Abstract
The title compound, C28H34N2O8S2, was synthesized as part of a project to develop synthetic routes to analogues of sporidesmins, a class of secondary metabolite produced by the filamentous fungi Chaetomium and Pithomyces sp. The complete molecule is generated by crystallographic inversion symmetry: the methoxy group is essentially coplanar with the benzene ring to which it is bonded, a mean plane fitted through the non-H atoms of the aromatic ring and the methoxy group having an r.m.s. deviation of 0.0140 Å. Similarly, the ester group is also essentially planar (r.m.s. deviation of a plane fitted through all non-H atoms is 0.0101 Å). There is only one independent C—H⋯O interaction, which links together adjacent molecules into a two-dimensional sheet in the bc plane.
Related literature
For background information on the biological activity of sporidesmins, see: Fujimoto et al. (2004 ▶); Gardiner et al. (2005 ▶); Li et al. (2006 ▶); Saito et al. (1988 ▶); Waksman & Bugie (1944 ▶). For a discussion on the anti-cancer activity of these compounds, see: Brewer et al. (1978 ▶); Hauser et al. (1970 ▶); Kung et al. (2004 ▶); McInnes et al. (1976 ▶); Waksman & Bugie (1944 ▶). For related crystal structures, see: Isaka et al. (2005 ▶), Dubey et al. (2009 ▶); Polaske et al. (2009 ▶). For synthetic details, see: Hino & Sato (1974 ▶); Kawamura et al. (1975 ▶).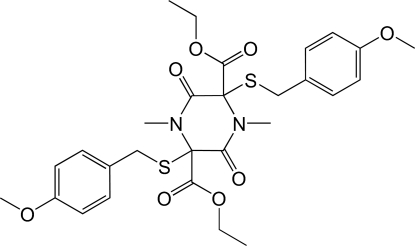
Experimental
Crystal data
C28H34N2O8S2
M r = 590.69
Monoclinic,

a = 11.290 (2) Å
b = 8.2259 (16) Å
c = 16.593 (3) Å
β = 109.704 (3)°
V = 1450.9 (5) Å3
Z = 2
Mo Kα radiation
μ = 0.24 mm−1
T = 150 K
0.32 × 0.30 × 0.10 mm
Data collection
Bruker SMART 1000 CCD diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.919, T max = 0.987
11556 measured reflections
3522 independent reflections
2778 reflections with I > 2σ(I)
R int = 0.028
Refinement
R[F 2 > 2σ(F 2)] = 0.036
wR(F 2) = 0.092
S = 1.03
3522 reflections
184 parameters
H-atom parameters constrained
Δρmax = 0.38 e Å−3
Δρmin = −0.24 e Å−3
Data collection: SMART (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶) and Mercury (Macrae et al., 2008 ▶); software used to prepare material for publication: SHELXTL, publCIF (Westrip, 2009 ▶) and local programs.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809022211/fj2226sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809022211/fj2226Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C5—H5⋯O4i | 0.95 | 2.43 | 3.2647 (19) | 147 |
Symmetry code: (i)  .
.
Acknowledgments
We thank the US National Science Foundation (CHE-0748838 & CHE-9610347) for support.
supplementary crystallographic information
Comment
Sporidesmins are an interesting class of secondary metabolites produced by the filamentous fungi Chaetomium and Pithomyces sp. This diverse class of natural products contains molecules with one or two epidithiodioxopiperazine rings that display a wide variety of biological activities (Waksman & Bugie, 1944; Saito et al., 1988; Fujimoto et al., 2004; Gardiner et al., 2005; Li et al., 2006). While toxic to mammalian cells, recent studies have suggested that certain sporidesmins may possess anticancer activity due to their ability to suppress neovascularization (Waksman & Bugie, 1944; Hauser et al., 1970; McInnes et al., 1976; Brewer et al., 1978; Kung et al., 2004). In the process of developing synthetic methodologies towards the synthesis of sporidesmin natural products, we came across a number of sulfenylated 2,5-piperazinediones whose structures could not be determined with confidence by NMR spectroscopy. Single crystal X-ray diffraction was found to be the only method available capable of unambiguously identifying the structures of these molecules. Herein we report the structure of the title compound (I).
