Abstract
The three-dimensional polymeric title compound, {[Zn(C9H4N2O4)]·H2O}n, contains one crystallographically independent ZnII atom, one fully deprotonated 1H-benzimidazole-5,6-dicarboxylate (bdc) ligand and one uncoordinated water molecule. The ZnII atom is four-coordinated by three O atoms and one N atom from the bdc ligands, giving a distorted tetrahedral coordination geometry. The uncoordinated water molecule is bound to the main structure through a strong bdc–water N—H⋯O hydrogen bond, and two much weaker water–bdc O—H⋯O interactions.
Related literature
For structures of other bdc complexes, see: Gao et al. (2008 ▶); Lo et al. (2007 ▶); Wei et al. (2008 ▶); Yao et al. (2008 ▶).
Experimental
Crystal data
[Zn(C9H4N2O4)]·H2O
M r = 287.55
Monoclinic,

a = 6.4735 (5) Å
b = 8.1836 (6) Å
c = 18.4407 (12) Å
β = 104.397 (2)°
V = 946.25 (12) Å3
Z = 4
Mo Kα radiation
μ = 2.61 mm−1
T = 298 K
0.35 × 0.26 × 0.18 mm
Data collection
Bruker APEXII area-detector diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 2004 ▶) T min = 0.444, T max = 0.625
4613 measured reflections
1665 independent reflections
1513 reflections with I > 2σ(I)
R int = 0.019
Refinement
R[F 2 > 2σ(F 2)] = 0.026
wR(F 2) = 0.072
S = 1.06
1665 reflections
154 parameters
H-atom parameters constrained
Δρmax = 0.54 e Å−3
Δρmin = −0.32 e Å−3
Data collection: APEX2 (Bruker, 2004 ▶); cell refinement: SAINT (Bruker, 2004 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: XP in SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809022107/bg2261sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809022107/bg2261Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N2—H2⋯O1W | 0.86 | 2.02 | 2.809 (3) | 152 |
| O1W—H1W⋯O1i | 0.93 | 2.45 | 3.211 (5) | 139 |
| O1W—H2W⋯O4ii | 0.91 | 2.29 | 3.095 (3) | 146 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Acknowledgments
This work was supported financially by the Nature and Science Foundation of Guangdong Province (grant No. 7005808), Guangdong Provincial Science and Technology Bureau (grant 2008B010600009) and the NSFC (grant No. U0734005).
supplementary crystallographic information
Comment
N-Heterocyclic carboxylic acids have atracted attention not only because their versatile coordination modes but also owing to their forming high-dimensional polymers through H-bonding interactions. 1H-benzimidazole-5,6-dicarboxylic acid (bdc), having six coordination points, is a good candidate for the generation of three-dimensional coordination polymers. However, up to now the complexes based on the bdc ligand are rare (Lo et al., 2007; Gao et al., 2008; Wei et al., 2008; Yao et al., 2008). Here we report the first three-dimensional Zn coordination polymer connected by bdc ligands, [Zn(C9H4N2O2)].H2O.
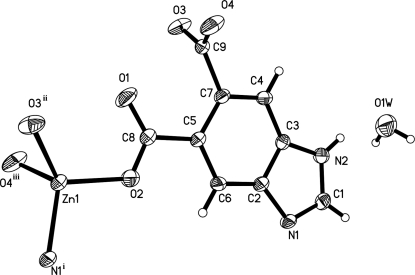
As is shown in Figure 1, the asymmetric unit consists of one Zn2+ cation, a fully deprotonated bdc2- ligand, and a free water moelcule. The cation has a tetrahedral coordination environment, and is surrounded by three oxygen and one nitrogen atoms from the bdc ligands. A packing diagram showing the 3D structure coming out from the tetradentate character of the bdc ligand is shown in Figure 2. To the best of our knowledge, the title compound is the first 3D transition metal coordination polymer based on the 1H-benzimidazole-5,6-dicarboxylic acid ligand.
Experimental
A mixture of bdc (0.0415 g, 0.20 mmol), Zn(NO3)2.6H2O (0.0594 g, 0.20 mmol) and water (10 ml) was heated up to 430 K for 72 h in a 23 ml Teflon-lined stainless-steel autoclave and then cooled down to room temperature in a 278 K/hour rate. Colourless prismatic crystals were collected and dried in air.
Refinement
H atoms attached to carbon and nitrogen were placed at calculated positions and treated as riding on their parent atoms with C—H = 0.93Å, C—N = 0.86Å. Those attached to oxygen were found from the Fourier maps, and allowed to ride without further refinement. In all cases Uĩso~(H) = 1.2 or 1.5 U~eq~(C, O).
Figures
Fig. 1.
