Abstract
In the title molecule, C15H11ClO2, the mean planes of the benzene and phenyl rings are inclined at 69.06 (11)° with respect to each other. The crystal structure is stablized by strong intermolecular O—H⋯O hydrogen bonds between the acid groups of pairs of molecules related by inversion centers.
Related literature
For background information, see: Canty & Van Koten (1995 ▶). For a related structure, see: Sadiq-ur-Rehman et al. (2006 ▶). For a description of the Cambridge Structural Database, see: Allen (2002 ▶).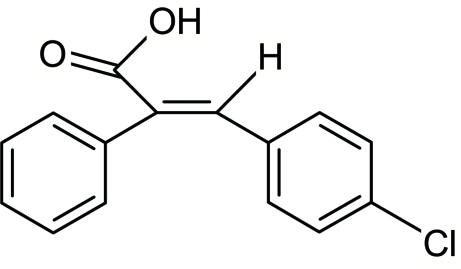
Experimental
Crystal data
C15H11ClO2
M r = 258.69
Monoclinic,

a = 14.405 (3) Å
b = 5.733 (9) Å
c = 15.416 (9) Å
β = 100.72 (3)°
V = 1251 (2) Å3
Z = 4
Mo Kα radiation
μ = 0.30 mm−1
T = 173 K
0.16 × 0.10 × 0.04 mm
Data collection
Nonius KappaCCD diffractometer
Absorption correction: multi-scan (SORTAV; Blessing, 1997 ▶) T min = 0.954, T max = 0.988
10074 measured reflections
2860 independent reflections
1431 reflections with I > 2σ(I)
R int = 0.100
Refinement
R[F 2 > 2σ(F 2)] = 0.047
wR(F 2) = 0.130
S = 0.96
2860 reflections
164 parameters
H-atom parameters constrained
Δρmax = 0.22 e Å−3
Δρmin = −0.33 e Å−3
Data collection: COLLECT (Hooft, 1998 ▶); cell refinement: DENZO (Otwinowski & Minor, 1997 ▶); data reduction: SCALEPACK (Otwinowski & Minor, 1997 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶); software used to prepare material for publication: SHELXTL (Sheldrick, 2008 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809021904/lh2835sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809021904/lh2835Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1⋯O2i | 0.84 | 1.82 | 2.658 (3) | 177 |
Symmetry code: (i)  .
.
Acknowledgments
Quaid-i-Azam University, Islamabad is gratefully acknowledged for financial support.
supplementary crystallographic information
Comment
Effort has been devoted to self assembly of organic and inorganic molecules in solid state because it extends a range of new solids with desirable physical and chemical properties (Canty & Van Koten, 1995). We report in this paper the crystal structure of the title compound (I) which has been synthesized in our laboratory.
The molecular structure of (I) is presented in Fig. 1. The benzene and phenyl rings are oriented at 69.06 (11)° with respect to each other. Molecules related by inversion ceneters form dimers via hydrogen bonds (Fig. 2); details of hydrogen bonding geometry have been given in Table 1. The molecular dimensions are normal (Allen, 2002). The cystal structure of a closely related compound has been previously reported from our laboratory (Sadiq-ur-Rehman et al., 2006).
Experimental
A mixture of the phenylacetic acid (0.15 mol), p-chlorobenzaldehyde (0.15 mol), anhydrous K2CO3 (0.095 mol) and acetic anhydride (0.38 mol) was slowly raised to the temperature 353–373 K and maintained for 24 h. To a hot solution were added, 200 ml of H2O and 100 ml of 10% HCl. The mixture was stirred at room temperature for 2 h and filtered. The solid mass obtained was recrystallized from commercial ethanol. Colorless crystals suitable for crystallographic study were obtained after three weeks.
Refinement
All the H-atoms were visible in the difference Fourier maps, they were included in the refinements at geometrically idealized positions with C—H and O—H distances = 0.95 and 0.84 Å, respectively, and Uiso = 1.5 and 1.2 times Ueq of the parent O and C-atoms respectively. The final difference map was free of chemically significant features.
Figures
Fig. 1.
ORTEP-3 (Farrugia, 1997) drawing of (I) with displacement ellipsoids plotted at 50% probability level.
Fig. 2.
Unit cell packing of (I) showing hydrogen bonding (dashed lines); H-atoms not involved in hydrogen bonding have been excluded for clarity.
