57684 (17) | 0.48961 (7) | −0.06421 (5) | 0.0150 (2) | |
| H1N | 0.5070 | 0.4514 | −0.0379 | 0.018* | |
| O1 | 0.50601 (15) | 0.85986 (6) | 0.14786 (4) | 0.0179 (2) | |
| O2 | 0.47768 (14) | 0.76214 (6) | 0.03689 (4) | 0.0166 (2) | |
| O3 | 0.50611 (14) | 0.73366 (6) | −0.06201 (4) | 0.0173 (2) | |
| C1 | 0.7548 (2) | 0.65406 (9) | 0.23912 (6) | 0.0186 (3) | |
| H1 | 0.8861 | 0.6573 | 0.2182 | 0.022* | |
| C2 | 0.7092 (2) | 0.72736 (9) | 0.28283 (6) | 0.0200 (3) | |
| H2A | 0.8339 | 0.7369 | 0.3093 | 0.024* | |
| H2B | 0.5881 | 0.7107 | 0.3090 | 0.024* | |
| C3 | 0.6567 (2) | 0.81361 (9) | 0.24889 (6) | 0.0187 (3) | |
| H3A | 0.5997 | 0.8572 | 0.2781 | 0.022* | |
| H3B | 0.7881 | 0.8382 | 0.2312 | 0.022* | |
| C4 | 0.4967 (2) | 0.79867 (9) | 0.19886 (6) | 0.0161 (3) | |
| C5 | 0.5848 (2) | 0.77075 (8) | 0.13995 (5) | 0.0150 (3) | |
| H5 | 0.7427 | 0.7662 | 0.1394 | 0.018* | |
| C6 | 0.4759 (2) | 0.71234 (9) | 0.09449 (5) | 0.0158 (3) | |
| H6 | 0.3272 | 0.6999 | 0.1075 | 0.019* | |
| C7 | 0.5921 (2) | 0.62571 (8) | 0.07898 (5) | 0.0151 (3) | |
| H7 | 0.7481 | 0.6367 | 0.0806 | 0.018* | |
| C8 | 0.5420 (2) | 0.54314 (9) | 0.11684 (6) | 0.0186 (3) | |
| H8A | 0.5512 | 0.4911 | 0.0898 | 0.022* | |
| H8B | 0.3937 | 0.5475 | 0.1311 | 0.022* | |
| C9 | 0.6848 (2) | 0.52658 (9) | 0.17255 (6) | 0.0191 (3) | |
| H9A | 0.6706 | 0.4642 | 0.1851 | 0.023* | |
| H9B | 0.8341 | 0.5364 | 0.1607 | 0.023* | |
| C10 | 0.6331 (2) | 0.58485 (9) | 0.22608 (6) | 0.0182 (3) | |
| C11 | 0.52671 (19) | 0.61351 (9) | 0.01203 (5) | 0.0146 (3) | |
| H11 | 0.3850 | 0.5841 | 0.0108 | 0.018* | |
| C12 | 0.50272 (19) | 0.70680 (9) | −0.01048 (5) | 0.0152 (3) | |
| C13 | 0.6820 (2) | 0.55958 (9) | −0.02661 (6) | 0.0155 (3) | |
| H13A | 0.7598 | 0.6001 | −0.0540 | 0.019* | |
| H13B | 0.7868 | 0.5313 | 0.0006 | 0.019* | |
| C14 | 0.4359 (2) | 0.55947 (10) | 0.26023 (6) | 0.0238 (3) | |
| H14A | 0.4091 | 0.6027 | 0.2924 | 0.036* | |
| H14B | 0.4549 | 0.5009 | 0.2783 | 0.036* | |
| H14C | 0.3154 | 0.5583 | 0.2322 | 0.036* | |
| C15 | 0.2737 (2) | 0.77859 (10) | 0.21831 (6) | 0.0209 (3) | |
| H15A | 0.2179 | 0.8281 | 0.2420 | 0.031* | |
| H15B | 0.2727 | 0.7251 | 0.2433 | 0.031* | |
| H15C | 0.1851 | 0.7695 | 0.1823 | 0.031* | |
| C16 | 0.7432 (2) | 0.43816 (10) | −0.09703 (6) | 0.0211 (3) | |
| H16A | 0.6762 | 0.3908 | −0.1204 | 0.032* | |
| H16B | 0.8420 | 0.4127 | −0.0675 | 0.032* | |
| H16C | 0.8204 | 0.4773 | −0.1247 | 0.032* | |
| C17 | 0.4184 (2) | 0.52542 (10) | −0.10767 (6) | 0.0221 (3) | |