Displacement ellipsoid plot (50% probability level) of the title compound, with atom numbering. Symmetry codes: (i) -x, -y+2, -z+2; (ii) -x, y+1/2, -z+3/2; (iii) x-1, y, z.
Fig. 2.
The packing diagram of the title compound, with H atoms omitted for clarity. Hydrogen bonds are shown as dashed lines.
Crystal data
| [Zn(C9H4N2O4)]·H2O | F(000) = 576 |
| Mr = 287.55 | Dx = 2.018 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 2639 reflections |
| a = 6.4735 (5) Å | θ = 2.3–27.6° |
| b = 8.1836 (6) Å | µ = 2.61 mm−1 |
| c = 18.4407 (12) Å | T = 298 K |
| β = 104.397 (2)° | Block, colorless |
| V = 946.25 (12) Å3 | 0.35 × 0.26 × 0.18 mm |
| Z = 4 |
Data collection
| Bruker APEXII area-detector diffractometer | 1665 independent reflections |
| Radiation source: fine-focus sealed tube | 1513 reflections with I > 2σ(I) |
| graphite | Rint = 0.019 |
| φ and ω scans | θmax = 25.0°, θmin = 2.3° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 2004) | h = −7→7 |
| Tmin = 0.444, Tmax = 0.625 | k = −9→8 |
| 4613 measured reflections | l = −21→16 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.026 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.072 | H-atom parameters constrained |
| S = 1.06 | w = 1/[σ2(Fo2) + (0.0407P)2 + 0.8076P] where P = (Fo2 + 2Fc2)/3 |
| 1665 reflections | (Δ/σ)max = 0.001 |
| 154 parameters | Δρmax = 0.54 e Å−3 |
| 0 restraints | Δρmin = −0.32 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.6805 (4) | 0.7618 (3) | 1.14674 (15) | 0.0244 (6) | |
| H1 | 0.7562 | 0.7565 | 1.1967 | 0.029* | |
| C2 | 0.4362 (4) | 0.8133 (3) | 1.04587 (14) | 0.0210 (6) | |
| C3 | 0.5993 (4) | 0.7274 (3) | 1.02485 (14) | 0.0209 (6) | |
| C4 | 0.5896 (4) | 0.6922 (3) | 0.95017 (14) | 0.0232 (6) | |
| H4 | 0.6986 | 0.6358 | 0.9364 | 0.028* | |
| C5 | 0.2433 (4) | 0.8286 (3) | 0.91858 (14) | 0.0210 (6) | |
| C6 | 0.2567 (4) | 0.8630 (3) | 0.99290 (14) | 0.0225 (6) | |
| H6 | 0.1474 | 0.9184 | 1.0070 | 0.027* | |
| C7 | 0.4108 (4) | 0.7447 (3) | 0.89738 (14) | 0.0198 (5) | |
| C8 | 0.0478 (4) | 0.8790 (3) | 0.85989 (15) | 0.0244 (6) | |
| C9 | 0.4056 (4) | 0.7163 (3) | 0.81632 (14) | 0.0200 (6) | |
| N1 | 0.4937 (3) | 0.8337 (3) | 1.12397 (12) | 0.0223 (5) | |
| N2 | 0.7500 (3) | 0.6973 (3) | 1.09061 (12) | 0.0255 (5) | |
| H2 | 0.8682 | 0.6459 | 1.0948 | 0.031* | |
| O1 | 0.0221 (4) | 0.8307 (4) | 0.79625 (13) | 0.0700 (10) | |
| O2 | −0.0815 (3) | 0.9719 (3) | 0.88154 (11) | 0.0371 (5) | |
| O3 | 0.3461 (3) | 0.5800 (3) | 0.78853 (10) | 0.0317 (5) | |
| O4 | 0.4720 (4) | 0.8254 (3) | 0.78041 (11) | 0.0360 (5) | |
| Zn1 | −0.35253 (5) | 1.02462 (4) | 0.814219 (16) | 0.02108 (13) | |
| O1W | 1.1216 (4) | 0.5187 (3) | 1.15400 (15) | 0.0533 (7) | |
| H1W | 1.0604 | 0.4556 | 1.1853 | 0.080* | |