Crystal data
| C15H11ClO2 | F(000) = 536 |
| Mr = 258.69 | Dx = 1.374 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2yn | Cell parameters from 10074 reflections |
| a = 14.405 (3) Å | θ = 1.8–27.5° |
| b = 5.733 (9) Å | µ = 0.30 mm−1 |
| c = 15.416 (9) Å | T = 173 K |
| β = 100.72 (3)° | Block, colourless |
| V = 1251 (2) Å3 | 0.16 × 0.10 × 0.04 mm |
| Z = 4 |
Data collection
| Nonius KappaCCD diffractometer | 2860 independent reflections |
| Radiation source: fine-focus sealed tube | 1431 reflections with I > 2σ(I) |
| graphite | Rint = 0.100 |
| φ and ω scans | θmax = 27.5°, θmin = 1.8° |
| Absorption correction: multi-scan (SORTAV; Blessing, 1997) | h = −18→18 |
| Tmin = 0.954, Tmax = 0.988 | k = −7→7 |
| 10074 measured reflections | l = −19→19 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.047 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.130 | H-atom parameters constrained |
| S = 0.96 | w = 1/[σ2(Fo2) + (0.06P)2] where P = (Fo2 + 2Fc2)/3 |
| 2860 reflections | (Δ/σ)max < 0.001 |
| 164 parameters | Δρmax = 0.22 e Å−3 |
| 0 restraints | Δρmin = −0.33 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | 0.44668 (5) | 0.75005 (11) | −0.15436 (5) | 0.0494 (2) | |
| O1 | 0.01817 (12) | −0.2417 (3) | −0.06692 (11) | 0.0418 (5) | |
| H1 | −0.0181 | −0.3550 | −0.0639 | 0.063* | |
| O2 | 0.09332 (13) | −0.3917 (3) | 0.06070 (11) | 0.0464 (5) | |
| C1 | 0.24216 (16) | −0.0735 (4) | 0.07913 (14) | 0.0301 (6) | |
| C2 | 0.25291 (17) | 0.1160 (4) | 0.13702 (15) | 0.0331 (6) | |
| H2 | 0.2109 | 0.2448 | 0.1257 | 0.040* | |
| C3 | 0.32420 (18) | 0.1175 (4) | 0.21066 (16) | 0.0390 (6) | |
| H3 | 0.3305 | 0.2456 | 0.2504 | 0.047* | |
| C4 | 0.38652 (18) | −0.0687 (5) | 0.22627 (16) | 0.0406 (7) | |
| H4 | 0.4364 | −0.0668 | 0.2762 | 0.049* | |
| C5 | 0.37624 (18) | −0.2559 (5) | 0.16958 (17) | 0.0400 (6) | |
| H5 | 0.4191 | −0.3830 | 0.1806 | 0.048* | |
| C6 | 0.30376 (17) | −0.2607 (4) | 0.09650 (16) | 0.0337 (6) | |
| H6 | 0.2963 | −0.3922 | 0.0583 | 0.040* | |
| C7 | 0.16421 (16) | −0.0712 (4) | 0.00023 (15) | 0.0306 (6) | |
| C8 | 0.15911 (17) | 0.0761 (4) | −0.06802 (15) | 0.0339 (6) | |
| H8 | 0.1017 | 0.0727 | −0.1100 | 0.041* | |
| C9 | 0.08865 (17) | −0.2473 (4) | 0.00101 (16) | 0.0339 (6) | |
| C10 | 0.23076 (17) | 0.2420 (4) | −0.08639 (15) | 0.0323 (6) | |
| C11 | 0.32781 (18) | 0.2055 (4) | −0.05820 (17) | 0.0373 (6) | |
| H11 | 0.3488 | 0.0719 | −0.0236 | 0.045* | |
| C12 | 0.39343 (19) | 0.3591 (4) | −0.07955 (17) | 0.0390 (6) | |
| H12 | 0.4590 | 0.3313 | −0.0602 | 0.047* | |
| C13 | 0.36306 (18) | 0.5541 (4) | −0.12935 (16) | 0.0370 (6) | |
| C14 | 0.26819 (18) | 0.5965 (4) | −0.15954 (16) | 0.0388 (6) | |