| H17A | 0.4871 | 0.5680 | −0.1347 | 0.033* | |
| H17B | 0.3047 | 0.5548 | −0.0851 | 0.033* | |
| H17C | 0.3589 | 0.4771 | −0.1318 | 0.033* | |
| O4 | −0.44376 (14) | 0.24575 (7) | 0.08148 (4) | 0.0203 (2) | |
| H4 | −0.4761 | 0.2890 | 0.0595 | 0.030* | |
| O5 | −0.15889 (15) | 0.19509 (7) | 0.12810 (4) | 0.0218 (2) | |
| O6 | 0.40728 (13) | 0.37319 (6) | 0.01876 (4) | 0.0173 (2) | |
| O7 | 0.12629 (15) | 0.46177 (7) | 0.01397 (5) | 0.0241 (2) | |
| C18 | −0.2399 (2) | 0.24937 (9) | 0.09497 (5) | 0.0161 (3) | |
| C19 | −0.1193 (2) | 0.32332 (9) | 0.06681 (6) | 0.0178 (3) | |
| H19 | −0.1953 | 0.3724 | 0.0512 | 0.021* | |
| C20 | 0.0892 (2) | 0.32310 (9) | 0.06284 (6) | 0.0183 (3) | |
| H20 | 0.1650 | 0.2766 | 0.0820 | 0.022* | |
| C21 | 0.21235 (19) | 0.39254 (9) | 0.02961 (6) | 0.0164 (3) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0157 (5) | 0.0132 (6) | 0.0161 (5) | −0.0008 (4) | 0.0006 (4) | 0.0002 (4) |
| O1 | 0.0244 (5) | 0.0109 (5) | 0.0184 (4) | 0.0016 (4) | 0.0014 (4) | 0.0005 (3) |
| O2 | 0.0190 (4) | 0.0151 (5) | 0.0159 (4) | 0.0024 (4) | 0.0009 (3) | 0.0014 (4) |
| O3 | 0.0150 (4) | 0.0191 (5) | 0.0177 (4) | 0.0008 (4) | −0.0013 (4) | 0.0027 (4) |
| C1 | 0.0177 (6) | 0.0190 (8) | 0.0191 (6) | 0.0032 (5) | −0.0013 (5) | 0.0031 (5) |
| C2 | 0.0217 (7) | 0.0186 (7) | 0.0197 (6) | −0.0007 (5) | −0.0042 (5) | 0.0006 (5) |
| C3 | 0.0212 (6) | 0.0147 (8) | 0.0201 (6) | −0.0005 (5) | 0.0013 (5) | −0.0025 (5) |
| C4 | 0.0188 (6) | 0.0114 (7) | 0.0182 (6) | 0.0029 (5) | 0.0024 (5) | 0.0007 (5) |
| C5 | 0.0163 (6) | 0.0101 (7) | 0.0184 (6) | 0.0010 (5) | 0.0017 (5) | 0.0006 (5) |
| C6 | 0.0159 (6) | 0.0162 (7) | 0.0153 (5) | 0.0008 (5) | 0.0018 (5) | 0.0026 (5) |
| C7 | 0.0147 (6) | 0.0146 (7) | 0.0159 (6) | −0.0003 (5) | 0.0011 (5) | 0.0007 (5) |
| C8 | 0.0233 (7) | 0.0144 (7) | 0.0181 (6) | −0.0019 (5) | 0.0005 (5) | 0.0000 (5) |
| C9 | 0.0247 (7) | 0.0124 (7) | 0.0202 (6) | 0.0019 (5) | 0.0004 (5) | 0.0028 (5) |
| C10 | 0.0221 (7) | 0.0161 (7) | 0.0163 (6) | 0.0036 (5) | −0.0017 (5) | 0.0034 (5) |
| C11 | 0.0134 (6) | 0.0136 (7) | 0.0167 (6) | −0.0002 (5) | 0.0009 (5) | −0.0004 (5) |
| C12 | 0.0086 (5) | 0.0179 (7) | 0.0192 (6) | 0.0002 (5) | −0.0012 (5) | −0.0014 (5) |
| C13 | 0.0138 (6) | 0.0149 (7) | 0.0179 (6) | −0.0003 (5) | −0.0009 (5) | −0.0011 (5) |
| C14 | 0.0297 (8) | 0.0201 (8) | 0.0215 (6) | −0.0038 (6) | 0.0035 (6) | −0.0005 (5) |
| C15 | 0.0181 (6) | 0.0243 (8) | 0.0202 (6) | 0.0021 (6) | 0.0026 (5) | −0.0001 (6) |
