Abstract
The asymmetric unit of the title compound, C5H6N3O2 +·HSeO4 −, contains two monoprotonated 2-amino-3-nitropyridinium cations and two hydrogen selenate anions which are connected through N—H⋯O and O—H⋯O hydrogen bonds, building chains parallel to the a direction. These chains are further connected to each other by weaker C—H⋯O hydrogen-bonding interactions, leading to the formation of a three-dimensional network.
Related literature
For related structures, see: Akriche et al. (2009 ▶); Fleck (2006 ▶); Le Fur, Masse & Nicoud (1998 ▶); Nicoud et al. (1997 ▶); Maalej et al. (2008 ▶).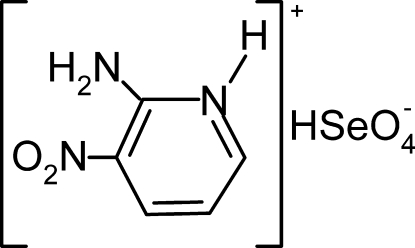
Experimental
Crystal data
C5H6N3O2 +·HSeO4 −
M r = 284.10
Monoclinic,
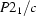
a = 9.090 (3) Å
b = 20.130 (2) Å
c = 10.434 (4) Å
β = 104.84 (2)°
V = 1845.6 (10) Å3
Z = 8
Mo Kα radiation
μ = 4.09 mm−1
T = 298 K
0.37 × 0.29 × 0.19 mm
Data collection
Enraf–Nonius Turbo-CAD-4 diffractometer
Absorption correction: multi-scan (Blessing, 1995 ▶) T min = 0.145, T max = 0.298 (expected range = 0.224–0.460)
7325 measured reflections
4433 independent reflections
2980 reflections with I > 2σ(I)
R int = 0.040
2 standard reflections frequency: 120 min intensity decay: 4%
Refinement
R[F 2 > 2σ(F 2)] = 0.047
wR(F 2) = 0.112
S = 1.01
4433 reflections
274 parameters
H-atom parameters constrained
Δρmax = 0.80 e Å−3
Δρmin = −0.75 e Å−3
Data collection: CAD-4 EXPRESS (Enraf–Nonius, 1994 ▶); cell refinement: CAD-4 EXPRESS; data reduction: XCAD4 (Harms & Wocadlo, 1995 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶) and DIAMOND (Brandenburg & Putz, 2005 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶).
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809022879/dn2458sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809022879/dn2458Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1⋯O6 | 0.82 | 1.75 | 2.565 (5) | 170 |
| O5—H5⋯O2i | 0.82 | 1.80 | 2.601 (5) | 167 |
| N1—H1A⋯O4 | 0.86 | 1.84 | 2.679 (5) | 166 |
| N2—H2A⋯O3 | 0.86 | 2.02 | 2.870 (6) | 172 |
| N2—H2B⋯O9 | 0.86 | 2.09 | 2.675 (6) | 124 |
| N2—H2B⋯O8ii | 0.86 | 2.28 | 2.933 (5) | 133 |
| N4—H4⋯O7 | 0.86 | 2.05 | 2.864 (5) | 157 |
| N5—H5A⋯O8 | 0.86 | 2.07 | 2.900 (5) | 163 |
| N5—H5B⋯O3ii | 0.86 | 2.11 | 2.798 (5) | 137 |
| N5—H5B⋯O11 | 0.86 | 2.12 | 2.693 (5) | 124 |
| C3—H3⋯O5iii | 0.93 | 2.54 | 3.443 (5) | 163 |
| C4—H4A⋯O12iv | 0.93 | 2.47 | 3.290 (6) | 148 |
| C8—H8⋯O6iii | 0.93 | 2.57 | 3.463 (6) | 162 |
| C10—H10⋯O4v | 0.93 | 2.35 | 3.228 (6) | 158 |
Symmetry codes: (i) 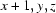 ; (ii)
; (ii) 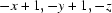 ; (iii)
; (iii) 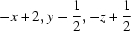 ; (iv)
; (iv) 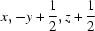 ; (v)
; (v) 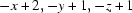 .