The stucture of (I) is shown in Figure 1. Molecular dimensions are unexceptional and the compound crystallizes with crystallographic inversion symmetry in an extended conformation composed of essentially planar components. The methoxy group is essentially coplanar with the benzene ring to which it is bonded and a mean plane fitted through the non-hydrogen atoms of the aromatic ring and the methoxy group has an r.m.s. deviation of 0.0140 Å. Similarly the ester moiety is also essentially planar (r.m.s. deviation of a plane fitted through all non-hydrogen atoms is 0.0101 Å). The crystal packing has few notable intermolecular interactions; there is only one C–H···O interaction (plus an equivalent related by inversion symmetry) which links together adjacent molecules into a thick two-dimensional sheet in the bc plane.
Experimental
To a dry flask equipped with a stir bar was added 1,4-dimethyl-3,6-diethoxycarbonyl-2,5-piperazinedione (Hino & Sato, 1974) (144 mg, 0.50 mmol), (DHQD)2PYR (88 mg, 0.10 mmol) and N-(4-methoxybenzylthio)succinimide (Kawamura et al., 1975) (504 mg, 2.0 mmol). The compounds were then dried under vacuum for 15 minutes, followed by the addition of CH2Cl2 (2.25 ml). The mixture was allowed to stir at room temperature for 5 days. Once complete, the reaction was quenched with 1M KHSO4 (3 ml) and extracted with CH2Cl2 (3 x 15 ml). The organic extracts were combined, dried over anhydrous MgSO4, filtered, and concentrated under reduced pressure. Purification by column chromatography (silica gel, hexane:CH2Cl2:EtOAc (5:4:1)) followed by recrystallization from ethanol yielded colorless prisms (236 mg, 80% yield). LRMS (FAB, [M+H]+) found 591.36, C28H35N2O8S2 requires 591.18.
Refinement
Hydrogen atoms were identified from a difference map and refined with Uiso(H)= 1.5Ueq(C) (methyl H atoms) and Uiso(H)= 1.2Ueq(C) for all others. Fixed C–H distances of 0.95Å (aryl), 0.98Å (methyl) and 0.99Å (methylene) were used.
Figures
Fig. 1.
The molecular structure of (I). Displacement ellipsoids are at the 50% probability level and hydrogen atoms are omitted. Labelled atoms denote the asymmetric unit; unlabelled atoms are related by inversion symmetry (symmetry operator -x, -y + 1, -z).
Fig. 2.
An a axis projection of the crystal packing of (I). Hydrogen bonding is indicated by dotted blue lines (dotted red lines indicate continuation of hydrogen bonding).
Crystal data
| C28H34N2O8S2 | F(000) = 624 |
| Mr = 590.69 | Dx = 1.352 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 5275 reflections |
| a = 11.290 (2) Å | θ = 2.3–28.3° |
| b = 8.2259 (16) Å | µ = 0.24 mm−1 |
| c = 16.593 (3) Å | T = 150 K |
| β = 109.704 (3)° | Plate, colourless |
| V = 1450.9 (5) Å3 | 0.32 × 0.30 × 0.10 mm |
| Z = 2 |
Data collection