| C16 | 0.0211 (7) | 0.0194 (8) | 0.0227 (6) | 0.0001 (6) | 0.0043 (6) | −0.0050 (5) |
| C17 | 0.0232 (7) | 0.0209 (8) | 0.0221 (6) | −0.0002 (6) | −0.0072 (5) | −0.0012 (6) |
| O4 | 0.0131 (4) | 0.0204 (5) | 0.0274 (5) | −0.0009 (4) | −0.0008 (4) | 0.0083 (4) |
| O5 | 0.0182 (5) | 0.0228 (6) | 0.0244 (5) | 0.0004 (4) | 0.0000 (4) | 0.0074 (4) |
| O6 | 0.0126 (4) | 0.0167 (5) | 0.0227 (5) | −0.0007 (4) | 0.0014 (4) | 0.0025 (4) |
| O7 | 0.0164 (5) | 0.0175 (6) | 0.0383 (5) | 0.0004 (4) | −0.0013 (4) | 0.0075 (4) |
| C18 | 0.0140 (6) | 0.0174 (7) | 0.0170 (6) | 0.0004 (5) | 0.0014 (5) | −0.0008 (5) |
| C19 | 0.0164 (6) | 0.0139 (7) | 0.0230 (7) | 0.0008 (5) | −0.0002 (5) | 0.0018 (5) |
| C20 | 0.0160 (6) | 0.0184 (8) | 0.0205 (6) | 0.0002 (5) | −0.0014 (5) | 0.0046 (5) |
| C21 | 0.0145 (6) | 0.0164 (7) | 0.0183 (6) | −0.0020 (5) | −0.0027 (5) | −0.0002 (5) |
Geometric parameters (Å, °)
| N1—C17 | 1.4870 (17) | C9—H9A | 0.9900 |
| N1—C16 | 1.4948 (17) | C9—H9B | 0.9900 |
| N1—C13 | 1.5005 (16) | C10—C14 | 1.505 (2) |
| N1—H1N | 0.9300 | C11—C12 | 1.5064 (19) |
| O1—C5 | 1.4507 (16) | C11—C13 | 1.5344 (17) |
| O1—C4 | 1.4574 (15) | C11—H11 | 1.0000 |
| O2—C12 | 1.3474 (15) | C13—H13A | 0.9900 |
| O2—C6 | 1.4754 (14) | C13—H13B | 0.9900 |
| O3—C12 | 1.2050 (15) | C14—H14A | 0.9800 |
| C1—C10 | 1.332 (2) | C14—H14B | 0.9800 |
| C1—C2 | 1.4980 (19) | C14—H14C | 0.9800 |
| C1—H1 | 0.9500 | C15—H15A | 0.9800 |
| C2—C3 | 1.5422 (19) | C15—H15B | 0.9800 |
| C2—H2A | 0.9900 | C15—H15C | 0.9800 |
| C2—H2B | 0.9900 | C16—H16A | 0.9800 |
| C3—C4 | 1.5114 (19) | C16—H16B | 0.9800 |
| C3—H3A | 0.9900 | C16—H16C | 0.9800 |
| C3—H3B | 0.9900 | C17—H17A | 0.9800 |
| C4—C5 | 1.4728 (17) | C17—H17B | 0.9800 |
| C4—C15 | 1.5032 (18) | C17—H17C | 0.9800 |
| C5—C6 | 1.5027 (18) | O4—C18 | 1.3224 (15) |
| C5—H5 | 1.0000 | O4—H4 | 0.8400 |
| C6—C7 | 1.5429 (18) | O5—C18 | 1.2128 (16) |
| C6—H6 | 1.0000 | O6—C21 | 1.2880 (16) |
| C7—C8 | 1.5368 (18) | O7—C21 | 1.2314 (17) |
| C7—C11 | 1.5411 (17) | C18—C19 | 1.4905 (19) |
| C7—H7 | 1.0000 | C19—C20 | 1.3199 (19) |
| C8—C9 | 1.5427 (18) | C19—H19 | 0.9500 |
| C8—H8A | 0.9900 | C20—C21 | 1.4996 (18) |
| C8—H8B | 0.9900 | C20—H20 | 0.9500 |
| C9—C10 | 1.5084 (18) | ||
| C17—N1—C16 | 110.68 (10) | C8—C9—H9B | 108.9 |
| C17—N1—C13 | 113.23 (10) | H9A—C9—H9B | 107.7 |
| C16—N1—C13 | 108.92 (10) | C1—C10—C14 | 124.88 (12) |
| C17—N1—H1N | 108.0 | C1—C10—C9 | 120.31 (12) |
| C16—N1—H1N | 108.0 | C14—C10—C9 | 114.79 (12) |