.
supplementary crystallographic information
Comment
The important advantage of hybrid organic inorganic salts is the opportunity offered by the chromophores that when anchored onto inorganic host matrices, lead to non-centrosymmetric frameworks suitable for NLO devices. The approach of this new engineering has been applied to 2-amino-3-nitropyridinium cation (2 A3NP+) encapsulated in various anionic subnetworks (Akriche et al., 2009, Nicoud et al.,1997, Le Fur et al., 1998). The attempt using (HSO4-) n polymeric anions has been successful with the cristallization of the non-centrosymmetric 2-Amino-3-nitropyridinium sulfate (Le Fur et al., 1998). The encapsulation of this cation in (HSeO4-)n polymeric anions leads to the title compound (I).
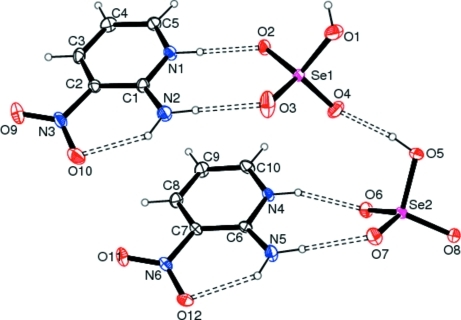
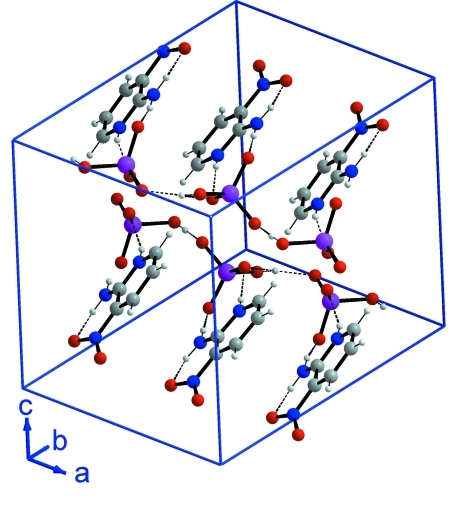
The asymmetric unit of (I) contains two monoprotonated 2-amino-3-nitropyridinium cations and two hydrogen selenate anions (Fig.1) which are connected through N—H···O, C—H···O and O—H···O hydrogen bonds. The hydrogen selenate anions are interconnected between themselves by H-bonds involving the proton of selenate groups leading to (HSeO4-) n chains parallel to the a axis (Fig. 2).
In this atomic arrangement the HSeO4- tetraedra are slightly distorted with Se—O distances from 1.594 (6) to 1.713 (4) Å. The Se—O bonds in selenate tetrahedra depends greatly on the nature of the O atoms acting as an acceptor or a donor atoms: the longer bonds (1.713 (4) and 1.692 (5) Å) involve oxygen atoms acting as a H-donor whereas shorter bonds ranging from 1.594 (6) to 1.623 (4) relate to oxygen atoms acting as H-acceptor participating in hydrogen bonds of type N—H···O and C—H···O. As expected, the geometrical features of anion agree with those previously observed for this group in other analogues (Fleck, 2006, Maalej et al., 2008).
The 2-amino-3-nitropyridinium cations are onchored onto anionic chains through short hydrogen bonds originating from the NH2 and NH+ groups. The unique inter-cation contact C4—H4A···O11(H4A···O11 = 2.45 Å) induces the aggregation of cations in pairs (2 A3NP+)2 elongated in -(a+c) direction. Two hydrogen bonds, N2—H2B···O10 (2.10 Å) and N5—H5B···O12 (2.11 Å) (see Table 1) ensure the intra-cation links. This situation is well observed in nitroaniline derivatives in which nitro and amino groups are ortho to one another which precludes the rotation of the nitro group with respect to the pyridinium rings. The diedral angles between the planes of the NO2 groups and the two pyridinium planes are 19.0 (3) and 15.9 (4) % indicating a distortion of the NO2 groups under the influence of C—H···O hydrogn bonds of neighbouring cations. This situation is alawys observed in other 2-amino-3-nitropyridinium salts (Nicoud et al.,1997, Le Fur et al., 1998). The bond lengths and angles in (I) are normal and comparable with the corresponding values observed in the related structure (Akriche et al., 2009, Nicoud et al.,1997, Le Fur et al., 1998).