| Bruker SMART 1000 CCD diffractometer | 3522 independent reflections |
| Radiation source: sealed tube | 2778 reflections with I > 2σ(I) |
| graphite | Rint = 0.028 |
| Thin–slice ω scans | θmax = 28.3°, θmin = 1.9° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1996) | h = −14→14 |
| Tmin = 0.919, Tmax = 0.987 | k = −10→10 |
| 11556 measured reflections | l = −21→22 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.036 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.092 | H-atom parameters constrained |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.0397P)2 + 0.678P] where P = (Fo2 + 2Fc2)/3 |
| 3522 reflections | (Δ/σ)max < 0.001 |
| 184 parameters | Δρmax = 0.38 e Å−3 |
| 0 restraints | Δρmin = −0.24 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S | 0.18848 (3) | 0.24066 (4) | 0.04288 (2) | 0.02157 (10) | |
| O1 | 0.59043 (11) | 0.03836 (16) | −0.14348 (8) | 0.0357 (3) | |
| O2 | −0.08416 (10) | 0.32864 (14) | 0.13178 (7) | 0.0271 (3) | |
| O3 | 0.07865 (11) | 0.15503 (14) | 0.16459 (7) | 0.0301 (3) | |
| O4 | −0.16577 (11) | 0.45732 (14) | −0.15608 (7) | 0.0283 (3) | |
| N | −0.05515 (11) | 0.34842 (14) | −0.02822 (7) | 0.0168 (2) | |
| C1 | 0.32030 (15) | 0.27272 (19) | −0.06556 (10) | 0.0251 (3) | |
| C2 | 0.44789 (16) | 0.2787 (2) | −0.01800 (11) | 0.0336 (4) | |
| H2 | 0.4754 | 0.3387 | 0.0339 | 0.040* | |
| C3 | 0.53537 (16) | 0.1986 (2) | −0.04519 (12) | 0.0363 (4) | |
| H3 | 0.6222 | 0.2038 | −0.0118 | 0.044* | |
| C4 | 0.49678 (14) | 0.1106 (2) | −0.12108 (10) | 0.0247 (3) | |
| C5 | 0.37024 (14) | 0.1019 (2) | −0.16888 (10) | 0.0256 (3) | |
| H5 | 0.3426 | 0.0411 | −0.2206 | 0.031* | |
| C6 | 0.28382 (15) | 0.1832 (2) | −0.14039 (11) | 0.0283 (4) | |
| H6 | 0.1970 | 0.1770 | −0.1734 | 0.034* | |
| C7 | 0.22463 (17) | 0.3623 (2) | −0.03733 (12) | 0.0313 (4) | |
| H7A | 0.1472 | 0.3810 | −0.0870 | 0.038* | |
| H7B | 0.2587 | 0.4692 | −0.0129 | 0.038* | |
| C8 | 0.55425 (18) | −0.0657 (2) | −0.21608 (12) | 0.0361 (4) | |
| H8A | 0.5000 | −0.1524 | −0.2077 | 0.054* | |
| H8B | 0.6295 | −0.1137 | −0.2233 | 0.054* | |
| H8C | 0.5084 | −0.0029 | −0.2673 | 0.054* | |
| C9 | 0.05188 (13) | 0.35374 (17) | 0.05104 (9) | 0.0167 (3) | |
| C10 | 0.01900 (14) | 0.26530 (18) | 0.12339 (9) | 0.0201 (3) | |
| C11 | −0.12546 (17) | 0.2576 (2) | 0.19906 (11) | 0.0353 (4) | |
| H11A | −0.0596 | 0.2720 | 0.2557 | 0.042* | |
| H11B | −0.1417 | 0.1399 | 0.1887 | 0.042* | |
| C12 | −0.24275 (16) | 0.3430 (2) | 0.19672 (11) | 0.0304 (4) | |
| H12A | −0.2258 | 0.4595 | 0.2063 | 0.046* | |
| H12B | −0.2721 | 0.2993 | 0.2416 | 0.046* | |
| H12C | −0.3077 | 0.3263 | 0.1407 | 0.046* | |
| C13 | −0.10631 (15) | 0.18707 (19) | −0.05932 (10) | 0.0258 (3) | |
| H13A | −0.1961 | 0.1967 | −0.0923 | 0.039* | |