| H14 | 0.2480 | 0.7306 | −0.1941 | 0.047* | |
| C15 | 0.20296 (18) | 0.4382 (4) | −0.13813 (15) | 0.0363 (6) | |
| H15 | 0.1376 | 0.4644 | −0.1594 | 0.044* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.0473 (4) | 0.0393 (4) | 0.0664 (5) | −0.0059 (3) | 0.0232 (3) | 0.0088 (4) |
| O1 | 0.0376 (11) | 0.0454 (11) | 0.0395 (10) | −0.0145 (9) | −0.0006 (8) | 0.0038 (9) |
| O2 | 0.0481 (12) | 0.0473 (12) | 0.0414 (11) | −0.0173 (9) | 0.0016 (8) | 0.0092 (10) |
| C1 | 0.0326 (14) | 0.0279 (13) | 0.0315 (13) | −0.0053 (11) | 0.0109 (11) | 0.0003 (11) |
| C2 | 0.0369 (15) | 0.0277 (14) | 0.0361 (14) | −0.0018 (11) | 0.0101 (12) | 0.0001 (11) |
| C3 | 0.0445 (17) | 0.0379 (15) | 0.0346 (15) | −0.0090 (13) | 0.0076 (13) | −0.0065 (12) |
| C4 | 0.0342 (15) | 0.0536 (17) | 0.0336 (14) | −0.0064 (13) | 0.0052 (12) | 0.0052 (14) |
| C5 | 0.0358 (16) | 0.0398 (15) | 0.0465 (16) | 0.0048 (13) | 0.0130 (13) | 0.0108 (14) |
| C6 | 0.0370 (15) | 0.0295 (13) | 0.0365 (14) | −0.0043 (12) | 0.0118 (12) | −0.0020 (12) |
| C7 | 0.0293 (14) | 0.0312 (14) | 0.0315 (13) | −0.0035 (11) | 0.0063 (11) | −0.0043 (12) |
| C8 | 0.0320 (15) | 0.0350 (14) | 0.0345 (14) | −0.0015 (11) | 0.0053 (11) | −0.0045 (12) |
| C9 | 0.0332 (14) | 0.0351 (14) | 0.0329 (14) | −0.0051 (12) | 0.0052 (11) | −0.0046 (13) |
| C10 | 0.0383 (15) | 0.0309 (13) | 0.0287 (13) | −0.0054 (12) | 0.0091 (11) | −0.0039 (11) |
| C11 | 0.0404 (16) | 0.0316 (15) | 0.0423 (15) | 0.0012 (11) | 0.0141 (12) | 0.0038 (11) |
| C12 | 0.0360 (16) | 0.0362 (14) | 0.0472 (16) | −0.0005 (12) | 0.0143 (12) | 0.0036 (13) |
| C13 | 0.0440 (17) | 0.0283 (13) | 0.0425 (15) | −0.0059 (12) | 0.0178 (12) | −0.0019 (12) |
| C14 | 0.0455 (18) | 0.0338 (15) | 0.0401 (14) | 0.0008 (12) | 0.0157 (13) | 0.0052 (12) |
| C15 | 0.0369 (15) | 0.0389 (15) | 0.0339 (14) | 0.0030 (12) | 0.0090 (11) | −0.0002 (12) |
Geometric parameters (Å, °)
| Cl1—C13 | 1.742 (3) | C6—H6 | 0.9500 |
| O1—C9 | 1.316 (3) | C7—C8 | 1.340 (3) |
| O1—H1 | 0.8400 | C7—C9 | 1.486 (3) |
| O2—C9 | 1.230 (3) | C8—C10 | 1.469 (3) |
| C1—C6 | 1.386 (3) | C8—H8 | 0.9500 |
| C1—C2 | 1.396 (3) | C10—C15 | 1.393 (4) |
| C1—C7 | 1.494 (3) | C10—C11 | 1.400 (4) |
| C2—C3 | 1.382 (3) | C11—C12 | 1.376 (4) |
| C2—H2 | 0.9500 | C11—H11 | 0.9500 |
| C3—C4 | 1.387 (4) | C12—C13 | 1.380 (4) |
| C3—H3 | 0.9500 | C12—H12 | 0.9500 |
| C4—C5 | 1.375 (4) | C13—C14 | 1.381 (3) |
| C4—H4 | 0.9500 | C14—C15 | 1.390 (4) |
| C5—C6 | 1.387 (4) | C14—H14 | 0.9500 |
| C5—H5 | 0.9500 | C15—H15 | 0.9500 |
| C9—O1—H1 | 109.5 | C7—C8—H8 | 115.8 |
| C6—C1—C2 | 119.2 (2) | C10—C8—H8 | 115.8 |
| C6—C1—C7 | 121.4 (2) | O2—C9—O1 | 122.5 (2) |
| C2—C1—C7 | 119.4 (2) | O2—C9—C7 | 121.7 (2) |