Experimental
The title compound (I) was cristallized by slow evaporation at room temperature of an aqueous solution (20 ml) of 2-amino-3-nitropyridine (4 mmol) and selenic acid (4 mmol) in a 1:1 stochiometric ratio.
Refinement
All H atoms attached to C, N and H atoms were fixed geometrically and treated as riding with C—H = 0.93 Å, N—H = 0.86Å and O—H = 0.82 Å with Uiso(H) = 1.2Ueq(C or N) and Uiso(H) = 1.5Ueq(O).
Figures
Fig. 1.
An ORTEP view of (I) with the atom-labelling scheme. Displacement ellipsoids are drawn at the 30% probability level. H atoms are represented as small spheres of arbitrary radii. Hydrogen bonds are represented as dashed lines.
Fig. 2.
A perspective view of (I) showing the (HSeO4-) n polymeric anions running along the a axis. The C—H···O bonds are omitted for clarity of figure.
Crystal data
| C5H6N3O2+·HO4Se− | F(000) = 1120 |
| Mr = 284.10 | Dx = 2.045 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 25 reflections |
| a = 9.090 (3) Å | θ = 9–11° |
| b = 20.130 (2) Å | µ = 4.09 mm−1 |
| c = 10.434 (4) Å | T = 298 K |
| β = 104.84 (2)° | Prism, yellow |
| V = 1845.6 (10) Å3 | 0.37 × 0.29 × 0.19 mm |
| Z = 8 |
Data collection
| Enraf–Nonius Turbo-CAD-4 diffractometer | 2980 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.040 |
| graphite | θmax = 28.0°, θmin = 2.0° |
| ω scans | h = −11→11 |
| Absorption correction: multi-scan (Blessing, 1995) | k = 0→26 |
| Tmin = 0.145, Tmax = 0.298 | l = −6→13 |
| 7325 measured reflections | 2 standard reflections every 120 min |
| 4433 independent reflections | intensity decay: 4% |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.047 | H-atom parameters constrained |
| wR(F2) = 0.112 | w = 1/[σ2(Fo2) + (0.0549P)2] where P = (Fo2 + 2Fc2)/3 |
| S = 1.01 | (Δ/σ)max = 0.001 |
| 4433 reflections | Δρmax = 0.80 e Å−3 |
| 274 parameters | Δρmin = −0.75 e Å−3 |
| 0 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.0106 (7) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Se1 | 0.39573 (5) | 0.57437 (2) | 0.26743 (5) | 0.03205 (15) | |
| Se2 | 0.88010 (5) | 0.61095 (2) | 0.28019 (5) | 0.02924 (14) | |
| O1 | 0.4598 (4) | 0.65134 (18) | 0.2463 (5) | 0.0680 (13) | |
| H1 | 0.5519 | 0.6530 | 0.2786 | 0.102* | |
| O3 | 0.3344 (5) | 0.5426 (2) | 0.1228 (4) | 0.0756 (14) | |
