Abstract
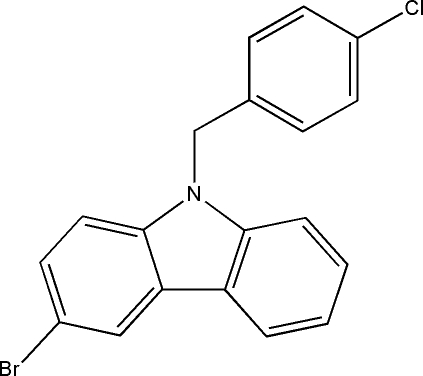
The title compound, C19H13BrClN, was synthesized by N-alkylation of 4-chloro-1-(chloromethyl)benzene with 3-bromo-9H-carbazole. The carbazole ring system is essentially planar, with a mean deviation of 0.028 Å, and it makes a dihedral angle of 91.2 (3) Å with the plane of the benzene ring.
Related literature
For the pharmaceutical properties of the title compound, see: Buu-Hoï & Royer (1950 ▶); Caulfield et al. (2002 ▶); Harfenist & Joyner (1983 ▶); Harper et al. (2002 ▶). For bond-length data, see Allen et al. (1987 ▶). For synthetic procedures, see: Duan et al. (2005a
▶,b
▶).
Experimental
Crystal data
C19H13BrClN
M r = 370.66
Orthorhombic,

a = 17.272 (4) Å
b = 15.789 (3) Å
c = 5.5948 (11) Å
V = 1525.7 (5) Å3
Z = 4
Mo Kα radiation
μ = 2.86 mm−1
T = 113 K
0.18 × 0.16 × 0.08 mm
Data collection
Rigaku Saturn CCD area-detector diffractometer
Absorption correction: multi-scan (CrystalClear; Rigaku, 2005 ▶) T min = 0.627, T max = 0.803
10796 measured reflections
2664 independent reflections
2401 reflections with I > 2σ(I)
R int = 0.031
Refinement
R[F 2 > 2σ(F 2)] = 0.023
wR(F 2) = 0.056
S = 1.04
2664 reflections
199 parameters
1 restraint
H-atom parameters constrained
Δρmax = 0.38 e Å−3
Δρmin = −0.47 e Å−3
Absolute structure: Flack (1983 ▶), 1163 Friedel pairs
Flack parameter: 0.014 (9)
Data collection: CrystalClear (Rigaku, 2005 ▶); cell refinement: CrystalClear; data reduction: CrystalClear; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809019448/dn2457sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809019448/dn2457Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Acknowledgments
This work was supported by the Hunan Natural Science Foundation (05 J J30198) and the Scientific Research Foundation of Hunan Province (2008 GK-037).
supplementary crystallographic information
Comment
Carbazole derivatives substituted by N-alkylation possess valuable pharmaceutical properties (Buu-Hoï & Royer, 1950; Harfenist & Joyner, 1983; Caulfield et al., 2002; Harper et al., 2002). In this paper, the structure of 3-bromo-9-(4-chlorobenzyl)-9H-carbazole (I), synthesized by N-alkylation of 1-(chloromethyl)-4-chloroobenzene with 3-bromo-9H-carbazole, is reported
The carbazole ring is essentially planar, with mean deviations of 0.0275 Å. The dihedral angle between the carbazole ring and the benzyl ring is 91.2° A. The C—Br distance is 1.909 (3) Å, consistent with the literature (Allen et al., 1987).
Experimental
The title compound was prepared according to the procedure of Duan et al. (2005a,b). A solution of potassium hydroxide (0.67 g) in dimethylformamide (8 ml) was stirred at room temperature for 20 min. 3-Bromo-9H-carbazole (1.0 g, 4 mmol) was added and the mixture stirred for a further 40 min. A solution of 1-(chloromethyl)-4-chlorobenzene (0.97 g, 6 mmol) in dimethylformamide (5 ml) was added dropwise with stirring. The resulting mixture was then stirred at room temperature for 12 h and poured into water (100 ml), yielding a white precipitate. The solid product was filtered off, washed with cold water and recrystallized from EtOH, giving crystals of (I). Yield: 1.26 g (85.2%); m.p. 431 - 433 K. Compound (I) (40 mg) was dissolved in mixture of chloroform (5 ml) and ethanol (5 ml) and the solution was kept at room temperature for 14 d. Natural evaporation of the solution gave colourless crystals suitable for X-Ray analysis.