| C3—C2—C1 | 120.5 (2) | O1—C9—C7 | 115.8 (2) |
| C3—C2—H2 | 119.8 | C15—C10—C11 | 117.5 (2) |
| C1—C2—H2 | 119.8 | C15—C10—C8 | 119.7 (2) |
| C2—C3—C4 | 119.7 (2) | C11—C10—C8 | 122.7 (2) |
| C2—C3—H3 | 120.1 | C12—C11—C10 | 121.4 (2) |
| C4—C3—H3 | 120.1 | C12—C11—H11 | 119.3 |
| C5—C4—C3 | 120.1 (2) | C10—C11—H11 | 119.3 |
| C5—C4—H4 | 120.0 | C11—C12—C13 | 119.4 (3) |
| C3—C4—H4 | 120.0 | C11—C12—H12 | 120.3 |
| C4—C5—C6 | 120.5 (2) | C13—C12—H12 | 120.3 |
| C4—C5—H5 | 119.7 | C14—C13—C12 | 121.4 (2) |
| C6—C5—H5 | 119.7 | C14—C13—Cl1 | 119.59 (19) |
| C1—C6—C5 | 120.0 (2) | C12—C13—Cl1 | 119.0 (2) |
| C1—C6—H6 | 120.0 | C13—C14—C15 | 118.4 (2) |
| C5—C6—H6 | 120.0 | C13—C14—H14 | 120.8 |
| C8—C7—C9 | 120.1 (2) | C15—C14—H14 | 120.8 |
| C8—C7—C1 | 124.6 (2) | C10—C15—C14 | 121.9 (2) |
| C9—C7—C1 | 115.3 (2) | C10—C15—H15 | 119.1 |
| C7—C8—C10 | 128.4 (2) | C14—C15—H15 | 119.1 |
| C6—C1—C2—C3 | 0.2 (3) | C1—C7—C9—O2 | −3.6 (3) |
| C7—C1—C2—C3 | 179.7 (2) | C8—C7—C9—O1 | −1.8 (3) |
| C1—C2—C3—C4 | 1.2 (4) | C1—C7—C9—O1 | 178.0 (2) |
| C2—C3—C4—C5 | −1.2 (4) | C7—C8—C10—C15 | −155.2 (3) |
| C3—C4—C5—C6 | 0.0 (4) | C7—C8—C10—C11 | 28.7 (4) |
| C2—C1—C6—C5 | −1.4 (3) | C15—C10—C11—C12 | 1.0 (3) |
| C7—C1—C6—C5 | 179.0 (2) | C8—C10—C11—C12 | 177.2 (2) |
| C4—C5—C6—C1 | 1.3 (4) | C10—C11—C12—C13 | 0.4 (4) |
| C6—C1—C7—C8 | −113.7 (3) | C11—C12—C13—C14 | −1.1 (4) |
| C2—C1—C7—C8 | 66.7 (3) | C11—C12—C13—Cl1 | 178.89 (19) |
| C6—C1—C7—C9 | 66.6 (3) | C12—C13—C14—C15 | 0.4 (4) |
| C2—C1—C7—C9 | −113.0 (3) | Cl1—C13—C14—C15 | −179.57 (18) |
| C9—C7—C8—C10 | −172.1 (2) | C11—C10—C15—C14 | −1.7 (3) |
| C1—C7—C8—C10 | 8.1 (4) | C8—C10—C15—C14 | −178.1 (2) |
| C8—C7—C9—O2 | 176.7 (2) | C13—C14—C15—C10 | 1.0 (4) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···O2i | 0.84 | 1.82 | 2.658 (3) | 177 |
Symmetry codes: (i) −x, −y−1, −z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: LH2835).
References
- Allen, F. H. (2002). Acta Cryst. B58, 380–388. [DOI] [PubMed]
- Blessing, R. H. (1997). J. Appl. Cryst.30, 421–426.
- Canty, A. J. & Van Koten, G. (1995). Acc. Chem. Res. 28, 406–413.
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Hooft, R. (1998). COLLECT Nonius BV, Delft, The Netherlands.
- Otwinowski, Z. & Minor, W. (1997). Methods in Enzymology, Vol. 276, Macromolecular Crystallography, Part A, edited by C. W. Carter Jr & R. M. Sweet, pp. 307–326. New York: Academic Press.
- Sadiq-ur-Rehman, Ali, S., Shahzadi, S. & Parvez, M. (2006). Acta Cryst. E62, o3313–o3315. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809021904/lh2835sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809021904/lh2835Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