| O2 | 0.2627 (4) | 0.58545 (19) | 0.3409 (4) | 0.0576 (11) | |
| O4 | 0.5374 (4) | 0.53198 (15) | 0.3555 (3) | 0.0411 (8) | |
| O5 | 1.0304 (3) | 0.66505 (15) | 0.3064 (4) | 0.0401 (8) | |
| H5 | 1.1091 | 0.6443 | 0.3109 | 0.060* | |
| O6 | 0.7514 (3) | 0.65653 (16) | 0.3186 (4) | 0.0433 (8) | |
| O7 | 0.9371 (4) | 0.54843 (15) | 0.3783 (3) | 0.0405 (8) | |
| O8 | 0.8397 (4) | 0.58792 (17) | 0.1281 (3) | 0.0478 (9) | |
| O9 | 0.4200 (5) | 0.2899 (2) | −0.0205 (4) | 0.0656 (12) | |
| O10 | 0.6241 (5) | 0.23103 (19) | 0.0156 (4) | 0.0672 (12) | |
| O11 | 0.8828 (4) | 0.33319 (18) | −0.0323 (4) | 0.0498 (9) | |
| O12 | 1.0837 (4) | 0.27232 (16) | −0.0052 (4) | 0.0503 (9) | |
| N1 | 0.6363 (4) | 0.41805 (19) | 0.2743 (4) | 0.0361 (9) | |
| H1A | 0.6010 | 0.4565 | 0.2869 | 0.043* | |
| N2 | 0.4378 (4) | 0.4105 (2) | 0.0899 (4) | 0.0442 (10) | |
| H2A | 0.4098 | 0.4494 | 0.1082 | 0.053* | |
| H2B | 0.3862 | 0.3897 | 0.0211 | 0.053* | |
| N3 | 0.5509 (5) | 0.2774 (2) | 0.0413 (5) | 0.0454 (10) | |
| N4 | 1.1029 (4) | 0.44386 (17) | 0.2922 (4) | 0.0333 (9) | |
| H4 | 1.0723 | 0.4821 | 0.3116 | 0.040* | |
| N5 | 0.9152 (4) | 0.4497 (2) | 0.0988 (4) | 0.0413 (10) | |
| H5A | 0.8914 | 0.4881 | 0.1236 | 0.050* | |
| H5B | 0.8653 | 0.4335 | 0.0241 | 0.050* | |
| N6 | 1.0111 (5) | 0.31667 (18) | 0.0290 (4) | 0.0348 (9) | |
| C1 | 0.5601 (5) | 0.3828 (2) | 0.1677 (5) | 0.0337 (10) | |
| C2 | 0.6231 (5) | 0.3199 (2) | 0.1535 (5) | 0.0339 (10) | |
| C3 | 0.7524 (5) | 0.2982 (2) | 0.2424 (5) | 0.0400 (12) | |
| H3 | 0.7922 | 0.2568 | 0.2303 | 0.048* | |
| C4 | 0.8245 (5) | 0.3366 (3) | 0.3493 (5) | 0.0427 (12) | |
| H4A | 0.9114 | 0.3214 | 0.4103 | 0.051* | |
| C5 | 0.7647 (5) | 0.3970 (2) | 0.3629 (5) | 0.0381 (11) | |
| H5C | 0.8120 | 0.4243 | 0.4333 | 0.046* | |
| C6 | 1.0279 (5) | 0.4158 (2) | 0.1752 (4) | 0.0288 (9) | |
| C7 | 1.0836 (5) | 0.3528 (2) | 0.1496 (4) | 0.0286 (9) | |
| C8 | 1.2031 (5) | 0.3237 (2) | 0.2393 (5) | 0.0387 (11) | |
| H8 | 1.2366 | 0.2819 | 0.2216 | 0.046* | |
| C9 | 1.2739 (6) | 0.3557 (3) | 0.3550 (5) | 0.0443 (12) | |
| H9 | 1.3562 | 0.3363 | 0.4150 | 0.053* | |
| C10 | 1.2217 (5) | 0.4159 (2) | 0.3799 (5) | 0.0402 (11) | |