| H13B | −0.0950 | 0.1145 | −0.0104 | 0.039* | |
| H13C | −0.0619 | 0.1424 | −0.0959 | 0.039* | |
| C14 | −0.09368 (13) | 0.47399 (17) | −0.08292 (9) | 0.0178 (3) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S | 0.02166 (18) | 0.01810 (18) | 0.02652 (19) | 0.00695 (14) | 0.01018 (14) | 0.00412 (15) |
| O1 | 0.0249 (6) | 0.0441 (7) | 0.0408 (7) | 0.0008 (5) | 0.0145 (5) | −0.0176 (6) |
| O2 | 0.0304 (6) | 0.0287 (6) | 0.0284 (6) | 0.0097 (5) | 0.0184 (5) | 0.0123 (5) |
| O3 | 0.0366 (6) | 0.0273 (6) | 0.0291 (6) | 0.0119 (5) | 0.0147 (5) | 0.0126 (5) |
| O4 | 0.0332 (6) | 0.0242 (6) | 0.0189 (5) | 0.0013 (5) | −0.0027 (5) | −0.0015 (4) |
| N | 0.0183 (6) | 0.0145 (6) | 0.0161 (6) | 0.0000 (4) | 0.0037 (5) | −0.0009 (5) |
| C1 | 0.0254 (8) | 0.0225 (8) | 0.0316 (8) | 0.0054 (6) | 0.0152 (7) | 0.0069 (6) |
| C2 | 0.0301 (9) | 0.0408 (10) | 0.0309 (9) | 0.0015 (7) | 0.0117 (7) | −0.0124 (8) |
| C3 | 0.0183 (8) | 0.0505 (11) | 0.0367 (10) | 0.0005 (7) | 0.0047 (7) | −0.0152 (8) |
| C4 | 0.0211 (7) | 0.0271 (8) | 0.0283 (8) | 0.0015 (6) | 0.0116 (6) | −0.0024 (7) |
| C5 | 0.0237 (8) | 0.0287 (8) | 0.0237 (8) | −0.0021 (6) | 0.0069 (6) | −0.0034 (6) |
| C6 | 0.0178 (7) | 0.0342 (9) | 0.0311 (8) | 0.0026 (6) | 0.0059 (6) | 0.0052 (7) |
| C7 | 0.0341 (9) | 0.0250 (8) | 0.0439 (10) | 0.0109 (7) | 0.0252 (8) | 0.0104 (7) |
| C8 | 0.0418 (10) | 0.0350 (10) | 0.0387 (10) | −0.0007 (8) | 0.0228 (8) | −0.0108 (8) |
| C9 | 0.0173 (7) | 0.0161 (7) | 0.0158 (6) | 0.0028 (5) | 0.0046 (5) | 0.0013 (5) |
| C10 | 0.0237 (7) | 0.0191 (7) | 0.0175 (7) | 0.0013 (6) | 0.0070 (6) | 0.0006 (6) |
| C11 | 0.0384 (10) | 0.0417 (10) | 0.0341 (9) | 0.0085 (8) | 0.0233 (8) | 0.0183 (8) |
| C12 | 0.0295 (8) | 0.0381 (10) | 0.0272 (8) | 0.0022 (7) | 0.0141 (7) | 0.0070 (7) |
| C13 | 0.0292 (8) | 0.0166 (7) | 0.0273 (8) | −0.0032 (6) | 0.0038 (7) | −0.0009 (6) |
| C14 | 0.0180 (7) | 0.0178 (7) | 0.0180 (7) | 0.0024 (5) | 0.0066 (6) | −0.0017 (5) |
Geometric parameters (Å, °)
| S—C7 | 1.8181 (17) | C5—C6 | 1.391 (2) |
| S—C9 | 1.8447 (14) | C6—H6 | 0.9500 |
| O1—C4 | 1.3689 (19) | C7—H7A | 0.9900 |
| O1—C8 | 1.421 (2) | C7—H7B | 0.9900 |
| O2—C10 | 1.3255 (18) | C8—H8A | 0.9800 |
| O2—C11 | 1.4682 (19) | C8—H8B | 0.9800 |
| O3—C10 | 1.1972 (18) | C8—H8C | 0.9800 |
| O4—C14 | 1.2201 (17) | C9—C10 | 1.552 (2) |
| N—C9 | 1.4560 (17) | C9—C14i | 1.531 (2) |
| N—C13 | 1.4698 (19) | C11—H11A | 0.9900 |
| N—C14 | 1.3469 (18) | C11—H11B | 0.9900 |
| C1—C2 | 1.391 (2) | C11—C12 | 1.488 (2) |
| C1—C6 | 1.382 (2) | C12—H12A | 0.9800 |
| C1—C7 | 1.507 (2) | C12—H12B | 0.9800 |
| C2—H2 | 0.9500 | C12—H12C | 0.9800 |
| C2—C3 | 1.383 (2) | C13—H13A | 0.9800 |
| C3—H3 | 0.9500 | C13—H13B | 0.9800 |
| C3—C4 | 1.389 (2) | C13—H13C | 0.9800 |