| C13—N1—H1N | 108.0 | C12—C11—C13 | 112.50 (10) |
| C5—O1—C4 | 60.85 (8) | C12—C11—C7 | 103.21 (10) |
| C12—O2—C6 | 110.27 (10) | C13—C11—C7 | 114.98 (10) |
| C10—C1—C2 | 127.72 (12) | C12—C11—H11 | 108.6 |
| C10—C1—H1 | 116.1 | C13—C11—H11 | 108.6 |
| C2—C1—H1 | 116.1 | C7—C11—H11 | 108.6 |
| C1—C2—C3 | 111.09 (10) | O3—C12—O2 | 121.29 (12) |
| C1—C2—H2A | 109.4 | O3—C12—C11 | 128.68 (12) |
| C3—C2—H2A | 109.4 | O2—C12—C11 | 110.03 (10) |
| C1—C2—H2B | 109.4 | N1—C13—C11 | 113.53 (10) |
| C3—C2—H2B | 109.4 | N1—C13—H13A | 108.9 |
| H2A—C2—H2B | 108.0 | C11—C13—H13A | 108.9 |
| C4—C3—C2 | 111.67 (11) | N1—C13—H13B | 108.9 |
| C4—C3—H3A | 109.3 | C11—C13—H13B | 108.9 |
| C2—C3—H3A | 109.3 | H13A—C13—H13B | 107.7 |
| C4—C3—H3B | 109.3 | C10—C14—H14A | 109.5 |
| C2—C3—H3B | 109.3 | C10—C14—H14B | 109.5 |
| H3A—C3—H3B | 107.9 | H14A—C14—H14B | 109.5 |
| O1—C4—C5 | 59.35 (8) | C10—C14—H14C | 109.5 |
| O1—C4—C15 | 112.71 (10) | H14A—C14—H14C | 109.5 |
| C5—C4—C15 | 123.12 (12) | H14B—C14—H14C | 109.5 |
| O1—C4—C3 | 115.99 (11) | C4—C15—H15A | 109.5 |
| C5—C4—C3 | 115.55 (11) | C4—C15—H15B | 109.5 |
| C15—C4—C3 | 116.71 (11) | H15A—C15—H15B | 109.5 |
| O1—C5—C4 | 59.80 (8) | C4—C15—H15C | 109.5 |
| O1—C5—C6 | 118.16 (10) | H15A—C15—H15C | 109.5 |
| C4—C5—C6 | 125.61 (11) | H15B—C15—H15C | 109.5 |
| O1—C5—H5 | 114.1 | N1—C16—H16A | 109.5 |
| C4—C5—H5 | 114.1 | N1—C16—H16B | 109.5 |
| C6—C5—H5 | 114.1 | H16A—C16—H16B | 109.5 |
| O2—C6—C5 | 105.46 (10) | N1—C16—H16C | 109.5 |
| O2—C6—C7 | 104.02 (9) | H16A—C16—H16C | 109.5 |
| C5—C6—C7 | 115.56 (10) | H16B—C16—H16C | 109.5 |
| O2—C6—H6 | 110.5 | N1—C17—H17A | 109.5 |
| C5—C6—H6 | 110.5 | N1—C17—H17B | 109.5 |
| C7—C6—H6 | 110.5 | H17A—C17—H17B | 109.5 |
| C8—C7—C11 | 111.41 (10) | N1—C17—H17C | 109.5 |
| C8—C7—C6 | 118.42 (10) | H17A—C17—H17C | 109.5 |
| C11—C7—C6 | 100.73 (10) | H17B—C17—H17C | 109.5 |
| C8—C7—H7 | 108.6 | C18—O4—H4 | 109.5 |
| C11—C7—H7 | 108.6 | O5—C18—O4 | 121.16 (12) |
| C6—C7—H7 | 108.6 | O5—C18—C19 | 123.02 (12) |
| C7—C8—C9 | 116.29 (11) | O4—C18—C19 | 115.82 (11) |
| C7—C8—H8A | 108.2 | C20—C19—C18 | 122.38 (13) |
| C9—C8—H8A | 108.2 | C20—C19—H19 | 118.8 |
| C7—C8—H8B | 108.2 | C18—C19—H19 | 118.8 |
| C9—C8—H8B | 108.2 | C19—C20—C21 | 123.24 (13) |
| H8A—C8—H8B | 107.4 | C19—C20—H20 | 118.4 |
| C10—C9—C8 | 113.48 (11) | C21—C20—H20 | 118.4 |
| C10—C9—H9A | 108.9 | O7—C21—O6 | 124.37 (12) |
| C8—C9—H9A | 108.9 | O7—C21—C20 | 120.40 (12) |
| C10—C9—H9B | 108.9 | O6—C21—C20 | 115.23 (12) |