| H2W | 1.1968 | 0.6024 | 1.1810 | 0.080* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0238 (14) | 0.0330 (16) | 0.0141 (13) | 0.0009 (12) | 0.0004 (11) | 0.0029 (11) |
| C2 | 0.0226 (13) | 0.0263 (14) | 0.0138 (13) | 0.0008 (11) | 0.0037 (11) | −0.0001 (10) |
| C3 | 0.0189 (13) | 0.0270 (15) | 0.0153 (13) | 0.0023 (11) | 0.0013 (11) | 0.0010 (11) |
| C4 | 0.0215 (13) | 0.0306 (15) | 0.0177 (14) | 0.0056 (11) | 0.0053 (11) | −0.0025 (11) |
| C5 | 0.0212 (13) | 0.0264 (15) | 0.0146 (13) | 0.0006 (11) | 0.0029 (11) | −0.0019 (10) |
| C6 | 0.0206 (13) | 0.0310 (15) | 0.0159 (13) | 0.0052 (11) | 0.0046 (11) | −0.0016 (11) |
| C7 | 0.0222 (13) | 0.0220 (14) | 0.0151 (13) | −0.0006 (11) | 0.0043 (10) | −0.0015 (10) |
| C8 | 0.0230 (14) | 0.0329 (16) | 0.0160 (14) | 0.0016 (12) | 0.0025 (11) | −0.0013 (11) |
| C9 | 0.0174 (12) | 0.0266 (15) | 0.0152 (12) | 0.0018 (10) | 0.0025 (10) | −0.0009 (11) |
| N1 | 0.0239 (12) | 0.0313 (13) | 0.0107 (11) | 0.0047 (10) | 0.0026 (9) | 0.0001 (9) |
| N2 | 0.0208 (12) | 0.0352 (14) | 0.0189 (12) | 0.0083 (10) | 0.0017 (9) | 0.0002 (10) |
| O1 | 0.0496 (15) | 0.125 (3) | 0.0230 (13) | 0.0460 (16) | −0.0143 (11) | −0.0294 (14) |
| O2 | 0.0285 (11) | 0.0578 (15) | 0.0209 (11) | 0.0200 (10) | −0.0012 (9) | −0.0072 (9) |
| O3 | 0.0511 (13) | 0.0269 (11) | 0.0190 (10) | −0.0077 (10) | 0.0125 (9) | −0.0057 (8) |
| O4 | 0.0532 (14) | 0.0376 (13) | 0.0185 (10) | −0.0200 (10) | 0.0117 (10) | −0.0050 (9) |
| Zn1 | 0.0221 (2) | 0.0270 (2) | 0.01325 (19) | 0.00140 (12) | 0.00268 (13) | −0.00017 (12) |
| O1W | 0.0410 (14) | 0.0652 (18) | 0.0468 (16) | 0.0148 (12) | −0.0022 (12) | −0.0085 (12) |
Geometric parameters (Å, °)
| C1—N1 | 1.316 (3) | C6—H6 | 0.9300 |
| C1—N2 | 1.336 (3) | C7—C9 | 1.505 (4) |
| C1—H1 | 0.9300 | C8—O1 | 1.210 (3) |
| C2—C6 | 1.380 (4) | C8—O2 | 1.266 (3) |
| C2—C3 | 1.401 (4) | C9—O3 | 1.247 (3) |
| C2—N1 | 1.405 (3) | C9—O4 | 1.250 (3) |
| C3—N2 | 1.376 (3) | N2—H2 | 0.8600 |
| C3—C4 | 1.393 (4) | Zn1—O2 | 1.929 (2) |
| C4—C7 | 1.383 (4) | Zn1—N1i | 1.999 (2) |
| C4—H4 | 0.9300 | Zn1—O3ii | 1.9587 (19) |
| C5—C6 | 1.381 (4) | Zn1—O4iii | 1.996 (2) |
| C5—C7 | 1.418 (4) | O1W—H1W | 0.9333 |
| C5—C8 | 1.504 (4) | O1W—H2W | 0.9127 |
| N1—C1—N2 | 112.9 (2) | O1—C8—C5 | 119.8 (3) |
| N1—C1—H1 | 123.6 | O2—C8—C5 | 116.2 (2) |
| N2—C1—H1 | 123.6 | O3—C9—O4 | 122.2 (2) |
| C6—C2—C3 | 120.8 (2) | O3—C9—C7 | 118.3 (2) |
| C6—C2—N1 | 130.8 (2) | O4—C9—C7 | 119.3 (2) |
| C3—C2—N1 | 108.5 (2) | C1—N1—C2 | 105.2 (2) |
| N2—C3—C4 | 133.0 (2) | C1—N1—Zn1i | 126.26 (18) |
| N2—C3—C2 | 105.3 (2) | C2—N1—Zn1i | 127.64 (17) |
| C4—C3—C2 | 121.7 (2) | C1—N2—C3 | 108.1 (2) |
| C7—C4—C3 | 117.1 (2) | C1—N2—H2 | 125.9 |
| C7—C4—H4 | 121.4 | C3—N2—H2 | 125.9 |
| C3—C4—H4 | 121.4 | C8—O2—Zn1 | 119.85 (18) |