Refinement
All H atoms were included in the riding model approximation with C—H distances = 0.93Å (benzene) and 0.97Å (methylene) with Uiso(H)= 1.2xUeq(C).
Figures
Fig. 1.
A view of the molecular structure of (I). Displacement ellipsoids are drawn at the 30% probability level (arbitrary spheres for H atoms).
Crystal data
| C19H13BrClN | Dx = 1.614 Mg m−3 |
| Mr = 370.66 | Melting point: 432 K |
| Orthorhombic, Pna21 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P 2c -2n | Cell parameters from 5012 reflections |
| a = 17.272 (4) Å | θ = 1.8–27.9° |
| b = 15.789 (3) Å | µ = 2.86 mm−1 |
| c = 5.5948 (11) Å | T = 113 K |
| V = 1525.7 (5) Å3 | Block, colorless |
| Z = 4 | 0.18 × 0.16 × 0.08 mm |
| F(000) = 744 |
Data collection
| Rigaku Saturn CCD area-detector diffractometer | 2664 independent reflections |
| Radiation source: rotating anode | 2401 reflections with I > 2σ(I) |
| confocal | Rint = 0.031 |
| Detector resolution: 7.31 pixels mm-1 | θmax = 25.0°, θmin = 1.8° |
| ω and φ scans | h = −20→19 |
| Absorption correction: multi-scan (CrystalClear; Rigaku, 2005) | k = −18→18 |
| Tmin = 0.627, Tmax = 0.803 | l = −6→6 |
| 10796 measured reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.023 | H-atom parameters constrained |
| wR(F2) = 0.056 | w = 1/[σ2(Fo2) + (0.0295P)2] where P = (Fo2 + 2Fc2)/3 |
| S = 1.04 | (Δ/σ)max = 0.001 |
| 2664 reflections | Δρmax = 0.38 e Å−3 |
| 199 parameters | Δρmin = −0.46 e Å−3 |
| 1 restraint | Absolute structure: Flack (1983), 1163 Friedel pairs |
| Primary atom site location: structure-invariant direct methods | Flack parameter: 0.014 (9) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Br1 | 0.258680 (12) | 0.185476 (15) | 1.32912 (10) | 0.02288 (9) | |
| Cl1 | 0.65120 (4) | 0.59416 (4) | 0.8259 (2) | 0.02847 (16) | |
| N1 | 0.53701 (13) | 0.18736 (14) | 0.6844 (4) | 0.0167 (5) | |
| C1 | 0.58534 (13) | 0.13162 (15) | 0.8041 (6) | 0.0152 (6) | |
| C2 | 0.66094 (15) | 0.10636 (17) | 0.7491 (5) | 0.0209 (7) | |
| H2 | 0.6862 | 0.1270 | 0.6143 | 0.025* | |
| C3 | 0.69681 (15) | 0.04979 (18) | 0.9013 (5) | 0.0224 (7) | |
| H3 | 0.7471 | 0.0322 | 0.8682 | 0.027* | |
| C4 | 0.65932 (16) | 0.01831 (19) | 1.1042 (6) | 0.0233 (7) | |
| H4 | 0.6848 | −0.0198 | 1.2038 | 0.028* | |
| C5 | 0.58446 (15) | 0.04349 (18) | 1.1579 (5) | 0.0186 (7) | |
| H5 | 0.5597 | 0.0227 | 1.2934 | 0.022* | |
| C6 | 0.54667 (15) | 0.10001 (16) | 1.0080 (5) | 0.0167 (6) | |
| C7 | 0.47140 (14) | 0.13996 (16) | 1.0130 (5) | 0.0135 (6) | |
| C8 | 0.40812 (14) | 0.13595 (18) | 1.1675 (5) | 0.0160 (6) | |
| H8 | 0.4082 | 0.1001 | 1.2993 | 0.019* | |