| C4—C5 | 1.382 (2) | C14—C9i | 1.531 (2) |
| C5—H5 | 0.9500 | ||
| C7—S—C9 | 100.12 (7) | H8A—C8—H8B | 109.5 |
| C4—O1—C8 | 117.65 (13) | H8A—C8—H8C | 109.5 |
| C10—O2—C11 | 116.04 (12) | H8B—C8—H8C | 109.5 |
| C9—N—C13 | 116.90 (11) | S—C9—N | 112.23 (9) |
| C9—N—C14 | 124.53 (12) | S—C9—C10 | 104.14 (9) |
| C13—N—C14 | 117.16 (12) | S—C9—C14i | 108.92 (9) |
| C2—C1—C6 | 117.82 (15) | N—C9—C10 | 110.02 (11) |
| C2—C1—C7 | 121.36 (16) | N—C9—C14i | 113.91 (11) |
| C6—C1—C7 | 120.82 (15) | C10—C9—C14i | 107.03 (11) |
| C1—C2—H2 | 119.5 | O2—C10—O3 | 125.65 (14) |
| C1—C2—C3 | 121.00 (16) | O2—C10—C9 | 110.17 (12) |
| H2—C2—C3 | 119.5 | O3—C10—C9 | 124.17 (13) |
| C2—C3—H3 | 119.9 | O2—C11—H11A | 110.2 |
| C2—C3—C4 | 120.24 (15) | O2—C11—H11B | 110.2 |
| H3—C3—C4 | 119.9 | O2—C11—C12 | 107.49 (13) |
| O1—C4—C3 | 115.94 (14) | H11A—C11—H11B | 108.5 |
| O1—C4—C5 | 124.42 (14) | H11A—C11—C12 | 110.2 |
| C3—C4—C5 | 119.64 (15) | H11B—C11—C12 | 110.2 |
| C4—C5—H5 | 120.4 | C11—C12—H12A | 109.5 |
| C4—C5—C6 | 119.20 (15) | C11—C12—H12B | 109.5 |
| H5—C5—C6 | 120.4 | C11—C12—H12C | 109.5 |
| C1—C6—C5 | 122.08 (14) | H12A—C12—H12B | 109.5 |
| C1—C6—H6 | 119.0 | H12A—C12—H12C | 109.5 |
| C5—C6—H6 | 119.0 | H12B—C12—H12C | 109.5 |
| S—C7—C1 | 108.65 (11) | N—C13—H13A | 109.5 |
| S—C7—H7A | 110.0 | N—C13—H13B | 109.5 |
| S—C7—H7B | 110.0 | N—C13—H13C | 109.5 |
| C1—C7—H7A | 110.0 | H13A—C13—H13B | 109.5 |
| C1—C7—H7B | 110.0 | H13A—C13—H13C | 109.5 |
| H7A—C7—H7B | 108.3 | H13B—C13—H13C | 109.5 |
| O1—C8—H8A | 109.5 | O4—C14—N | 122.68 (13) |
| O1—C8—H8B | 109.5 | O4—C14—C9i | 118.20 (13) |
| O1—C8—H8C | 109.5 | N—C14—C9i | 119.02 (12) |
| C6—C1—C2—C3 | 0.5 (3) | C14—N—C9—C10 | 138.48 (13) |
| C7—C1—C2—C3 | −178.77 (17) | C14—N—C9—C14i | 18.3 (2) |
| C1—C2—C3—C4 | 0.1 (3) | C7—S—C9—N | 64.78 (12) |
| C8—O1—C4—C3 | 173.97 (17) | C7—S—C9—C10 | −176.24 (10) |
| C8—O1—C4—C5 | −6.6 (2) | C7—S—C9—C14i | −62.31 (11) |
| C2—C3—C4—O1 | 178.72 (17) | C11—O2—C10—O3 | 1.3 (2) |
| C2—C3—C4—C5 | −0.7 (3) | C11—O2—C10—C9 | −179.23 (13) |
| O1—C4—C5—C6 | −178.71 (16) | S—C9—C10—O2 | −175.74 (10) |
| C3—C4—C5—C6 | 0.7 (3) | S—C9—C10—O3 | 3.69 (18) |
| C2—C1—C6—C5 | −0.5 (2) | N—C9—C10—O2 | −55.27 (15) |
| C7—C1—C6—C5 | 178.73 (15) | N—C9—C10—O3 | 124.16 (15) |
| C4—C5—C6—C1 | 0.0 (3) | C14i—C9—C10—O2 | 68.99 (14) |
| C2—C1—C7—S | −81.68 (18) | C14i—C9—C10—O3 | −111.58 (16) |
| C6—C1—C7—S | 99.06 (17) | C10—O2—C11—C12 | −178.98 (14) |
| C9—S—C7—C1 | −169.77 (12) | C9—N—C14—O4 | 164.45 (14) |
| C13—N—C9—S | 59.98 (15) | C9—N—C14—C9i | −19.2 (2) |
| C13—N—C9—C10 | −55.48 (16) | C13—N—C14—O4 | −1.6 (2) |
| C13—N—C9—C14i | −175.65 (12) | C13—N—C14—C9i | 174.82 (12) |
| C14—N—C9—S | −106.05 (13) |
Symmetry codes: (i) −x, −y+1, −z.