| H10 | 1.2680 | 0.4382 | 0.4580 | 0.048* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Se1 | 0.0214 (2) | 0.0312 (2) | 0.0401 (3) | 0.00508 (17) | 0.00162 (18) | −0.0022 (2) |
| Se2 | 0.0249 (2) | 0.0232 (2) | 0.0358 (3) | −0.00061 (17) | 0.00088 (18) | −0.00011 (18) |
| O1 | 0.0320 (19) | 0.040 (2) | 0.129 (4) | 0.0059 (17) | 0.015 (2) | 0.030 (2) |
| O3 | 0.079 (3) | 0.075 (3) | 0.049 (3) | 0.045 (2) | −0.026 (2) | −0.021 (2) |
| O2 | 0.040 (2) | 0.055 (2) | 0.088 (3) | 0.0112 (18) | 0.034 (2) | 0.009 (2) |
| O4 | 0.0365 (17) | 0.0333 (17) | 0.045 (2) | 0.0110 (14) | −0.0050 (15) | −0.0029 (15) |
| O5 | 0.0281 (16) | 0.0327 (17) | 0.057 (2) | −0.0049 (13) | 0.0061 (16) | −0.0020 (16) |
| O6 | 0.0299 (17) | 0.0338 (17) | 0.066 (2) | 0.0023 (14) | 0.0125 (16) | −0.0058 (17) |
| O7 | 0.0416 (18) | 0.0311 (16) | 0.048 (2) | 0.0056 (14) | 0.0099 (16) | 0.0142 (15) |
| O8 | 0.055 (2) | 0.0375 (18) | 0.041 (2) | 0.0050 (17) | −0.0043 (17) | −0.0050 (16) |
| O9 | 0.054 (2) | 0.061 (3) | 0.071 (3) | −0.007 (2) | −0.004 (2) | −0.023 (2) |
| O10 | 0.084 (3) | 0.046 (2) | 0.074 (3) | 0.004 (2) | 0.026 (2) | −0.023 (2) |
| O11 | 0.0348 (18) | 0.057 (2) | 0.050 (2) | −0.0048 (17) | −0.0036 (17) | −0.0155 (18) |
| O12 | 0.066 (2) | 0.0294 (18) | 0.057 (2) | 0.0063 (17) | 0.018 (2) | −0.0091 (16) |
| N1 | 0.037 (2) | 0.031 (2) | 0.038 (2) | 0.0060 (17) | 0.0050 (18) | −0.0043 (18) |
| N2 | 0.039 (2) | 0.041 (2) | 0.046 (3) | 0.0080 (19) | 0.000 (2) | −0.004 (2) |
| N3 | 0.055 (3) | 0.029 (2) | 0.055 (3) | −0.006 (2) | 0.019 (2) | −0.0050 (19) |
| N4 | 0.039 (2) | 0.0234 (17) | 0.032 (2) | 0.0039 (16) | −0.0007 (17) | −0.0015 (15) |
| N5 | 0.046 (2) | 0.036 (2) | 0.033 (2) | 0.0113 (18) | −0.0072 (19) | −0.0061 (17) |
| N6 | 0.043 (2) | 0.0291 (19) | 0.034 (2) | −0.0076 (17) | 0.0117 (19) | −0.0040 (16) |
| C1 | 0.033 (2) | 0.032 (2) | 0.037 (3) | 0.0022 (19) | 0.010 (2) | 0.003 (2) |
| C2 | 0.034 (2) | 0.031 (2) | 0.038 (3) | −0.0001 (19) | 0.012 (2) | 0.000 (2) |
| C3 | 0.042 (3) | 0.027 (2) | 0.055 (3) | 0.008 (2) | 0.020 (2) | 0.007 (2) |
| C4 | 0.037 (3) | 0.047 (3) | 0.042 (3) | 0.009 (2) | 0.005 (2) | 0.014 (2) |
| C5 | 0.035 (2) | 0.041 (3) | 0.035 (3) | 0.002 (2) | 0.003 (2) | −0.001 (2) |
| C6 | 0.032 (2) | 0.029 (2) | 0.023 (2) | −0.0036 (18) | 0.0041 (19) | 0.0005 (18) |