| C9 | 0.34553 (15) | 0.18694 (17) | 1.1181 (5) | 0.0174 (6) | |
| C10 | 0.34293 (15) | 0.24129 (18) | 0.9223 (5) | 0.0210 (7) | |
| H10 | 0.2993 | 0.2746 | 0.8960 | 0.025* | |
| C11 | 0.40475 (15) | 0.24581 (18) | 0.7676 (5) | 0.0202 (7) | |
| H11 | 0.4040 | 0.2824 | 0.6373 | 0.024* | |
| C12 | 0.46844 (13) | 0.19405 (14) | 0.8117 (6) | 0.0147 (5) | |
| C13 | 0.55872 (15) | 0.24502 (16) | 0.4941 (5) | 0.0193 (7) | |
| H13A | 0.5160 | 0.2495 | 0.3822 | 0.023* | |
| H13B | 0.6026 | 0.2214 | 0.4087 | 0.023* | |
| C14 | 0.57980 (15) | 0.33317 (17) | 0.5812 (5) | 0.0151 (6) | |
| C15 | 0.62228 (15) | 0.34401 (18) | 0.7921 (6) | 0.0227 (7) | |
| H15 | 0.6358 | 0.2971 | 0.8835 | 0.027* | |
| C16 | 0.64432 (14) | 0.42472 (17) | 0.8658 (6) | 0.0215 (7) | |
| H16 | 0.6729 | 0.4319 | 1.0052 | 0.026* | |
| C17 | 0.62343 (16) | 0.49356 (17) | 0.7311 (5) | 0.0187 (6) | |
| C18 | 0.58085 (15) | 0.48457 (18) | 0.5212 (6) | 0.0223 (7) | |
| H18 | 0.5671 | 0.5318 | 0.4313 | 0.027* | |
| C19 | 0.55924 (15) | 0.40353 (17) | 0.4486 (5) | 0.0192 (7) | |
| H19 | 0.5306 | 0.3967 | 0.3091 | 0.023* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Br1 | 0.01586 (13) | 0.02907 (15) | 0.02370 (15) | 0.00028 (9) | 0.0041 (2) | −0.0042 (2) |
| Cl1 | 0.0313 (3) | 0.0172 (3) | 0.0370 (4) | −0.0025 (2) | 0.0062 (6) | −0.0066 (5) |
| N1 | 0.0162 (12) | 0.0175 (14) | 0.0164 (13) | −0.0024 (9) | 0.0028 (10) | 0.0012 (10) |
| C1 | 0.0157 (11) | 0.0141 (13) | 0.0159 (16) | −0.0043 (9) | −0.0006 (15) | −0.0046 (15) |
| C2 | 0.0167 (14) | 0.0249 (17) | 0.0210 (17) | −0.0076 (12) | 0.0033 (11) | −0.0064 (12) |
| C3 | 0.0131 (14) | 0.0229 (17) | 0.031 (2) | 0.0027 (11) | −0.0004 (12) | −0.0099 (13) |
| C4 | 0.0196 (16) | 0.0219 (17) | 0.0283 (18) | 0.0007 (13) | −0.0074 (13) | −0.0015 (14) |
| C5 | 0.0187 (16) | 0.0179 (16) | 0.0193 (16) | −0.0019 (12) | −0.0037 (13) | −0.0021 (12) |
| C6 | 0.0161 (14) | 0.0147 (16) | 0.0192 (16) | −0.0050 (11) | −0.0006 (12) | −0.0059 (13) |
| C7 | 0.0121 (13) | 0.0118 (15) | 0.0166 (15) | −0.0036 (10) | −0.0011 (12) | −0.0024 (12) |
| C8 | 0.0184 (15) | 0.0137 (16) | 0.0159 (16) | −0.0045 (11) | −0.0051 (12) | −0.0018 (12) |
| C9 | 0.0128 (13) | 0.0187 (16) | 0.0208 (17) | −0.0032 (12) | 0.0023 (12) | −0.0078 (13) |
| C10 | 0.0161 (15) | 0.0222 (17) | 0.0248 (17) | 0.0011 (11) | −0.0055 (12) | −0.0029 (13) |
| C11 | 0.0232 (14) | 0.0199 (15) | 0.017 (2) | −0.0010 (11) | −0.0014 (12) | 0.0006 (12) |
| C12 | 0.0155 (11) | 0.0150 (13) | 0.0137 (14) | −0.0051 (9) | 0.0017 (18) | −0.0022 (16) |
| C13 | 0.0193 (15) | 0.0235 (17) | 0.0151 (15) | −0.0050 (12) | 0.0030 (13) | −0.0026 (14) |