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C5—H5···O4ii | 0.95 | 2.43 | 3.2647 (19) | 147 |
Symmetry codes: (ii) −x, y−1/2, −z−1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: FJ2226).
References
- Brewer, D., McInnes, A. G., Smith, D. G., Taylor, A., Walter, J. A., Loosli, H. R. & Kis, Z. L. (1978). J. Chem. Soc. Perkin Trans. 1, pp. 1248–1251.
- Bruker (2007). SMART and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Dubey, R., Polaske, N. W., Nichol, G. S. & Olenyuk, B. (2009). Tetrahedron Lett 50, 4310–4313. [DOI] [PMC free article] [PubMed]
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Fujimoto, H., Sumino, M., Okuyama, E. & Ishibashi, M. (2004). J. Nat. Prod.67, 98–102. [DOI] [PubMed]
- Gardiner, D. M., Waring, P. & Howlett, B. J. (2005). Microbiology, 151, 1021–1032. [DOI] [PubMed]
- Hauser, D., Weber, H. P. & Sigg, H. P. (1970). Helv. Chim. Acta, 53, 1061–1063. [DOI] [PubMed]
- Hino, T. & Sato, T. (1974). Chem. Pharm. Bull.22, 2866–2874.
- Isaka, M., Palasarn, S., Rachtawee, P., Vimuttipong, S. & Kongsaeree, P. (2005). Org. Lett.7, 2257–2260. [DOI] [PubMed]
- Kawamura, S., Horii, T., Nakabayashi, T. & Hamada, M. (1975). Bull. Chem. Soc. Jpn, 48, 2993–2994.
- Kung, A. L., Zabludoff, S. D., France, D. S., Freedman, S. J., Tanner, E. A., Vieira, A., Cornell-Kennon, S., Lee, J., Wang, B., Wang, J., Memmert, K., Naegeli, H.-U., Petersen, F., Eck, M. J., Bair, K. W., Wood, A. W. & Livingston, D. M. (2004). Cancer Cell, 6, 33–43. [DOI] [PubMed]
- Li, G.-Y., Li, B.-G., Yang, T., Yan, J.-F., Liu, G.-Y. & Zhang, G.-L. (2006). J. Nat. Prod 69, 1374–1376. [DOI] [PubMed]
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst.41, 466–470.
- McInnes, A. G., Taylor, A. & Walter, J. A. (1976). J. Am. Chem. Soc.98, 6741. [DOI] [PubMed]
- Polaske, N. W., Nichol, G. S., Szabo, L. J. & Olenyuk, B. (2009). Cryst. Growth Des.9, 2191–2197. [DOI] [PMC free article] [PubMed]
- Saito, T., Suzuki, Y., Koyama, K., Natori, S., Iitaka, Y. & Kinoshita, T. (1988). Chem. Pharm. Bull.36, 1942–1956.
- Sheldrick, G. M. (1996). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Waksman, S. A. & Bugie, E. (1944). J. Bacteriol 48, 527–530. [DOI] [PMC free article] [PubMed]
- Westrip, S. P. (2009). publCIF In preparation.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809022211/fj2226sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809022211/fj2226Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