| C10—C1—C2—C3 | −107.32 (15) | C7—C8—C9—C10 | 76.93 (14) |
| C1—C2—C3—C4 | 47.74 (15) | C2—C1—C10—C14 | −9.6 (2) |
| C5—O1—C4—C15 | −116.14 (13) | C2—C1—C10—C9 | 168.47 (12) |
| C5—O1—C4—C3 | 105.60 (13) | C8—C9—C10—C1 | −103.08 (15) |
| C2—C3—C4—O1 | −152.71 (11) | C8—C9—C10—C14 | 75.20 (15) |
| C2—C3—C4—C5 | −86.02 (14) | C8—C7—C11—C12 | 157.79 (10) |
| C2—C3—C4—C15 | 70.72 (15) | C6—C7—C11—C12 | 31.31 (11) |
| C4—O1—C5—C6 | 116.91 (12) | C8—C7—C11—C13 | −79.31 (13) |
| C15—C4—C5—O1 | 98.58 (13) | C6—C7—C11—C13 | 154.20 (11) |
| C3—C4—C5—O1 | −106.34 (12) | C6—O2—C12—O3 | 179.12 (11) |
| O1—C4—C5—C6 | −104.76 (14) | C6—O2—C12—C11 | −1.67 (13) |
| C15—C4—C5—C6 | −6.2 (2) | C13—C11—C12—O3 | 34.87 (18) |
| C3—C4—C5—C6 | 148.90 (12) | C7—C11—C12—O3 | 159.39 (13) |
| C12—O2—C6—C5 | 144.48 (10) | C13—C11—C12—O2 | −144.26 (10) |
| C12—O2—C6—C7 | 22.45 (12) | C7—C11—C12—O2 | −19.74 (13) |
| O1—C5—C6—O2 | 54.19 (13) | C17—N1—C13—C11 | 59.61 (14) |
| C4—C5—C6—O2 | 125.63 (13) | C16—N1—C13—C11 | −176.80 (10) |
| O1—C5—C6—C7 | 168.46 (10) | C12—C11—C13—N1 | −109.70 (12) |
| C4—C5—C6—C7 | −120.11 (13) | C7—C11—C13—N1 | 132.54 (11) |
| O2—C6—C7—C8 | −154.30 (11) | O5—C18—C19—C20 | 17.7 (2) |
| C5—C6—C7—C8 | 90.61 (13) | O4—C18—C19—C20 | −162.07 (13) |
| O2—C6—C7—C11 | −32.63 (11) | C18—C19—C20—C21 | 174.10 (11) |
| C5—C6—C7—C11 | −147.72 (10) | C19—C20—C21—O7 | 13.5 (2) |
| C11—C7—C8—C9 | 151.72 (11) | C19—C20—C21—O6 | −165.65 (13) |
| C6—C7—C8—C9 | −92.20 (14) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N···O6 | 0.93 | 1.83 | 2.7563 (14) | 172 |
| O4—H4···O6i | 0.84 | 1.73 | 2.5544 (13) | 169 |
Symmetry codes: (i) x−1, y, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HG2516).
References
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
- Bruker–Nonius (2006). APEX2 andSAINT Bruker–Nonius AXS Inc., Madison, Wisconsin, USA.
- Crooks, P. A., Jordan, C. T. & Wei, X. (2007). US Patent No. 7 312 242.
- Flack, H. D. (1983). Acta Cryst. A39, 876–881.
- Heptinstall, S., Groenewegen, W. A., Spangenberg, P. & Lösche, W. (1988). Folia Haematol. Int. Mag. Klin. Morphol. Blutforsch 115, 447–449. [PubMed]
- Nasim, S., Parkin, S. & Crooks, P. A. (2007a). Acta Cryst. E63, o3922.
- Nasim, S., Parkin, S. & Crooks, P. A. (2007b). Acta Cryst. E63, o4274.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809021941/hg2516sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809021941/hg2516Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report