| C7 | 0.035 (2) | 0.023 (2) | 0.029 (2) | −0.0039 (17) | 0.0094 (19) | −0.0007 (18) |
| C8 | 0.043 (3) | 0.027 (2) | 0.047 (3) | 0.006 (2) | 0.012 (2) | 0.005 (2) |
| C9 | 0.041 (3) | 0.045 (3) | 0.038 (3) | 0.013 (2) | −0.005 (2) | 0.005 (2) |
| C10 | 0.044 (3) | 0.038 (3) | 0.031 (3) | −0.003 (2) | −0.005 (2) | 0.000 (2) |
Geometric parameters (Å, °)
| Se1—O3 | 1.602 (4) | N4—C10 | 1.347 (5) |
| Se1—O2 | 1.604 (3) | N4—C6 | 1.359 (5) |
| Se1—O4 | 1.620 (3) | N4—H4 | 0.8600 |
| Se1—O1 | 1.689 (4) | N5—C6 | 1.316 (5) |
| Se2—O8 | 1.603 (3) | N5—H5A | 0.8600 |
| Se2—O6 | 1.616 (3) | N5—H5B | 0.8600 |
| Se2—O7 | 1.621 (3) | N6—C7 | 1.456 (5) |
| Se2—O5 | 1.713 (3) | C1—C2 | 1.412 (6) |
| O1—H1 | 0.8200 | C2—C3 | 1.369 (7) |
| O5—H5 | 0.8200 | C3—C4 | 1.377 (7) |
| O9—N3 | 1.225 (5) | C3—H3 | 0.9300 |
| O10—N3 | 1.216 (5) | C4—C5 | 1.355 (7) |
| O11—N6 | 1.224 (5) | C4—H4A | 0.9300 |
| O12—N6 | 1.217 (5) | C5—H5C | 0.9300 |
| N1—C1 | 1.352 (6) | C6—C7 | 1.417 (6) |
| N1—C5 | 1.357 (6) | C7—C8 | 1.370 (6) |
| N1—H1A | 0.8600 | C8—C9 | 1.374 (7) |
| N2—C1 | 1.321 (6) | C8—H8 | 0.9300 |
| N2—H2A | 0.8600 | C9—C10 | 1.351 (7) |
| N2—H2B | 0.8600 | C9—H9 | 0.9300 |
| N3—C2 | 1.463 (6) | C10—H10 | 0.9300 |
| O3—Se1—O2 | 112.5 (2) | O11—N6—C7 | 118.4 (4) |
| O3—Se1—O4 | 111.03 (18) | N2—C1—N1 | 117.2 (4) |
| O2—Se1—O4 | 112.89 (19) | N2—C1—C2 | 127.9 (5) |
| O3—Se1—O1 | 106.9 (3) | N1—C1—C2 | 114.9 (4) |
| O2—Se1—O1 | 105.2 (2) | C3—C2—C1 | 121.1 (4) |
| O4—Se1—O1 | 107.86 (17) | C3—C2—N3 | 119.1 (4) |
| O8—Se2—O6 | 114.36 (18) | C1—C2—N3 | 119.8 (4) |
| O8—Se2—O7 | 110.84 (17) | C2—C3—C4 | 121.1 (4) |
| O6—Se2—O7 | 114.76 (17) | C2—C3—H3 | 119.4 |
| O8—Se2—O5 | 108.25 (19) | C4—C3—H3 | 119.4 |
| O6—Se2—O5 | 101.41 (16) | C5—C4—C3 | 117.9 (4) |
| O7—Se2—O5 | 106.24 (17) | C5—C4—H4A | 121.0 |
| Se1—O1—H1 | 109.5 | C3—C4—H4A | 121.0 |
| Se2—O5—H5 | 109.5 | C4—C5—N1 | 120.4 (5) |
| C1—N1—C5 | 124.5 (4) | C4—C5—H5C | 119.8 |
| C1—N1—H1A | 117.7 | N1—C5—H5C | 119.8 |
| C5—N1—H1A | 117.7 | N5—C6—N4 | 117.6 (4) |
| C1—N2—H2A | 120.0 | N5—C6—C7 | 127.6 (4) |
| C1—N2—H2B | 120.0 | N4—C6—C7 | 114.8 (4) |
| H2A—N2—H2B | 120.0 | C8—C7—C6 | 120.8 (4) |
| O10—N3—O9 | 123.6 (5) | C8—C7—N6 | 118.7 (4) |
| O10—N3—C2 | 117.8 (4) | C6—C7—N6 | 120.4 (4) |
| O9—N3—C2 | 118.6 (4) | C7—C8—C9 | 120.8 (4) |