| C14 | 0.0138 (14) | 0.0174 (15) | 0.0142 (15) | −0.0024 (11) | 0.0050 (12) | 0.0002 (12) |
| C15 | 0.0235 (13) | 0.0193 (14) | 0.025 (2) | −0.0009 (10) | −0.0009 (16) | 0.0049 (15) |
| C16 | 0.0220 (13) | 0.0251 (16) | 0.017 (2) | −0.0035 (10) | 0.0012 (14) | −0.0007 (14) |
| C17 | 0.0181 (14) | 0.0149 (16) | 0.0229 (16) | −0.0008 (11) | 0.0056 (12) | −0.0012 (12) |
| C18 | 0.0201 (15) | 0.0194 (17) | 0.0273 (18) | 0.0039 (12) | 0.0024 (14) | 0.0017 (14) |
| C19 | 0.0145 (14) | 0.0261 (18) | 0.0172 (16) | 0.0001 (11) | 0.0007 (12) | 0.0013 (13) |
Geometric parameters (Å, °)
| Br1—C9 | 1.909 (3) | C8—H8 | 0.9300 |
| Cl1—C17 | 1.742 (3) | C9—C10 | 1.392 (4) |
| N1—C1 | 1.385 (3) | C10—C11 | 1.376 (4) |
| N1—C12 | 1.386 (3) | C10—H10 | 0.9300 |
| N1—C13 | 1.450 (3) | C11—C12 | 1.393 (4) |
| C1—C2 | 1.399 (3) | C11—H11 | 0.9300 |
| C1—C6 | 1.413 (4) | C13—C14 | 1.519 (4) |
| C2—C3 | 1.381 (4) | C13—H13A | 0.9700 |
| C2—H2 | 0.9300 | C13—H13B | 0.9700 |
| C3—C4 | 1.398 (4) | C14—C19 | 1.382 (4) |
| C3—H3 | 0.9300 | C14—C15 | 1.400 (4) |
| C4—C5 | 1.386 (4) | C15—C16 | 1.392 (4) |
| C4—H4 | 0.9300 | C15—H15 | 0.9300 |
| C5—C6 | 1.388 (4) | C16—C17 | 1.371 (4) |
| C5—H5 | 0.9300 | C16—H16 | 0.9300 |
| C6—C7 | 1.445 (3) | C17—C18 | 1.393 (4) |
| C7—C8 | 1.395 (4) | C18—C19 | 1.393 (4) |
| C7—C12 | 1.415 (4) | C18—H18 | 0.9300 |
| C8—C9 | 1.376 (4) | C19—H19 | 0.9300 |
| C1—N1—C12 | 108.4 (2) | C9—C10—H10 | 119.9 |
| C1—N1—C13 | 126.7 (2) | C10—C11—C12 | 118.1 (3) |
| C12—N1—C13 | 123.4 (2) | C10—C11—H11 | 121.0 |
| N1—C1—C2 | 129.6 (3) | C12—C11—H11 | 121.0 |
| N1—C1—C6 | 109.2 (2) | N1—C12—C11 | 129.0 (3) |
| C2—C1—C6 | 121.2 (3) | N1—C12—C7 | 109.4 (2) |
| C3—C2—C1 | 117.9 (3) | C11—C12—C7 | 121.6 (3) |
| C3—C2—H2 | 121.1 | N1—C13—C14 | 113.7 (2) |
| C1—C2—H2 | 121.1 | N1—C13—H13A | 108.8 |
| C2—C3—C4 | 121.5 (3) | C14—C13—H13A | 108.8 |
| C2—C3—H3 | 119.2 | N1—C13—H13B | 108.8 |
| C4—C3—H3 | 119.2 | C14—C13—H13B | 108.8 |
| C5—C4—C3 | 120.4 (3) | H13A—C13—H13B | 107.7 |
| C5—C4—H4 | 119.8 | C19—C14—C15 | 119.3 (3) |
| C3—C4—H4 | 119.8 | C19—C14—C13 | 120.2 (3) |
| C4—C5—C6 | 119.5 (3) | C15—C14—C13 | 120.5 (3) |
| C4—C5—H5 | 120.3 | C16—C15—C14 | 120.3 (3) |
| C6—C5—H5 | 120.3 | C16—C15—H15 | 119.8 |
| C5—C6—C1 | 119.5 (2) | C14—C15—H15 | 119.8 |
| C5—C6—C7 | 133.8 (3) | C17—C16—C15 | 119.4 (3) |
| C1—C6—C7 | 106.7 (2) | C17—C16—H16 | 120.3 |
| C8—C7—C12 | 119.5 (2) | C15—C16—H16 | 120.3 |
| C8—C7—C6 | 134.2 (3) | C16—C17—C18 | 121.4 (3) |
| C12—C7—C6 | 106.3 (2) | C16—C17—Cl1 | 118.9 (2) |
| C9—C8—C7 | 117.7 (3) | C18—C17—Cl1 | 119.7 (2) |
| C9—C8—H8 | 121.2 | C17—C18—C19 | 118.8 (3) |