| C6—C5—C7 | 120.6 (2) | C9—O3—Zn1iv | 121.73 (18) |
| C6—C5—C8 | 119.5 (2) | C9—O4—Zn1v | 131.46 (18) |
| C7—C5—C8 | 119.9 (2) | O2—Zn1—O3ii | 116.08 (9) |
| C2—C6—C5 | 118.5 (2) | O2—Zn1—O4iii | 111.99 (10) |
| C2—C6—H6 | 120.8 | O3ii—Zn1—O4iii | 91.96 (9) |
| C5—C6—H6 | 120.8 | O2—Zn1—N1i | 103.56 (9) |
| C4—C7—C5 | 121.3 (2) | O3ii—Zn1—N1i | 122.67 (9) |
| C4—C7—C9 | 117.4 (2) | O4iii—Zn1—N1i | 110.27 (9) |
| C5—C7—C9 | 121.3 (2) | H1W—O1W—H2W | 109.2 |
| O1—C8—O2 | 124.0 (3) | ||
| C6—C2—C3—N2 | −179.7 (3) | C5—C7—C9—O3 | −99.2 (3) |
| N1—C2—C3—N2 | 0.4 (3) | C4—C7—C9—O4 | −92.4 (3) |
| C6—C2—C3—C4 | 1.5 (4) | C5—C7—C9—O4 | 84.8 (3) |
| N1—C2—C3—C4 | −178.4 (3) | N2—C1—N1—C2 | 0.3 (3) |
| N2—C3—C4—C7 | −178.8 (3) | N2—C1—N1—Zn1i | −169.63 (19) |
| C2—C3—C4—C7 | −0.5 (4) | C6—C2—N1—C1 | 179.7 (3) |
| C3—C2—C6—C5 | −1.1 (4) | C3—C2—N1—C1 | −0.4 (3) |
| N1—C2—C6—C5 | 178.8 (3) | C6—C2—N1—Zn1i | −10.6 (4) |
| C7—C5—C6—C2 | −0.2 (4) | C3—C2—N1—Zn1i | 169.32 (18) |
| C8—C5—C6—C2 | 179.0 (3) | N1—C1—N2—C3 | 0.0 (3) |
| C3—C4—C7—C5 | −0.9 (4) | C4—C3—N2—C1 | 178.4 (3) |
| C3—C4—C7—C9 | 176.3 (2) | C2—C3—N2—C1 | −0.2 (3) |
| C6—C5—C7—C4 | 1.3 (4) | O1—C8—O2—Zn1 | 7.0 (5) |
| C8—C5—C7—C4 | −178.0 (3) | C5—C8—O2—Zn1 | −173.02 (19) |
| C6—C5—C7—C9 | −175.8 (3) | O4—C9—O3—Zn1iv | 0.2 (4) |
| C8—C5—C7—C9 | 4.9 (4) | C7—C9—O3—Zn1iv | −175.59 (17) |
| C6—C5—C8—O1 | −170.2 (3) | O3—C9—O4—Zn1v | −160.8 (2) |
| C7—C5—C8—O1 | 9.1 (5) | C7—C9—O4—Zn1v | 14.9 (4) |
| C6—C5—C8—O2 | 9.8 (4) | C8—O2—Zn1—O3ii | −44.1 (3) |
| C7—C5—C8—O2 | −170.9 (3) | C8—O2—Zn1—O4iii | 59.7 (2) |
| C4—C7—C9—O3 | 83.6 (3) | C8—O2—Zn1—N1i | 178.5 (2) |
Symmetry codes: (i) −x, −y+2, −z+2; (ii) −x, y+1/2, −z+3/2; (iii) x−1, y, z; (iv) −x, y−1/2, −z+3/2; (v) x+1, y, z.
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H2···O1W | 0.86 | 2.02 | 2.809 (3) | 152 |
| O1W—H1W···O1vi | 0.93 | 2.45 | 3.211 (5) | 139 |
| O1W—H2W···O4vii | 0.91 | 2.29 | 3.095 (3) | 146 |
Symmetry codes: (vi) −x+1, −y+1, −z+2; (vii) x+1, −y+3/2, z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: BG2261).
References
- Bruker (2004). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Gao, Q., Gao, W.-H., Zhang, C.-Y. & Xie, Y.-B. (2008). Acta Cryst. E64, m928. [DOI] [PMC free article] [PubMed]
- Lo, Y.-L., Wang, W.-C., Lee, G.-A. & Liu, Y.-H. (2007). Acta Cryst. E63, m2657–m2658.
- Sheldrick, G. M. (2004). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Wei, Y.-Q., Yu, Y.-F. & Wu, K.-C. (2008). Cryst. Growth Des.8, 2087–2089.
- Yao, Y.-L., Che, Y.-X. & Zheng, J.-M. (2008). Cryst. Growth Des.8, 2299–2306.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809022107/bg2261sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809022107/bg2261Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report