| C10—N4—C6 | 124.6 (4) | C7—C8—H8 | 119.6 |
| C10—N4—H4 | 117.7 | C9—C8—H8 | 119.6 |
| C6—N4—H4 | 117.7 | C10—C9—C8 | 118.7 (4) |
| C6—N5—H5A | 120.0 | C10—C9—H9 | 120.6 |
| C6—N5—H5B | 120.0 | C8—C9—H9 | 120.6 |
| H5A—N5—H5B | 120.0 | N4—C10—C9 | 120.3 (4) |
| O12—N6—O11 | 124.2 (4) | N4—C10—H10 | 119.9 |
| O12—N6—C7 | 117.5 (4) | C9—C10—H10 | 119.9 |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···O6 | 0.82 | 1.75 | 2.565 (5) | 170 |
| O5—H5···O2i | 0.82 | 1.80 | 2.601 (5) | 167 |
| N1—H1A···O4 | 0.86 | 1.84 | 2.679 (5) | 166 |
| N2—H2A···O3 | 0.86 | 2.02 | 2.870 (6) | 172 |
| N2—H2B···O9 | 0.86 | 2.09 | 2.675 (6) | 124 |
| N2—H2B···O8ii | 0.86 | 2.28 | 2.933 (5) | 133 |
| N4—H4···O7 | 0.86 | 2.05 | 2.864 (5) | 157 |
| N5—H5A···O8 | 0.86 | 2.07 | 2.900 (5) | 163 |
| N5—H5B···O3ii | 0.86 | 2.11 | 2.798 (5) | 137 |
| N5—H5B···O11 | 0.86 | 2.12 | 2.693 (5) | 124 |
| C3—H3···O5iii | 0.93 | 2.54 | 3.443 (5) | 163 |
| C4—H4A···O12iv | 0.93 | 2.47 | 3.290 (6) | 148 |
| C8—H8···O6iii | 0.93 | 2.57 | 3.463 (6) | 162 |
| C10—H10···O4v | 0.93 | 2.35 | 3.228 (6) | 158 |
Symmetry codes: (i) x+1, y, z; (ii) −x+1, −y+1, −z; (iii) −x+2, y−1/2, −z+1/2; (iv) x, −y+1/2, z+1/2; (v) −x+2, −y+1, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: DN2458).
References
- Akriche, S. & Rzaigui, M. (2009). Acta Cryst. E65, o793. [DOI] [PMC free article] [PubMed]
- Blessing, R. H. (1995). Acta Cryst. A51, 33–38. [DOI] [PubMed]
- Brandenburg, K. & Putz, H. (2005). DIAMOND Crystal Impact GbR, Bonn, Germany.
- Enraf–Nonius (1994). CAD-4 EXPRESS Enraf–Nonius, Delft, The Netherlands.
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst.32, 837–838.
- Fleck, M. (2006). Acta Cryst. E62, o4939–o4941.
- Harms, K. & Wocadlo, S. (1995). XCAD4 University of Marburg, Germany.
- Le Fur, Y., Masse, R. & Nicoud, J. F. (1998). New J. Chem. pp. 159–163.
- Maalej, W., Elaoud, Z., Mhiri, T., Daoud, A. & Driss, A. (2008). Acta Cryst. E64, o2172. [DOI] [PMC free article] [PubMed]
- Nicoud, J. F., Masse, R., Bourgogne, C. & Evans, C. (1997). J. Mater. Chem.7, 35-39.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809022879/dn2458sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809022879/dn2458Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report