| C7—C8—H8 | 121.2 | C17—C18—H18 | 120.6 |
| C8—C9—C10 | 123.0 (3) | C19—C18—H18 | 120.6 |
| C8—C9—Br1 | 119.1 (2) | C14—C19—C18 | 120.8 (3) |
| C10—C9—Br1 | 117.9 (2) | C14—C19—H19 | 119.6 |
| C11—C10—C9 | 120.1 (3) | C18—C19—H19 | 119.6 |
| C11—C10—H10 | 119.9 | ||
| C12—N1—C1—C2 | −178.2 (3) | C9—C10—C11—C12 | 0.8 (4) |
| C13—N1—C1—C2 | −12.0 (4) | C1—N1—C12—C11 | 176.2 (3) |
| C12—N1—C1—C6 | 1.6 (3) | C13—N1—C12—C11 | 9.5 (4) |
| C13—N1—C1—C6 | 167.8 (2) | C1—N1—C12—C7 | −1.7 (3) |
| N1—C1—C2—C3 | 179.4 (3) | C13—N1—C12—C7 | −168.4 (2) |
| C6—C1—C2—C3 | −0.4 (4) | C10—C11—C12—N1 | −179.6 (3) |
| C1—C2—C3—C4 | 0.1 (4) | C10—C11—C12—C7 | −1.9 (4) |
| C2—C3—C4—C5 | −0.1 (4) | C8—C7—C12—N1 | −179.7 (2) |
| C3—C4—C5—C6 | 0.3 (4) | C6—C7—C12—N1 | 1.2 (3) |
| C4—C5—C6—C1 | −0.6 (4) | C8—C7—C12—C11 | 2.2 (4) |
| C4—C5—C6—C7 | −178.3 (3) | C6—C7—C12—C11 | −177.0 (2) |
| N1—C1—C6—C5 | −179.2 (2) | C1—N1—C13—C14 | −93.3 (3) |
| C2—C1—C6—C5 | 0.6 (4) | C12—N1—C13—C14 | 70.8 (3) |
| N1—C1—C6—C7 | −0.9 (3) | N1—C13—C14—C19 | −142.8 (3) |
| C2—C1—C6—C7 | 178.9 (2) | N1—C13—C14—C15 | 39.3 (4) |
| C5—C6—C7—C8 | −1.2 (5) | C19—C14—C15—C16 | −0.7 (4) |
| C1—C6—C7—C8 | −179.1 (3) | C13—C14—C15—C16 | 177.2 (2) |
| C5—C6—C7—C12 | 177.8 (3) | C14—C15—C16—C17 | 0.6 (4) |
| C1—C6—C7—C12 | −0.1 (3) | C15—C16—C17—C18 | −0.3 (4) |
| C12—C7—C8—C9 | −1.3 (4) | C15—C16—C17—Cl1 | 179.8 (2) |
| C6—C7—C8—C9 | 177.6 (3) | C16—C17—C18—C19 | 0.1 (4) |
| C7—C8—C9—C10 | 0.2 (4) | Cl1—C17—C18—C19 | 180.0 (2) |
| C7—C8—C9—Br1 | −178.51 (19) | C15—C14—C19—C18 | 0.6 (4) |
| C8—C9—C10—C11 | 0.0 (4) | C13—C14—C19—C18 | −177.4 (2) |
| Br1—C9—C10—C11 | 178.8 (2) | C17—C18—C19—C14 | −0.2 (4) |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: DN2457).
References
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
- Buu-Hoï, N. P. & Royer, R. (1950). J. Org. Chem.15, 123–130.
- Caulfield, T., Cherrier, M. P., Combeau, C. & Mailliet, P. (2002). Eur. Patent EP 1253141.
- Duan, X. M., Han, J., Chen, L. G., Xu, Y. J. & Li, Y. (2005a). Fine Chem.22, 39–40.
- Duan, X. M., Han, J., Chen, L. G., Xu, Y. J. & Li, Y. (2005b). Fine Chem.22, 52.
- Flack, H. D. (1983). Acta Cryst. A39, 876–881.
- Harfenist, M. & Joyner, C. T. (1983). US Patent No. 4 379 160.
- Harper, R. W., Lin, H. S. & Richett, M. E. (2002). World Patent WO2002079154.
- Rigaku (2005). CrystalClear Rigaku Americas Corporation, The Woodlands, Texas, USA.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809019448/dn2457sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809019448/dn2457Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report



